Abstract
The growth characteristics and underlying metabolism of microbial production hosts are critical to the productivity of metabolically engineered pathways. Production in parallel with growth often leads to biomass/bio-product competition for carbon. The growth arrest phenotype associated with the Saccharomyces cerevisiae pheromone-response is potentially an attractive production phase because it offers the possibility of decoupling production from population growth. However, little is known about the metabolic phenotype associated with the pheromone-response, which has not been tested for suitability as a production phase. Analysis of extracellular metabolite fluxes, available transcriptomic data, and heterologous compound production (para-hydroxybenzoic acid) demonstrate that a highly active and distinct metabolism underlies the pheromone-response. These results indicate that the pheromone-response is a suitable production phase, and that it may be useful for informing synthetic biology design principles for engineering productive stationary phase phenotypes.
Abbreviations: PHBA, para-hydroxybenzoic acid; GFP, Green Fluorescent Protein; MAPK, Mitogen Activated Protein Kinase
Keywords: Synthetic biology, Mating, Metabolic productivity, Yeast, Shikimate, Dynamic regulation, Metabolic engineering, Stationary phase
Highlights
-
•
Separating metabolic productivity from growth is highly desirable for bioprocesses.
-
•
The Saccharomyces cerevisiae mating phenotype involves a cell cycle arrest.
-
•
This unique phenotype was investigated for potential as a production phase.
-
•
Gene expression capacity and central metabolic fluxes were high.
-
•
Metabolism was more respiratory even in the presence of excess glucose.
1. Introduction
Microorganisms can be used to manufacture products ranging from therapeutic proteins and industrial enzymes through to metabolites for the replacement of existing petrochemicals and fossil fuels (Woolston et al., 2013). A major challenge for all bioprocesses is balancing resources between biomass accumulation and product formation, as both outcomes require the same cellular resources such as carbon precursors, energy in the form of ATP, and reducing power in the form of NADH and NADPH. Biomass accumulation is essential to achieve the volumetric productivity required for commercial processes; however, excess biomass accumulation limits product yields. Moreover, the product or its intermediates may be toxic to the host organism, again limiting biomass production. Dynamic regulatory systems can be used to trigger the expression of a production pathway after the completion of a growth phase (Venayak et al., 2015). However most non-growth associated phenotypes are poor production phases due to the depletion of available resources and the subsequent induction of stress response mechanisms (Albers et al., 2007, Chubukov and Sauer, 2014).
The yeast Saccharomyces cerevisiae is a widely used industrial host microbe, and has growth characteristics that typify the limitations of normal growth based physiology in industrial microorganisms. S. cerevisiae populations undergo an exponential growth phase where carbon and nitrogen resources are rapidly consumed until they limit biomass production. During exponential growth, approximately 90% of cellular energy is directed towards ribosome biogenesis (Warner et al., 2001). Carbon- or nitrogen-limited populations cease rapid growth and enter a ‘stationary phase’, which is characterised by the induction of stress survival mechanisms and a drastic reduction in the overall rate of protein synthesis relative to the exponential phase (Werner-Washburne et al., 1993). In the case of carbon starvation, there is no substrate left for conversion into product; and under nitrogen starvation, stress signalling severely limits metabolic productivity even in the presence of excess carbon (Albers et al., 2007). An ideal scenario for bio-production would involve a rapid growth phase where biomass (or ‘catalyst’) accumulates to a level that enables high volumetric productivity, before switching to a metabolically active stationary phase. This phase would then be maintained even in the presence of high concentrations of cellular resources such as carbon and nitrogen. With cells metabolically active but not growing and dividing, a much greater proportion of carbon could be directed towards target metabolites. Such a strategy would also open up the possibility of implementing growth limiting genetic modifications such as the silencing of essential genes using dynamic regulatory mechanisms (Williams et al., 2015a, Williams et al., 2015b). Stationary phase production is also very attractive because it enables the formation of products that are normally toxic to growth, and therefore limiting to production (Holtz and Keasling, 2010, Keasling, 2008).
The cell-cycle arrest phenotype of the yeast mating system represents a unique phase in the life-cycle of S. cerevisiae, which could be useful as a production phase for metabolic engineering where metabolic productivity is decoupled from growth-based physiology. The mating system has evolved to facilitate the synchronisation of the cell cycle and the fusion of two cells of opposite mating type to form a diploid. Briefly, haploid yeast cells of each mating type (a or α) secrete specific peptide pheromones that they use to detect the proximity of a potential mating partner of the opposite mating type. Binding of pheromone to specific G-protein coupled membrane receptors triggers an intracellular mitogen activated protein kinase signalling event which results in the de-repression of the Ste12p transcription factor and the initiation of the pheromone-response (Bardwell, 2005). Activation of the mating phenotype results in polarized growth, remodelling of cellular morphology and global transcription patterns, and arrest of growth in the G1 phase of the cell cycle (Bardwell, 2005), similarly to entry into stationary phase during carbon or nitrogen starvation. This phenotype can also be triggered via the addition of purified mating peptide to laboratory yeast cultures.
The S. cerevisiae mating system has become a cornerstone of eukaryotic synthetic biology (Furukawa and Hohmann, 2013). The pheromone communication system has been utilised for synthetic quorum sensing (Williams et al., 2015a, Williams et al., 2013), signal amplification (Groß et al., 2011), intercellular and interspecies communication (Hennig et al., 2015, Jahn et al., 2013), and biological computation (Regot et al., 2011). Furthermore, the depth of knowledge surrounding the mitogen activated protein kinase (MAPK) signal transduction machinery has enabled the construction and fine-tuning of a multitude of synthetic regulatory circuits (Bashor et al., 2008, Ingolia and Murray, 2007, O’Shaughnessy et al., 2011, Tanaka and Yi, 2009). In addition to relevance as a potential production phase, knowledge of the pheromone-response metabolism will be invaluable for future design of MAPK related synthetic regulatory systems. However, despite extensive utilisation of the mating system in synthetic biology, almost nothing is known about aspects of the phenotype that are not specifically related to mating.
Activation of the pheromone-response could result in a number of different scenarios with respect to metabolic engineering outcomes for a specific product. These include: an unproductive phenotype similar to the G1 arrest of the carbon- or nitrogen-limited stationary phases; higher productivity due to the limitation of carbon flux towards biomass; or no overall effect on cellular productivity due to the diversion of cellular resources towards the mating phenotype. In addition to considerations of general metabolic productivity, it is also important to identify any fundamental differences in metabolism, as they can help to decide which heterologous products will be favoured by the natural fluxes in the network. For example, specific anabolic pathways could be up-regulated in response to mating pheromone, suggesting that industrial products which are derived from these pathways would have higher yields during the pheromone-response.
The concept of limiting biomass formation to enhance cellular productivity has received some attention in the field of therapeutic protein production in mammalian cell cultures (Kumar et al., 2007). In particular, the manipulation of the eukaryotic cell cycle to induce a growth arrest phenotype has been successfully used to improve heterologous protein production. For example, the over-expression of the cyclin dependent kinase inhibitor p21and its inducer C/EBPα in a Chinese Hamster Ovary cell line resulted in stable cell-cycle arrest in the G1 phase and a 10–15 fold higher protein productivity per cell (Fussenegger et al., 1998). Similarly, the overexpression of the p21 cyclin inhibitor in an NS0 mouse myeloma cell line increased protein productivity ~4 fold (Watanabe et al., 2002). The increased productivity due to p21 mediated cell-cycle arrest has been attributed to higher mitochondrial membrane potential providing more ATP for peptide bond formation, and increased ribosomal biogenesis (Bi et al., 2004, Khoo and Al-Rubeai, 2009). It is possible that the cell cycle arrest phenotype of the S. cerevisiae pheromone-response could result in similar productivity improvements.
In this work, we have investigated the pheromone-response in S. cerevisiae as a growth arrest phase for metabolic engineering and synthetic biology applications. Fundamental metabolic differences in pheromone-treated populations were identified by comparing external metabolite fluxes, metabolic and global gene expression patterns, and the production capacity of a heterologous compound of industrial importance, para-hydroxybenzoic acid (PHBA).
2. Materials and methods
2.1. Growth media
Strains were grown in chemically defined CBS medium with 5 g/L ammonium sulfate, 20 g/L glucose, vitamins and trace elements (Verduyn et al., 1992) solidified with 20 g/L agar when solid medium was required. During strain construction auxotrophies were complemented with purified amino acids (Sigma) in CBS agar plates, while YPD (Yeast extract 10 g/L, Bacteriological Peptone 20 g/L, glucose 20 g/L) or YPG (galactose in place of glucose) supplemented with appropriate antibiotics was used during gene deletion procedures. E. coli DH5α was used for plasmid propagation/storage and was grown in LB medium with appropriate antibiotics.
2.2. Strains and plasmids
Primers, plasmids, and strains used in this study are shown in Tables S1, Table 1, Table 2 respectively. DNA manipulation and propagation were carried out using standard techniques (Sambrook and Russell, 2001) unless stated otherwise. All S. cerevisiae transformations were carried out using the lithium acetate method (Gietz and Schiestl, 2007). Yeast cells can die if the pheromone-response is induced and a mating partner is not found, and this effect can be mitigated by deleting the FUS1 gene (Zhang et al., 2006). Deletion of the FUS1 gene was performed with the reusable LoxP-KanMX-LoxP cassette as described previously (Güldener et al., 1996) using the FUS1KOF and FUS1KOR primers to amplify the geneticin deletion cassette from pUG6 (Gueldener et al., 2002) and the FUS1DCF and FUS1DCR primers to check the chromosomal locus for deletion. Strains transformed with yeast integrating plasmids were screened for correct integration using PCR as previously described (Stansfield and Stark, 2007). In order to reduce the capacity of yeast cells to become desensitized to mating pheromone we deleted the BAR1 gene which encodes for a secreted alpha-pheromone protease (Chan and Otte, 1982). The BAR1 ORF was replaced with the phleomycin resistance cassette from pUG66 using the same method as for FUS1 except with the BAR1 primers.
Table 1.
Plasmids.
| Name | Details | Origin |
|---|---|---|
| pRS406 | URA3 integrating vector | (Sikorski and Hieter, 1989), Euroscarf |
| yEGFPCLN2PEST-pRS406 | Destabilized GFP gene in pRS406 | (Williams et al., 2013) |
| pSF019 | pTEF1 driven lacZ expression | (Partow et al., 2010) |
| pTEF1-yEGFPCLN2PEST | TEF1 promoter driven GFP expression | This Study |
| pUG6 | Contains geneticin resistance marker gene | (Güldener et al., 1996), Euroscarf |
| pUG66 | Contains phleomycin resistance marker gene | (Güldener et al., 1996), Euroscarf |
| pTCW022 | pFUS1J2-UBiC-CYC1t-pFUS1J2-ARO4-CYC1t-pFUS1J2-TKL1-CYC1t-pRS406 | (Williams et al., 2015a) |
| PHBA01 | pTEF1-UBiC-CYC1t-pRS406 | This study |
| PHBA02 | pTEF1-UBiC-CYC1t-pTEF1-ARO4K229L-CYC1t-pRS406 | This study |
| PHBA03 | pTEF1-UBiC-CYC1t-pTEF1-ARO4K229L-CYC1t-pTEF1-TKL1-CYC1t-pRS406 | This study |
Table 2.
S. cerevisiae strains.
| Name | Genotype | Notes | Origin |
|---|---|---|---|
| CEN.PK113-5D | MATa; ura3-52; MAL2-8C; SUC2 | Haploid MATa lab strain | Euroscarf |
| PSP01 | CEN.PK113-5D, bar1::phleo | BAR1 gene deleted | This study |
| PSP02 | CEN.PK113-5D, bar1::phleo, fus1::KanMX | BAR1 and FUS1 deleted | This study |
| PSP03 | CEN.PK113-5D, bar1::phleo, fus1::KanMX, ura3-52::pTEF1-GFPCLN2PEST-ADH1t-pRS406 | PSP02+constitutive destabilized GFP expression | This study |
| PSP04 | CEN.PK113-5D, bar1::phleo, fus1::KanMX, ura3-52::pRS406 | Prototrophic control strain | This study |
| PSP05 | CEN.PK113-5D, bar1::phleo, fus1::KanMX, ura3-52: :pTEF1-UBiC-CYC1t-pTEF1-ARO4K229L-CYC1t-pTEF1-TKL1-CYC1t-pRS406 | para-hydroxybenzoic acid producing strain | This study |
The pTEF1-yEGFPCLN2PEST-416 plasmid was made by inserting the TEF1 promoter amplified from pSF-019 (Partow et al., 2010) using primers 9/10 into yEGFPCLN2PEST-pRS406 (Williams et al., 2013) digested with XhoI/EcoRI. The gene – CYC1 terminators for UBiC, ARO4K229L, and TKL1 were amplified from the pTCW022 (Williams et al., 2015a) plasmid using primers 13/14, 17/18, and 20/21. TEF1 promoters were used to control the expression of UBiC, ARO4K229L, and TKL1 genes. The TEF1 promoter region was PCR amplified from the pSF019 plasmid using primers that have 5′ extensions to create 40 bp homologous overlap junctions for each gene - CYC1 terminator cassette using primers 11/12 (UBiC), 15/16 (ARO4), and 19/20 (ARO4K229L). Overlap extension PCR (Horton et al., 1989) was used to assemble the TEF1 promoters 5′ of their respective ORF-CYC1 terminators. The pTEF1-UBiC-CYC1t cassette from the overlap assembly process was then PCR-amplified using primer pair 23/24 and inserted into pRS406 (Sikorski and Hieter, 1989) using XhoI and EcoRI to make plasmid PHBA01. Similarly the ARO4 expression cassette was amplified (primers 25/26) and inserted into PHBA01 using EcoRI and NotI to make PHBA02. The TKL1 expression cassette generated by overlap assembly was amplified with primer pair 27/28 and inserted into PHBA02 using NotI cut sites at both ends to make PHBA03. All constructs were sequenced to check for PCR errors.
2.3. Growth conditions
Shake-flask fermentations were carried out at 30 °C, 200 rpm with aluminium foil used to cover flask tops and medium making up 10% of the baffled flask volume. Single colonies from solid CBS agar plates that had been streaked with glycerol-stocked strains were used to inoculate 10 mL of liquid CBS medium. After 24 h growth, cells were passaged into a second pre-culture (25 mL) and grown to mid to late log phase (OD660 nm of 1–5) prior to inoculation of the experimental culture (50 mL) at an OD660 nm of 0.4. Synthetic alpha-pheromone (Genscript, Piscataway, NJ, USA) was added to flasks at a final concentration of 1 µM at indicated time points. Samples for analysis of extracellular metabolites were obtained by centrifugation of 1 mL of culture at 13,000×g for 7 min at 4 °C and storing the supernatant at −20 °C until analysis. Population density was measured using absorbance at 660 nm (OD660nm) on a spectrophotometer (LibraS4, Biochrom UK). OD660 nm values were converted to biomass using a conversion factor of 0.243 g dry cell weight per 1 OD unit (determined using exponentially growing CEN.PK113-5D populations).
2.4. Analytics
Extracellular glucose, ethanol, glycerol, and acetate concentrations were determined using HPLC as previously described (Dietmair et al., 2010). Metabolite concentration were normalised to carbon moles along with biomass, and these values were used to estimate CO2 production and O2 consumption as described previously (Stephanopoulos et al., 1998). PHBA concentrations were measured in extracellular supernatants as previously described (Williams et al., 2015a). GFP measurement was carried out as described previously (Williams et al., 2013).
2.5. Transcriptome analysis
Although transcriptomics has previously been carried out on yeast populations responding to pheromone (Roberts et al., 2000), the data analyses and interpretation were oriented specifically towards mating related processes. As we were interested other changes that are more peripheral to the canonical pheromone-response, the existing transcriptome data were re-analysed with known mating related genes excluded. The log normalised fold changes in gene expression and corresponding p values for populations treated with 50 nM alpha pheromone for 2 h were obtained from a previous study (Roberts et al., 2000). The gene names were assigned to gene ontology (GO) terms using the Saccharomyces Genome Database (SGD) GO slim mapper (http://www.yeastgenome.org/cgi-bin/GO/goSlimMapper.pl), and any genes under the categories of ‘sexual reproduction’ and/or ‘conjugation’ were removed from the data set. Genes with p values≤0.01 and fold changes≥2 were then used for GO term analysis. For central carbon metabolic gene analysis, transcripts with p values≤0.05 and fold changes≤−1.5 and ≥+1.5 were considered, and manually mapped to central carbon metabolism using an adaptation of the metabolic map presented in (Oliveira et al., 2012). Figures were constructed using Adobe Illustrator (Adobe Systems Software Ireland Ltd).
2.6. Statistical analysis
All experiments were conducted in biological triplicate (pre-cultures initiated using separate colonies from solid media). Means, standard deviations, and p-values were calculated in GraphPad Prism 6 using two-sided student's t-tests with equal variance. Biomass and extracellular metabolite rates were calculated using linear regression (LINEST equation in Microsoft Excel) individually for each biological replicate, only using data points after the addition of pheromone to cultures at 4.5 h. In the case of biomass, data were natural log transformed (Ln) prior to regression analysis. The mean rates from triplicate experiments were reported along with the pooled standard errors.
3. Results and discussion
3.1. Gene expression capacity and growth characteristics of the pheromone-response
An ideal bio-production phase has a high general capacity for gene expression such that heterologous enzymes can be expressed to a high level. A production phase should also have high metabolic activity through central carbon metabolism to allow efficient supply of carbon precursors to products. These factors are particularly important to assess for the pheromone-response because all of the other known types of growth arrest in yeast result in a ‘stationary phase’ which is characterised by low gene expression and metabolic activity (Albers et al., 2007, Herman, 2002, Werner-Washburne et al., 1993).
To address the question of gene expression capacity of the cell-cycle arrest associated with the pheromone-response, GFP expression levels between cultures treated with and without synthetic α-pheromone were compared (strain PSP03, Table 2). The TEF1 promoter was used to control GFP expression because the promoter is constitutively active (Da Silva and Srikrishnan, 2012) and TEF1 mRNA levels are unaffected by pheromone treatment (Roberts et al., 2000). Consequently, the level of GFP should reflect the overall gene expression capacity of the populations.
To induce the mating response, alpha-pheromone was added to one set of parallel cultures at 4.5 h. A rapid reduction in growth rate was observed within two hours relative to the non-pheromone-treated control, consistent with the cell cycle arrest phenotype of the pheromone-response (Bardwell, 2005) (Fig. 1a). GFP fluorescence showed a similar pattern in both treatments throughout the experiment (Fig. 1a). Fluorescence initially increased up until 4 h, then declined over the remainder of the culture. The decreasing expression from the TEF1 promoter is consistent with our recent findings (Peng et al., 2015, Williams et al., 2015b), and is explained by the use of a highly destabilized version of GFP with a 20-min half-life (Mateus and Avery, 2000). This destabilized GFP is highly sensitive to decreases in expression, which may have been masked by the stability (7 h half-life) of the GFP protein used in previous analyses of the TEF1 promoter in yeast (Partow et al., 2010, Sun et al., 2012). Although this decline occurred after pheromone addition in the treated culture, the fluorescence response was the same in the untreated culture, indicating that this is a generic pattern during cultivation rather than being pheromone-specific. The declining GFP expression rate in the treated strain (−782±104 au/cell/hr) was very similar to the control strain (−934±54 au/cell/hr) over the remainder of the cultivation time following initiation of culture growth arrest. These data suggest that gene expression capacity remains as active during G1 phase growth arrest as it is in exponentially growing populations, thus demonstrating the potential for this phase to be useful for production.
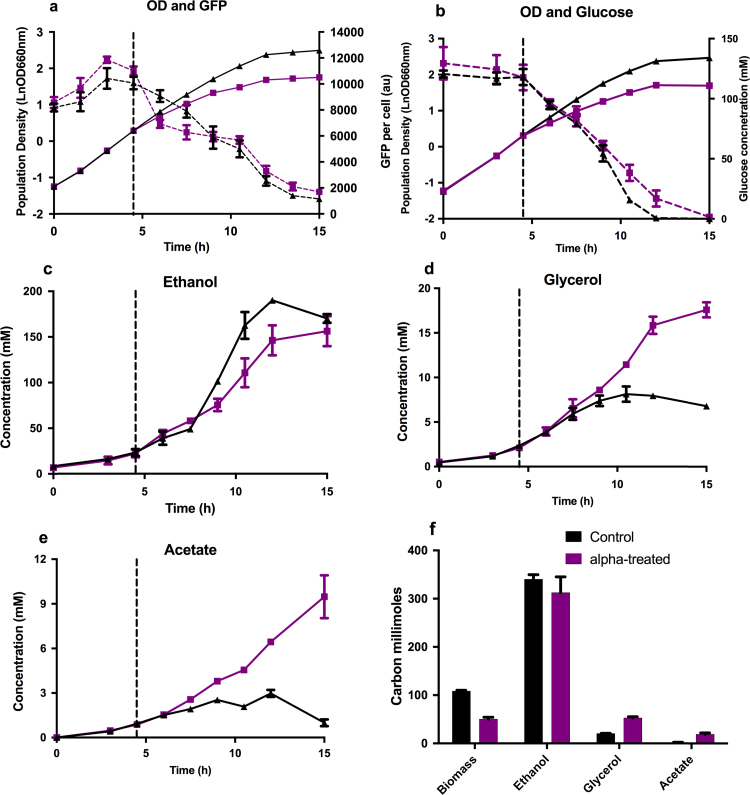
Fig. 1.
A strain with constitutive expression of GFP (PSP03, a), and a strain with no GFP expression (PSP04, b-e) were grown for 15 h in shake-flasks with (squares, purple lines) and without (triangles, black lines) 1 µM alpha pheromone added at 4.5 h (vertical dashed lines). GFP expression per cell (dashed lines) and population density (solid lines) were measured (a) along with extracellular glucose (dashed lines in b) and extracellular metabolite concentrations (b-e). Relative carbon concentrations were calculated as described in (Stephanopoulos et al., 1998) after 15 h of growth (f), where an S. cerevisiae carbon content of 24.6 g/C-mol with 7% ash (Stephanopoulos et al., 1998) was used along with a conversion factor of 0.243 to convert OD660 nm to grams dry cell weight. All measurements were carried out in biological triplicate with error bars representing ±1 standard deviation.
In addition to gene expression capacity, another principle requirement for metabolic engineering applications is to have an active central carbon metabolic network. This can be investigated generally by examining biomass accumulation in parallel with carbon source consumption and the accumulation of the predominant extracellular metabolites produced by S. cerevisiae. Parallel measurements of glucose uptake, along with biomass, acetate, ethanol, and glycerol production throughout 15 h of shake-flask cultivation clearly demonstrated that populations treated with pheromone were at least as metabolically active as the non-treated control populations (Fig. 1b-h). As with the GFP expression strains, pheromone addition at 4.5 h resulted in a characteristic decrease in growth rate (Fig. 1b), but surprisingly a very similar glucose consumption profile (Fig. 1c) with rates of 15.40±1.52 mmol g−1 h−1 and 12.55±1.47 mmol g−1 h−1 for pheromone treated and non-treated respectively (Table 3). In contrast to the relatively small differences in glucose consumption and ethanol production, glycerol and acetate production were markedly increased in pheromone-treated cultures (Fig. 1, Table 3).
Table 3.
Summary of external metabolite flux rates.
| µ (h−1)* | Glucose uptake (mmol g−1 h−1) | Ethanol (mmol g−1 h−1) | Glycerol* (mmol g−1 h−1) | Acetate* (mmol g−1 h−1) | Respiratory quotient | |
|---|---|---|---|---|---|---|
| Control | 0.26±0.03 | 12.55±1.47 | 20.52±2.58 | 0.53±0.17 | 0.16±0.06 | 4 |
| Treated | 0.17±0.02 | 15.40±1.52 | 19.65±2.85 | 2.17±0.29 | 0.89±0.11 | 2 |
Average growth and extracellular product secretion rates are shown for cultures treated with alpha-pheromone (treated) and without (control). All rates were calculated using data points after the addition of pheromone at 4.5 h, including in the control populations. Mean values of biological triplicates are presented with errors representing ±standard error. * denotes significant difference between control and pheromone treated groups (two sided students t-test with equal variance with p≤0.05).
When summarized with end-point metabolite concentration values converted to carbon moles (Fig. 1f) it is clear that the carbon not used for biomass production is directed towards side-product formation in the form of glycerol, acetate, and CO2 (calculated as 296 and 273 carbon moles for pheromone treated and non-treated respectively). When carbon balance values were used to infer CO2 and O2 production and consumption rates, the respiratory quotients obtained (Table 3) strongly suggest that the pheromone-response entails a shift towards a more respiratory metabolism compared to the fermentative metabolism of the control cultures. The current understanding of the shift between fermentative and respiratory metabolism in yeast is that a reduced glucose uptake rate results in de-repression of TCA cycle enzymes and a corresponding increase in TCA cycle flux, oxidative phosphorylation, and a decrease in fermentative flux towards ethanol (Blank and Sauer, 2004, Dijken et al., 1993, Heyland et al., 2009). The slightly reduced ethanol production rate of pheromone treated populations is consistent with this understanding, but it is interesting that the specific glucose uptake rates are not significantly different between the groups. These data support the concept that cell cycle arrest in response to pheromone results in an active and distinct metabolic phenotype, as compared to ‘standard’ carbon/nitrogen-limited stationary phases (Albers et al., 2007).
3.2. Transcriptome analysis
In addition to assessing the general gene expression capacity and external metabolic fluxes during the pheromone-response, it is also important to consider global changes in gene expression. Most starvation based stationary phases are characterised by the induction of stress resistance modules at the transcriptional level, and the hypothesis that the pheromone-response growth arrest phenotype is distinct from these phases can be tested using transcriptomics. Global changes in S. cerevisiae gene expression in response to pheromone treatment have previously been reported (Roberts et al., 2000). This study elegantly demonstrated the complexity of the pheromone-response pathway and the degree to which its signalling components are related to other MAPK modules. However, the data interpretation/analysis did not include transcriptional changes outside of the signalling and effector components of the pheromone-response and related MAPK modules. To gain insight into other changes of relevance to metabolic engineering, the data were re-analysed after excluding any genes primarily involved in the pheromone-response (GO terms ‘sexual reproduction’ and ‘conjugation’). A 99% confidence interval and minimum 2-fold change were used as selection criteria to identify up-regulated (Table S2) and down-regulated (Table S3) genes.
Structural processes, including cell wall and cytoskeletal organization, were up-regulated in pheromone-treated populations. This likely relates to the characteristic ‘shmoo’ cell morphology of the mating phenotype. Many up-regulated genes were assigned to categories associated with control of the cell cycle, mitosis, budding, and cytokinesis. These genes regulate the characteristic G1 phase cell-cycle arrest of the mating phenotype. The same categories involving cell-cycle related genes that were up-regulated also featured in the down-regulated gene GO terms (Table S3). This reflects the complexity of regulation required to arrest cell division, with the coordinated up- and down-regulation of a multitude of genes needed to elicit such fine control.
The GO term ‘Transposition’ refers to the movement of DNA between non-homologous sites and includes many retrotransposon genes. It has previously been shown that Ty3 retrotransposons are up-regulated in mating populations (Kinsey and Sandmeyer, 1995), and it was interesting to see that genes more generally involved in transposition were significantly up-regulated (Table S2). Transposition during the pheromone-response might provide a mechanism to increase genetic variation in the population prior to mating and, given that the process appears to be regulated by the host (not the transposons), could serve as an example of symbiotic retrotransposition.
The most notable down-regulated genes were involved in ribosomal RNA biogenesis and processing. This is consistent with the fact that much of the cellular resources of an exponentially growing population are directed towards ribosome synthesis (Warner et al., 2001), and demonstrates a down-regulation of this process during the pheromone-response phase. Although a decrease in ribosome biogenesis could be thought of as limiting the protein expression capacity of the cell, there are still very high expression levels of genes which are switched on during the pheromone-response (Bardwell, 2005), and we have previously demonstrated sustained induction of a pheromone regulated metabolic pathway (Williams et al., 2015a). Given the drain that ribosome biogenesis imposes on ATP supply (Warner et al., 2001), and the fact that pheromone regulated genes can still be highly expressed, a reduction in ribosome synthesis can be viewed as a principle requirement for a ‘productive stationary phase’. Many down-regulated genes were involved in DNA replication/repair and chromosome segregation, again reflecting the arrested state of cell division. It is interesting to note that these processes are also typical of the starvation responses associated with stress induced stationary phases (Wu et al., 2004), but that the yeast which were used for the pheromone-response transcriptome analysis were cultured in rich YPD media, and were not starving (Roberts et al., 2000). This observation highlights the unique nature of the pheromone mediated growth-arrest, and validates the idea of attempting to use it as a production phase where flux towards biomass is limited while nutrients are still abundant.
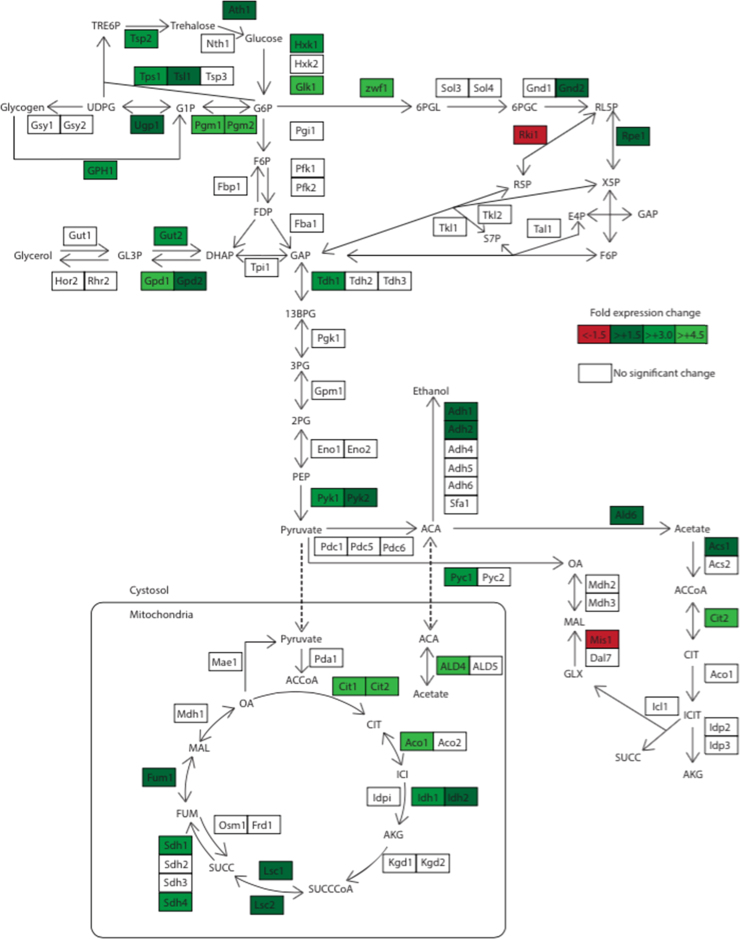
Changes in the expression levels of metabolic genes are of direct relevance to the metabolic component of the pheromone-response. Therefore in addition to analysing global transcriptional changes, central carbon metabolism-specific changes were analysed by mapping the expression levels of significantly changed (95% confidence intervals) metabolic genes along with the reactions they encode (Fig. 2). Significant increases in the transcript levels of a multitude of metabolic enzymes were observed, further suggesting that the mating phenotype has a distinct and active metabolism. In particular, expression levels of genes involved trehalose and glycerol synthesis, the TCA cycle, and the pentose phosphate pathway were significantly up-regulated (Fig. 2). The high expression levels observed for multiple central carbon metabolic genes, along with the active secretion of metabolic side-products (Fig. 1) act as a strong indication that central carbon metabolism is active during the growth-arrest phenotype. There were also a number of interesting trends in regards to specific metabolic processes that are worth speculating on.
Fig. 2.
Transcript levels for genes encoding central metabolic enzymes are shown with up-regulation in green and down-regulation in red. Metabolites are connected by arrows representing reactions catalysed by enzymes (gene names in boxes). Single headed arrows represent one way reactions and double headed denote reversible reactions. The metabolic map and abbreviations were adapted from Oliveira et al. (2012).
Trehalose acts as a major storage carbohydrate in yeast, and increased synthesis has been associated with exposure to thermal, osmotic, and ethanol stress (Pereira et al., 2001). Given that these experiments were carried out at 30 °C (Roberts et al., 2000) and considering that the hyper osmolarity glycerol (HOG) response to osmotic stress and the pheromone-response are insulated from one another (O’Rourke and Herskowitz, 1998) it is possible that ethanol stress during the pheromone-response may be linked to the observed up-regulation of trehalose synthesis genes (Fig. 2). It has previously been reported that trehalose synthesis is required to enable endocytosis at relatively low ethanol concentrations (Lucero et al., 2000). Endocytosis is a process where cells internalise their plasma membrane proteins from the extracellular environment, and is integral to the pheromone-response in yeast where pheromone bound membrane receptor proteins are internalised (Marsh et al., 1991). Consistent with this idea, genes involved in endocytosis were highly up-regulated in response to pheromone (Table S2). It is therefore possible that the up-regulation of trehalose biosynthetic genes in response to pheromone evolved as a mechanism to protect cells from ethanol stress during pheromone bound receptor endocytosis. An alternative explanation is that there is actually a low level of osmotic stress associated with the morphological changes that occur during the pheromone-response, which is responsible for the up-regulation of storage carbohydrate synthesis (see below).
Up-regulation of glycerol synthesis genes (Fig. 2) is consistent with the much higher levels of glycerol accumulation observed in pheromone-treated cells (Fig. 1). It has been proposed that mating yeast cells require a precise osmotic balance prior to cell wall degradation and membrane fusion, and that this balance is achieved with the export of glycerol from the cell via the FPS1 transporter (Philips and Herskowitz, 1997). Recent work has further demonstrated the capacity of yeast responding to pheromone to excrete glycerol, demonstrating that the HOG pathway is actually partially activated by the pheromone-response (Baltanas et al., 2013). The results presented here strongly support these findings, and support the role of trehalose synthesis as an osmoprotectant rather than solely to enable endocytosis under ethanol stress.
Respiratory metabolism requires a greater flux through the TCA cycle for the generation of reducing power to drive oxidative phosphorylation through the electron transport chain. The strong up-regulation of TCA cycle genes suggests that pheromone treatment results in a more highly respiratory metabolism. The extracellular metabolite accumulation rates and respiratory quotients independently support this finding (Table 3). The oxidative branch of the pentose phosphate pathway (PPP) is initiated by glucose-6-phosphate dehydrogenase (ZWF1) in an irreversible step (Nogae and Johnston, 1990). The PPP is responsible for producing NADPH, a critical source of reduction potential required by many anabolic pathways (Minard et al., 1998). The PPP also plays a major role in mitigating the effects of oxidative stress by supplying NADPH to glutathione- and thioredoxin-dependent enzymes (Slekar et al., 1996). The strong up-regulation of PPP genes (ZWF1, GND2, RPE1) in response to pheromone (Fig. 2) could occur as a consequence of the increased rate of respiration initiated by pheromone (Table 3) and the subsequent increase in oxygen radicals. In concordance with this idea was the up-regulation of a multitude of genes involved in both oxidative stress and respiration (Table S2).
It is important to note that the cultures which were used for RNA extraction in the original transcriptome study (Roberts et al., 2000) were carried out under different conditions than the cultures used here to calculate extracellular flux rates (rich YPD rather than minimal medium, Fig. 1, Table 3). Therefore the trends in metabolic gene expression levels which correspond to the flux data should be considered as independent, but consistent observations.
3.3. Engineered pathway productivity during the pheromone-response
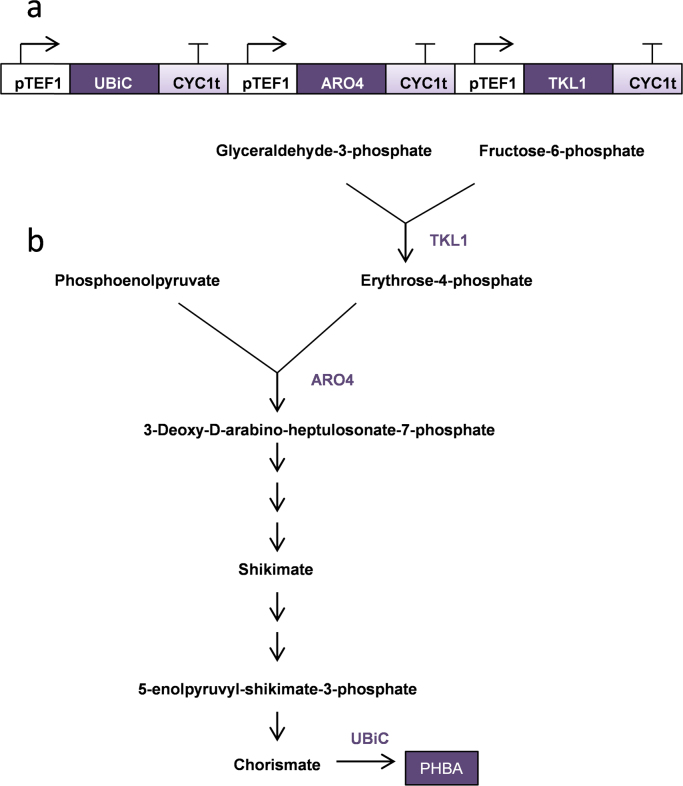
To directly test the hypothesis that the pheromone-response is suitable as a production phase, heterologous compound titers from a strain engineered to produce PHBA±pheromone were compared. PHBA is an important industrial chemical used in liquid crystal polymers (Krömer et al., 2012) that can be derived from the shikimate pathway (Averesch and Krömer, 2014, Winter et al., 2014), which is now also being developed for the synthesis of important products such as codeine and morphine (DeLoache et al., 2015, Galanie et al., 2015, Thodey et al., 2014). PHBA productivity under pheromone exposure was here used to assess the suitability of the mating phenotype as a production phase. A minimally engineered PHBA producing strain was constructed as in Fig. 3 using manipulations previously shown to be effective at increasing shikimate pathway flux (Curran et al., 2013). The constitutive TEF1 promoter was used to drive heterologous expression of the UBiC gene and over-expression of both the TKL1 and the feedback resistant ARO4K229L (Luttik et al., 2008) genes in the engineered pathway; this ensured that gene expression levels would be consistent between treated and untreated populations.
Fig. 3.
Engineered PHBA production pathway. Gene expression constructs (a) and the role of the expressed enzymes (TKL1, ARO4, UBiC) are shown for PHBA production from the shikimate pathway (b).
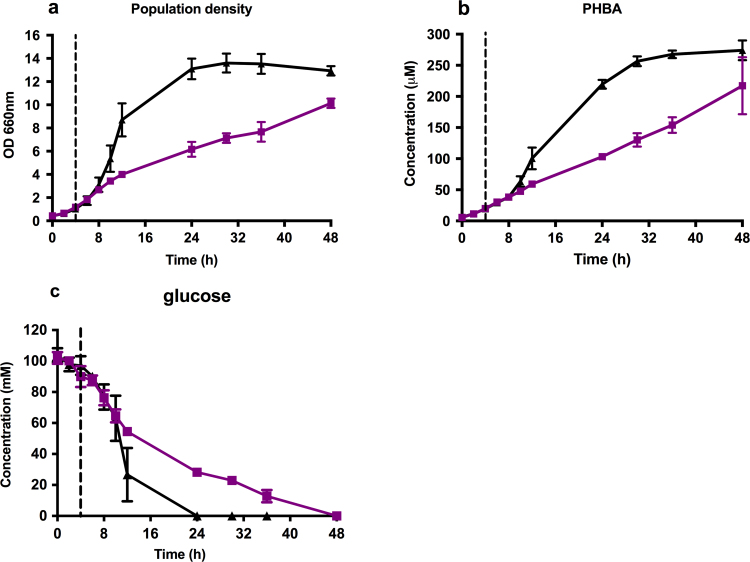
The PHBA producing strain (PSP05) was grown with and without 1 µM alpha pheromone treatment in early log phase (OD660 nm ~1, 4 h). As expected, growth rate slowed significantly after α-pheromone treatment, indicating that the pheromone-response had initiated (Fig. 4a). After 48 h, population densities were significantly different, with the control group at an OD660 nm of 12.93±0.40, and the pheromone-treated group at 10.13±0.40. PHBA concentrations were measured during 48 h of fermentation (Fig. 4b), with no significant difference (p=0.11) between the two groups after 48 h (control=274±16 μM, alpha=217±46 μM). In order to more directly assess the capacity for metabolic productivity, the biomass-specific PHBA production yields and rates between pheromone-mediated growth-arrest at 6 h and glucose exhaustion at 48 h were compared with the control population rate and yield during the glucose consumption phase between 0 and 24 h. There was a significant decrease in the biomass specific PHBA production rate during the glucose consumption phase with the control group producing 9.4±1.95 μM g−1 h−1 and the pheromone treated group producing 3.35±0.58 μM g−1 h−1. Interestingly, the biomass specific PHBA yield was significantly higher in the pheromone treated group during this phase at 93.2±16.2 μM gDCW−1 compared to 62.3±1.3 μM gDCW−1 (p=0.03). Pheromone treatment also resulted in a significantly higher glucose specific PHBA yield during the respective glucose consumption phases (1.64±0.11 μMPHBA compared to 1.19±0.05 μMPHBA p=0.003). Although the PHBA production rate was lower during pheromone-mediated growth-arrest, both the biomass- and glucose-specific PHBA yields were higher. This suggests that the growth arrest phenotype of the S. cerevisiae pheromone response system results in a more productive glucose consumption phase where carbon otherwise directed towards cell growth becomes available for metabolite production. The titer was not greater in the pheromone treated group due to the lower amount of biomass, and the fact that the growth-arrest phenotype was not initiated until the mid-exponential growth phase.
Fig. 4.
PHBA production in pheromone treated cultures. A strain minimally engineered to produce PHBA (strain PHBA03) was grown with (purple lines, squares) and without (black lines, triangles) 1 µM alpha pheromone treatment at early exponential phase (OD of 1, at 4 h, indicated by vertical dashed lines). (a) Population density (OD660 nm), (b) extracellular PHBA concentration, and (c) extracellular glucose concentrations were measured for shake-flask growth with and without pheromone over 48 h.(For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.4. Practical considerations for growth-arrested metabolic productivity
The current data are promising in that we demonstrate the pheromone mediated cell-cycle arrest phenotype is an active phase, but further optimisation of the system would be required to move towards the idealised ‘productive stationary phase’ that was outlined in the introduction. Cells responding to pheromone expend resources executing the mating program, which involves extensive cell-wall remodelling and cell morphology changes. Minimising this energy/resource drain through further metabolic engineering might improve overall pheromone-phase productivity of specific engineered pathways. Moreover, use of a fed-batch approach along with titration of pheromone induction time might demonstrate greater improvements in the pheromone-response productivity compared to the control. A potential issue with using the pheromone-response as a production phase is the capacity for cells to become desensitized to pheromone after prolonged exposure through a variety of well-understood mechanisms (Bardwell, 2005). This phenomenon was not observed during the time-frame of these experiments (population density was still reduced after 48 h of cultivation; Fig. 4), but may need to be prevented through engineering for any longer-term applications. Another consideration is that although it is possible to use the mating phenotype as a production phase, adding purified pheromone to a large-scale fermentation is likely to be prohibitively expensive, depending on the value of the fermentation product. To circumvent this limitation, we recently developed a synthetic quorum sensing circuit where cells produce and respond to their own pheromone in a population density dependent manner (Williams et al., 2013). This quorum sensing network has since been coupled to a recently developed yeast RNA interference module (Crook et al., 2014, Drinnenberg et al., 2009, Si et al., 2014, Williams et al., 2015b) to successfully dynamically regulate the production of PHBA (Williams et al., 2015a). This auto-response system could be applied to ensure sufficient pheromone is available during cultivation.
There are many other methods for arresting the cell cycle under nutrient rich conditions, and it is possible that they could be used as inducers of stationary phase metabolic productivity. For example hydroxyurea can be used to inhibit dNTP synthesis and arrest the cell cycle in S phase (Koç et al., 2004), and nocodazole to disrupt microtubule polymerisation and arrest cells in metaphase (Arber, 2000). While these methods have proved invaluable in elucidating cell-cycle related processes in basic research, they are yet to be explored as inducers of productive growth-arrest phenotypes in biotechnology. These inducers would have the advantage of not involving the initiation of a complex mating phenotype and the associated energetic cost. However, they indirectly induce stress response mechanisms that are likely to impose their own limitations on metabolic productivity. Furthermore, unlike with our synthetic pheromone quorum-sensing circuits (Williams et al., 2015a, Williams et al., 2013) there are no available mechanisms for autoinduction using chemical inducers such as hydroxyurea and nocodazole. In an excellent recent example of chemically induced growth-limitation, pantothenate (vitamin B12) was used to regulate growth and farnesene (jet-fuel) productivity in yeast (Sandoval et al., 2014). In this system the removal of pantothenate from the growth medium of a producer strain resulted in a 70% reduction in farnesene yield and a concomitant increase in growth rate. This enabled pantothenate to be used as an inducer whereby a rapid biomass formation phase in the absence of pantothenate was followed by a switch to a growth-limited production mode upon pantothenate addition to the medium. There are likely to be many analogous systems for other engineered pathways and metabolic networks that hold great promise as inducers of stationary phase metabolic productivity.
It is possible that the growth arrest induced by pheromone in S. cerevisiae could also be a useful production phase in other yeast species that use sexual pheromones to coordinate mating. However, although some species such as Schizosaccharomyces pombe use pheromones, they only mate under nutrient starvation conditions (Yamamoto et al., 1997). Clearly the growth arrest phenotype would not be effective as a production phase under these conditions. Given the drawbacks of using a naturally evolved mating phenotype for production, a much grander solution is to try and reverse engineer the principle features of a ‘productive stationary phase’. In this scenario a synthetic regulatory circuit could be employed as a ‘master controller’ that can simultaneously arrest the cell cycle, reduce ribosomal biogenesis, and switch on the expression of production pathway enzymes while maintaining high central carbon metabolism fluxes. The fact that growth fluxes are no longer required in this scenario opens up the possibility of silencing the expression of ‘essential’ genes, and producing metabolites or proteins that are normally highly toxic to growth. Although the specific mechanisms required to implement such a dramatic regulatory and metabolic shift would differ between organisms, it is possible that the general features are universal.
4. Conclusions
The S. cerevisiae pheromone-response leads to a distinct and active metabolic phenotype that is suitable for the production of PHBA, and for bioprocesses in general. The key metabolic differences which result from pheromone treatment were identified as a glucose uptake rate that is comparable to exponentially growing populations, increased by-product formation, up-regulation of storage carbohydrate synthesis genes associated with osmolarity and oxidative stress responses, and increased respiratory activity. It is conceivable that the pheromone-response could be used with particularly good effect to increase flux towards metabolites of interest that are associated with the metabolic changes that underlie the phenotype.
Although the pheromone-response shows promise as a production phase, it is associated with a complex mating phenotype. The concept of using a growth-arrest as a production phase would be far more effective if it could be reverse engineered and streamlined so that it is decoupled from the unnecessary aspects of the mating phenotype. The ability to engineer a switch from rapid growth, to growth-arrested production in the presence of abundant carbon and nitrogen sources would become an essential design feature of industrial microorganisms. Further investigation of the S. cerevisiae pheromone-response may provide a means for understanding how to coordinate and engineer such a powerful synthetic biology module.
Acknowledgements
TCW was supported by the Australian Postgraduate Award and the AIBN Top-up scholarship. CEV was supported by Queensland Government Smart State and Accelerate fellowships. We thank Dr Manuel Plan and the Metabolomics Australia Queensland node for technical assistance with HPLC.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j. meteno.2016.05.001.
Contributor Information
Thomas C. Williams, Email: tom.williams@mq.edu.au.
Bingyin Peng, Email: bingyin.peng@uq.net.au.
Claudia E. Vickers, Email: c.vickers@uq.edu.au.
Lars K. Nielsen, Email: lars.nielsen@uq.edu.au.
Appendix A. Supplementary material
Supplementary material
.
References
- Albers E., Larsson C., Andlid T., Walsh M.C., Gustafsson L. Effect of nutrient starvation on the cellular composition and metabolic capacity of Saccharomyces cerevisiae. Appl. Environ. Microbiol. 2007;73:4839–4848. doi: 10.1128/AEM.00425-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arber W. Genetic variation: molecular mechanisms and impact on microbial evolution. FEMS Microbiol. Rev. 2000;24:1–7. doi: 10.1111/j.1574-6976.2000.tb00529.x. [DOI] [PubMed] [Google Scholar]
- Averesch N.J.H., Krömer J.O. Tailoring strain construction strategies for muconic acid production in S. cerevisiae and E. coli. Metab. Eng. Commun. 2014;1:19–28. doi: 10.1016/j.meteno.2014.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baltanas R., Bush A., Couto A., Durrieu L., Hohmann S., Colman-Lerner A. Pheromone-induced morphogenesis improves osmoadaptation capacity by activating the HOG MAPK pathway. Sci. Signal. 2013;6 doi: 10.1126/scisignal.2003312. ra26- [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardwell L. A walk-through of the yeast mating pheromone response pathway. Peptides. 2005;26:339–350. doi: 10.1016/j.peptides.2004.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashor C.J., Helman N.C., Yan S., Lim W.A. Using engineered scaffold interactions to reshape MAP kinase pathway signaling dynamics. Science. 2008;319:1539–1543. doi: 10.1126/science.1151153. [DOI] [PubMed] [Google Scholar]
- Bi J.-X., Shuttleworth J., Al-Rubeai M. Uncoupling of cell growth and proliferation results in enhancement of productivity in p21CIP1-arrested CHO cells. Biotechnol. Bioeng. 2004;85:741–749. doi: 10.1002/bit.20025. [DOI] [PubMed] [Google Scholar]
- Blank L.M., Sauer U. TCA cycle activity in Saccharomyces cerevisiae is a function of the environmentally determined specific growth and glucose uptake rates. Microbiology. 2004;150:1085–1093. doi: 10.1099/mic.0.26845-0. [DOI] [PubMed] [Google Scholar]
- Chan R.K., Otte C.A. Physiological characterization of Saccharomyces cerevisiae mutants supersensitive to G1 arrest by a factor and alpha factor pheromones. Mol. Cell. Biol. 1982;2:21–29. doi: 10.1128/mcb.2.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chubukov V., Sauer U. Environmental dependence of stationary-phase metabolism in Bacillus subtilis and Escherichia coli. Appl. Environ. Microbiol. 2014;80:2901–2909. doi: 10.1128/AEM.00061-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crook N.C., Schmitz A.C., Alper H.S. Optimization of a yeast RNA interference system for controlling gene expression and enabling rapid metabolic engineering. ACS Synth. Biol. 2014;3:307–313. doi: 10.1021/sb4001432. [DOI] [PubMed] [Google Scholar]
- Curran K.A., Leavitt J.M., Karim A.S., Alper H.S. Metabolic engineering of muconic acid production in Saccharomyces cerevisiae. Metab. Eng. 2013;15:55–66. doi: 10.1016/j.ymben.2012.10.003. [DOI] [PubMed] [Google Scholar]
- Da Silva N.A., Srikrishnan S. Introduction and expression of genes for metabolic engineering applications in Saccharomyces cerevisiae. FEMS Yeast Res. 2012;12:197–214. doi: 10.1111/j.1567-1364.2011.00769.x. [DOI] [PubMed] [Google Scholar]
- DeLoache W.C., Russ Z.N., Narcross L., Gonzales A.M., Martin V.J.J., Dueber J.E. An enzyme-coupled biosensor enables (S)-reticuline production in yeast from glucose. Nat. Chem. Biol. 2015;11:465–471. doi: 10.1038/nchembio.1816. [DOI] [PubMed] [Google Scholar]
- Dietmair S., Timmins N.E., Gray P.P., Nielsen L.K., Krömer J.O. Towards quantitative metabolomics of mammalian cells: development of a metabolite extraction protocol. Anal. Biochem. 2010;404:155–164. doi: 10.1016/j.ab.2010.04.031. [DOI] [PubMed] [Google Scholar]
- Dijken J., Weusthuis R., Pronk J. Kinetics of growth and sugar consumption in yeasts. Antonie Van. Leeuwenhoek. 1993;63:343–352. doi: 10.1007/BF00871229. [DOI] [PubMed] [Google Scholar]
- Drinnenberg I.A., Weinberg D.E., Xie K.T., Mower J.P., Wolfe K.H., Fink G.R., Bartel D.P. RNAi in budding yeast. Science. 2009;326:544–550. doi: 10.1126/science.1176945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa K., Hohmann S. Synthetic biology: lessons from engineering yeast MAPK signalling pathways. Mol. Microbiol. 2013;88:5–19. doi: 10.1111/mmi.12174. [DOI] [PubMed] [Google Scholar]
- Fussenegger M., Schlatter S., Datwyler D., Mazur X., Bailey J.E. Controlled proliferation by multigene metabolic engineering enhances the productivity of Chinese hamster ovary cells. Nat. Biotechnol. 1998;16:468–472. doi: 10.1038/nbt0598-468. [DOI] [PubMed] [Google Scholar]
- Galanie S., Thodey K., Trenchard I.J., Filsinger Interrante M., Smolke C.D. Complete biosynthesis of opioids in yeast. Science. 2015;349:1095–1100. doi: 10.1126/science.aac9373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz R.D., Schiestl R.H. Quick and easy yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat. Protoc. 2007;2:35–37. doi: 10.1038/nprot.2007.14. [DOI] [PubMed] [Google Scholar]
- Groß A., Rödel G., Ostermann K. Application of the yeast pheromone system for controlled cell–cell communication and signal amplification. Lett. Appl. Microbiol. 2011;52:521–526. doi: 10.1111/j.1472-765X.2011.03035.x. [DOI] [PubMed] [Google Scholar]
- Gueldener U., Heinisch J., Koehler G.J., Voss D., Hegemann J.H. A second set of loxP marker cassettes for Cre-mediated multiple gene knockouts in budding yeast. Nucleic Acids Res. 2002;30:e23. doi: 10.1093/nar/30.6.e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Güldener U., Heck S., Fiedler T., Beinhauer J., Hegemann J.H. A new efficient gene disruption cassette for repeated use in budding yeast. Nucleic Acids Res. 1996;24:2519–2524. doi: 10.1093/nar/24.13.2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennig S., Clemens A., Rödel G., Ostermann K. A yeast pheromone-based inter-species communication system. Appl. Microbiol. Biotechnol. 2015;99:1299–1308. doi: 10.1007/s00253-014-6133-5. [DOI] [PubMed] [Google Scholar]
- Herman P.K. Stationary phase in yeast. Curr. Opin. Microbiol. 2002;5:602–607. doi: 10.1016/s1369-5274(02)00377-6. [DOI] [PubMed] [Google Scholar]
- Heyland J., Fu J., Blank L.M. Correlation between TCA cycle flux and glucose uptake rate during respiro-fermentative growth of Saccharomyces cerevisiae. Microbiology. 2009;155:3827–3837. doi: 10.1099/mic.0.030213-0. [DOI] [PubMed] [Google Scholar]
- Holtz W.J., Keasling J.D. Engineering static and dynamic control of synthetic pathways. Cell. 2010;140:19–23. doi: 10.1016/j.cell.2009.12.029. [DOI] [PubMed] [Google Scholar]
- Horton R.M., Hunt H.D., Ho S.N., Pullen J.K., Pease L.R. Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene. 1989;77:61–68. doi: 10.1016/0378-1119(89)90359-4. [DOI] [PubMed] [Google Scholar]
- Ingolia N.T., Murray A.W. Positive-feedback loops as a flexible biological module. Curr. Biol. 2007;17:668–677. doi: 10.1016/j.cub.2007.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahn M., Mölle A., Rödel G., Ostermann K. Temporal and spatial properties of a yeast multi-cellular amplification system based on signal molecule diffusion. Sensors. 2013;13:14511–14522. doi: 10.3390/s131114511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keasling J.D. Synthetic biology for synthetic chemistry. ACS Chem. Biol. 2008;3:64–76. doi: 10.1021/cb7002434. [DOI] [PubMed] [Google Scholar]
- Khoo S.H.G., Al-Rubeai M. Metabolic characterization of a hyper-productive state in an antibody producing NS0 myeloma cell line. Metab. Eng. 2009;11:199–211. doi: 10.1016/j.ymben.2009.02.001. [DOI] [PubMed] [Google Scholar]
- Kinsey P.T., Sandmeyer S.B. Ty3 transposes in mating populations of yeast - a novel transposition assay for Ty3. Genetics. 1995;139:81–94. doi: 10.1093/genetics/139.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koç A., Wheeler L.J., Mathews C.K., Merrill G.F. Hydroxyurea arrests DNA replication by a mechanism that preserves basal dNTP pools. J. Biol. Chem. 2004;279:223–230. doi: 10.1074/jbc.M303952200. [DOI] [PubMed] [Google Scholar]
- Krömer J.O., Nunez-Bernal D., Averesch N.J.H., Hampe J., Varela J., Varela C. Production of aromatics in Saccharomyces cerevisiae—a feasibility study. J. Biotechnol. 2012 doi: 10.1016/j.jbiotec.2012.04.014. [DOI] [PubMed] [Google Scholar]
- Kumar N., Gammell P., Clynes M. Proliferation control strategies to improve productivity and survival during CHO based production culture. Cytotechnology. 2007;53:33–46. doi: 10.1007/s10616-007-9047-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucero P., Peñalver E., Moreno E., Lagunas R. Internal trehalose protects endocytosis from inhibition by ethanol in Saccharomyces cerevisiae. Appl. Environ. Microbiol. 2000;66:4456–4461. doi: 10.1128/aem.66.10.4456-4461.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luttik M.A.H., Vuralhan Z., Suir E., Braus G.H., Pronk J.T., Daran J.M. Alleviation of feedback inhibition in Saccharomyces cerevisiae aromatic amino acid biosynthesis: quantification of metabolic impact. Metab. Eng. 2008;10:141–153. doi: 10.1016/j.ymben.2008.02.002. [DOI] [PubMed] [Google Scholar]
- Marsh L., Neiman A.M., Herskowitz I. Signal transduction during pheromone response in yeast. Annu. Rev. Cell Biol. 1991;7:699–728. doi: 10.1146/annurev.cb.07.110191.003411. [DOI] [PubMed] [Google Scholar]
- Mateus C., Avery S.V. Destabilized green fluorescent protein for monitoring dynamic changes in yeast gene expression with flow cytometry. Yeast. 2000;16:1313–1323. doi: 10.1002/1097-0061(200010)16:14<1313::AID-YEA626>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- Minard K.I., Jennings G.T., Loftus T.M., Xuan D., McAlister-Henn L. Sources of NADPH and expression of mammalian NADP+-specific isocitrate dehydrogenases in Saccharomyces cerevisiae. J. Biol. Chem. 1998;273:31486–31493. doi: 10.1074/jbc.273.47.31486. [DOI] [PubMed] [Google Scholar]
- Nogae I., Johnston M. Isolation and characterization of the ZWF1 gene of Saccharomyces cerevisiae, encoding glucose-6-phosphate dehydrogenase. Gene. 1990;96:161–169. doi: 10.1016/0378-1119(90)90248-p. [DOI] [PubMed] [Google Scholar]
- O’Shaughnessy E.C., Palani S., Collins J.J., Sarkar C.A. Tunable signal processing in synthetic MAP kinase cascades. Cell. 2011;144:119–131. doi: 10.1016/j.cell.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Rourke S.M., Herskowitz I. The Hog1 MAPK prevents cross talk between the HOG and pheromone response MAPK pathways in Saccharomyces cerevisiae. Genes Dev. 1998;12:2874–2886. doi: 10.1101/gad.12.18.2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira A.P., Ludwig C., Picotti P., Kogadeeva M., Aebersold R., Sauer U. Regulation of yeast central metabolism by enzyme phosphorylation. Mol. Syst. Biol. 2012;8 doi: 10.1038/msb.2012.55. (n/a-n/a) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partow S., Siewers V., Bjørn S., Nielsen J., Maury J. Characterization of different promoters for designing a new expression vector in Saccharomyces cerevisiae. Yeast. 2010;27:955–964. doi: 10.1002/yea.1806. [DOI] [PubMed] [Google Scholar]
- Peng B., Williams T.C., Henry M., Nielsen L.K., Vickers C.E. Controlling heterologous gene expression in yeast cell factories on different carbon substrates and across the diauxic shift: a comparison of yeast promoter activities. Microb. Cell Factor. 2015;14:91. doi: 10.1186/s12934-015-0278-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira M., Eleutherio E., Panek A. Acquisition of tolerance against oxidative damage in Saccharomyces cerevisiae. BMC Microbiol. 2001;1:11. doi: 10.1186/1471-2180-1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philips J., Herskowitz I. Osmotic balance regulates cell fusion during mating in Saccharomyces cerevisiae. J. Cell Biol. 1997;138:961–974. doi: 10.1083/jcb.138.5.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regot S., Macia J., Conde N., Furukawa K., Kjellen J., Peeters T., Hohmann S., de Nadal E., Posas F., Sole R. Distributed biological computation with multicellular engineered networks. Nature. 2011;469:207–211. doi: 10.1038/nature09679. [DOI] [PubMed] [Google Scholar]
- Roberts C.J., Nelson B., Marton M.J., Stoughton R., Meyer M.R., Bennett H.A., He Y.D., Dai H., Walker W.L., Hughes T.R., Tyers M., Boone C., Friend S.H. Signaling and Circuitry of multiple MAPK pathways revealed by a matrix of global gene expression profiles. Science. 2000;287:873–880. doi: 10.1126/science.287.5454.873. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., Russell, D.W., 2001. Molecular Cloning: A Laboratory Manual (3-Volume Set). Cold Spring Harbor Laboratory Press.
- Sandoval C.M., Ayson M., Moss N., Lieu B., Jackson P., Gaucher S.P., Horning T., Dahl R.H., Denery J.R., Abbott D.A., Meadows A.L. Use of pantothenate as a metabolic switch increases the genetic stability of farnesene producing Saccharomyces cerevisiae. Metab. Eng. 2014;25:215–226. doi: 10.1016/j.ymben.2014.07.006. [DOI] [PubMed] [Google Scholar]
- Si T., Luo Y., Bao Z., Zhao H. RNAi-assisted genome evolution in Saccharomyces cerevisiae for complex phenotype engineering. ACS Synth. Biol. 2014 doi: 10.1021/sb500074a. [DOI] [PubMed] [Google Scholar]
- Sikorski R.S., Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slekar K.H., Kosman D.J., Culotta V.C. The yeast copper/zinc superoxide dismutase and the pentose phosphate pathway play overlapping roles in oxidative stress protection. J. Biol. Chem. 1996;271:28831–28836. doi: 10.1074/jbc.271.46.28831. [DOI] [PubMed] [Google Scholar]
- Stansfield I., Stark M.J.R. 2 yeast genetics and strain construction. In: Ian S., Michael J.R.S., editors. Methods in Microbiology. vol. Academic Press; Cambridge: 2007. pp. 23–43. [Google Scholar]
- Stephanopoulos G., Aristidou A.A., Nielsen J. Elsevier Science; Amsterdam: 1998. Metabolic Engineering: Principles and Methodologies. [Google Scholar]
- Sun J., Shao Z., Zhao H., Nair N., Wen F., Xu J.H., Zhao H. Cloning and characterization of a panel of constitutive promoters for applications in pathway engineering in Saccharomyces cerevisiae. Biotechnol. Bioeng. 2012;109:2082–2092. doi: 10.1002/bit.24481. [DOI] [PubMed] [Google Scholar]
- Tanaka H., Yi T.M. Synthetic morphology using alternative inputs. PLoS ONE. 2009;4:e6946. doi: 10.1371/journal.pone.0006946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thodey K., Galanie S., Smolke C.D. A microbial biomanufacturing platform for natural and semisynthetic opioids. Nat. Chem. Biol. 2014;10:837–844. doi: 10.1038/nchembio.1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venayak N., Anesiadis N., Cluett W.R., Mahadevan R. Engineering metabolism through dynamic control. Curr. Opin. Biotechnol. 2015;34:142–152. doi: 10.1016/j.copbio.2014.12.022. [DOI] [PubMed] [Google Scholar]
- Verduyn C., Postma E., Scheffers W.A., Van Dijken J.P. Effect of benzoic acid on metabolic fluxes in yeasts: a continuous-culture study on the regulation of respiration and alcoholic fermentation. Yeast. 1992;8:501–517. doi: 10.1002/yea.320080703. [DOI] [PubMed] [Google Scholar]
- Warner J.R., Vilardell J., Sohn J.H. Economics of Ribosome Biosynthesis. Cold Spring Harb. Symp. Quant. Biol. 2001;66:567–574. doi: 10.1101/sqb.2001.66.567. [DOI] [PubMed] [Google Scholar]
- Watanabe S., Shuttleworth J., Al-Rubeai M. Regulation of cell cycle and productivity in NS0 cells by the over-expression of p21CIP1. Biotechnol. Bioeng. 2002;77:1–7. doi: 10.1002/bit.10112. [DOI] [PubMed] [Google Scholar]
- Werner-Washburne M., Braun E., Johnston G.C., Singer R.A. Stationary phase in the yeast Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 1993;57:383–401. doi: 10.1128/mr.57.2.383-401.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams T.C., Averesch N.J.H., Winter G., Plan M.R., Vickers C.E., Nielsen L.K., Krömer J.O. Quorum-sensing linked RNA interference for dynamic metabolic pathway control in Saccharomyces cerevisiae. Metab. Eng. 2015;29:124–134. doi: 10.1016/j.ymben.2015.03.008. [DOI] [PubMed] [Google Scholar]
- Williams T.C., Espinosa M.I., Nielsen L.K., Vickers C.E. Dynamic regulation of gene expression using sucrose responsive promoters and RNA interference in Saccharomyces cerevisiae. Microb. Cell Factor. 2015;14:43. doi: 10.1186/s12934-015-0223-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams T.C., Nielsen L.K., Vickers C.E. Engineered quorum sensing using pheromone-mediated cell-to-cell communication in Saccharomyces cerevisiae. ACS Synth. Biol. 2013;2:136–149. doi: 10.1021/sb300110b. [DOI] [PubMed] [Google Scholar]
- Winter G., Averesch N.J., Nunez-Bernal D., Kromer J.O. In vivo instability of chorismate causes substrate loss during fermentative production of aromatics. Yeast. 2014;31:333–341. doi: 10.1002/yea.3025. [DOI] [PubMed] [Google Scholar]
- Woolston B.M., Edgar S., Stephanopoulos G. Metabolic engineering: past and future. Ann. Rev. Chem. Biomol. Eng. 2013;4:259–288. doi: 10.1146/annurev-chembioeng-061312-103312. [DOI] [PubMed] [Google Scholar]
- Wu J., Zhang N., Hayes A., Panoutsopoulou K., Oliver S.G. Global analysis of nutrient control of gene expression in Saccharomyces cerevisiae during growth and starvation. Proc. Natl. Acad. Sci. USA. 2004;101:3148–3153. doi: 10.1073/pnas.0308321100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto M., Imai Y., Watanabe Y. 12 Mating and Sporulation in Schizosaccharomyces pombe. Cold Spring Harb. Monogr. Arch. 1997;21:1037–1106. [Google Scholar]
- Zhang N.-N., Dudgeon D.D., Paliwal S., Levchenko A., Grote E., Cunningham K.W. Multiple signaling pathways regulate yeast cell death during the response to mating pheromones. Mol. Biol. Cell. 2006;17:3409–3422. doi: 10.1091/mbc.E06-03-0177. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material