Abstract
Succinate produced by microorganisms can replace currently used petroleum-based succinate but typically requires mono- or poly-saccharides as a feedstock. The cyanobacterium Synechocystis sp. PCC6803 can produce organic acids such as succinate from CO2 not supplemented with sugars under dark anoxic conditions using an unknown metabolic pathway. The TCA cycle in cyanobacteria branches into oxidative and reductive routes. Time-course analyses of the metabolome, transcriptome and metabolic turnover described here revealed dynamic changes in the metabolism of Synechocystis sp. PCC6803 cultivated under dark anoxic conditions, allowing identification of the carbon flow and rate-limiting steps in glycogen catabolism. Glycogen biosynthesized from CO2 assimilated during periods of light exposure is catabolized to succinate via glycolysis, the anaplerotic pathway, and the reductive TCA cycle under dark anoxic conditions. Expression of the phosphoenolpyruvate (PEP) carboxylase gene (ppc) was identified as a rate-limiting step in succinate biosynthesis and this rate limitation was alleviated by ppc overexpression, resulting in improved succinate excretion. The sugar-free succinate production was further enhanced by the addition of bicarbonate. In vivo labeling with NaH13CO3 clearly showed carbon incorporation into succinate via the anaplerotic pathway. Bicarbonate is in equilibrium with CO2. Succinate production by Synechocystis sp. PCC6803 therefore holds significant promise for CO2 capture and utilization.
Keywords: Autofermentation, Cyanobacteria, Dynamic metabolic profiling, Metabolomics, Succinate, Synechocystis
Highlights
-
•
The cyanobacterium Synechocystis produces succinate under dark anoxic condition.
-
•
Multi-omics revealed dynamic change in metabolism under dark anoxic condition.
-
•
Carbon flow and rate-limiting steps in glycogen catabolism was elucidated.
-
•
PEP carboxylase gene overexpression improved Synechocystis succinate production.
-
•
Bicarbonate addition to medium dramatically improved succinate production.
1. Introduction
The issue of fossil resource depletion and global warming has prompted research on the sustainable production of environmentally benign fuels and chemicals. In particular, the utilization of biomass as a starting material for the production of fuels and chemicals has attracted considerable recent attention (Bozell and Peterson, 2010, Choi et al., 2015). The “biorefinery” is a manufacturing process for producing a wide variety of chemicals from biomass and is a promising alternative to conventional oil refinery processes. The use of bio-based chemicals can help reduce the amount of CO2 emitted by fossil fuel combustion.
Photosynthetic algae are of increasing interest for the sustainable production of bio-based fuels and chemicals because the cultivation of algae does not directly compete with terrestrial agricultural resources such as productive land and fresh water (Dismukes et al., 2008, Wijffels et al., 2013). Several species of microalgae and cyanobacteria can store significant amounts of energy-rich compounds such as lipids and polysaccharides (starch and glycogen) that can be utilized for the production of distinct bio-fuels, including bio-diesel and bio-ethanol (Aikawa et al., 2013, Aikawa et al., 2014). In particular, cyanobacteria are attractive because they are responsible for a substantial proportion of biomass in the hydrosphere (Partensky et al., 1999), and their metabolic pathways are more readily amenable to genetic modifications designed to enhance photosynthetic activity than are those of eukaryotic algae (Hasunuma et al., 2014). The genome of the unicellular cyanobacterium Synechocystis sp. PCC6803 (hereafter Synechocystis 6803) was sequenced in 1996 (Kaneko et al., 1996) and Synechocystis 6803 is one of the most extensively investigated cyanobacteria. Synechocystis 6803 was recently shown to directly convert CO2 into commodity chemicals such as acetone, 2,3-butanediol, 3-hydroxypropionic acid, isobutanol and n-butanol following the application of metabolic engineering strategies such as overexpression, gene deletion, and tuning of inherent and heterologous metabolic pathways (Anfelt et al., 2015, Savakis et al., 2013, Varman et al., 2013, Wang et al., 2016, Zhou et al., 2012).
Succinic acid (1,2-ethanedicarboxylic acid) is likely to become an important chemical intermediate for the production of biodegradable plastics, polybutylene succinate, polyester polyols and polyurethanes (Cukalovic and Stevens, 2008, Delhomme et al., 2009). As a platform chemical, succinic acid can be chemically converted to other valuable chemicals such as 1,4-butanediol, γ-butyrolactone, tetrahydrofuran and N-methylpyrrolidone, which can be used as surfactants and ion chelators, as well as being used in the food and pharmaceutical industries (Akhtar et al., 2014). The current market size for succinate is about 30,000–50,000 t per year, while the market potential is expected to be more than 700,000 t per year by 2020 (Choi et al., 2015). Succinate can be produced from renewable resources, including sugars and starchy/lignocellulosic biomass, by biotechnological approaches (Akhtar et al., 2014, Chen and Nielsen, 2013, Tsuge et al., 2013). Bio-based succinate has been generated using native producers such as rumen bacteria (e. g., Actinobacillus succinogenes, Anaerobiospirillum succiniciproducens, and Bacillus fragilis) and certain fungi (e. g., Fusarium, Aspergillus and Penicillium species), and metabolically engineered microorganisms such as Escherichia coli, Corynebacterium glutamicum and Saccharomyces cerevisiae (Beauprez et al., 2010, Chen and Nielsen, 2013, Lee et al., 2014). However, such heterotrophic microorganisms need sugars or biomass feedstocks as carbon sources for succinate fermentation, and thus the supply of fermentation substrates remains a bottleneck to realizing the economical and energy-saving production of bio-based chemicals.
Synechocystis 6803 can excrete succinate in the absence of sugars by autofermentation under dark anoxic conditions (Osanai et al., 2015). Glycogen, the primary storage polysaccharide in cyanobacteria (Ball and Morell, 2003), is biosynthesized from CO2 under phototrophic conditions, which might be catabolized to form a range of fermentation products under dark anoxic conditions. In other words, succinate can be directly produced from CO2 without the addition of any other feedstock. However, metabolic flow during sugar catabolism has never been directly observed, and little is known about the mechanism that regulates succinate biosynthesis in cyanobacteria under dark anoxic conditions.
Metabolomics is the comprehensive analysis of a wide range of intracellular metabolites and has identified metabolites that play important roles in specific biological processes (Camañes et al., 2015, Diamond et al., 2015, Link et al., 2013). Recently, dynamic metabolic profiling, which directly measures the turnover of metabolic intermediates in cyanobacteria, was developed by combining in vivo 13C-labeling of metabolites, metabolomics, and mass distribution analysis using mass spectrometry (MS) (Hasunuma et al., 2013). Such profiling enables kinetic visualization of carbon metabolism in central metabolic pathways such as glycolysis and the pentose phosphate pathway under phototrophic conditions (Hasunuma et al., 2014).
The present study investigated the organic acids released into the fermentation medium during dark anoxic cultivation of Synechocystis 6803. Time-course analysis of Synechocystis metabolomics and dynamic metabolic profiling were performed during cultivation to isolate the rate-limiting steps of cyanobacterial succinate biosynthesis. Moreover, the limiting step was removed by overexpression of the rate-limiting step gene and by controlling the components in the medium to improve succinic acid production.
2. Materials and methods
2.1. Strains and culture conditions
A GT strain of Synechocystis sp. PCC6803 (Williams, 1988), which was adapted to utilize glucose as a carbon source, was grown in BG11 liquid medium (Rippka et al., 1979). Cells were pre-cultivated for 4 days in 500-mL flasks containing 150 mL BG11 medium and 20 mM Hepes-KOH (pH 7.8) in an NC350-HC plant chamber (Nippon Medical and Chemical Instruments Co., Ltd., Osaka, Japan) under continuous irradiation with photosynthetically-active 50 µmol m–2 s–1 white light photons and with 100-rpm agitation at 30 °C. The preculture was adjusted to an initial optical density at 750 nm (OD750) of 0.1. Cultivation was performed in a closed double-deck flask: the first stage contained 50 mL of 2 M NaHCO3/Na2CO3 buffer at the pH required to obtain a CO2 concentration of 1% (v/v), and the second stage contained 70 mL modified-BG11 medium (BG11 medium containing 5 mM NH4Cl or 5 mM NaNO3 as a nitrogen source and 50 mM Hepes-KOH, pH 7.8) (Hasunuma et al., 2014). The pre-cultured cells were inoculated into fresh modified-BG11 medium at a biomass concentration of 0.1 g dry cell weight (DCW) L−1 (OD750 of 0.4) and cultivated for 3 days under continuous irradiation with 100–110 µmol m–2 s–1 white light photons with 100-rpm agitation at 30 °C. The light intensity was measured in the center of the medium using an LI-250A light meter equipped with an LI-190SA quantum sensor (LI-COR, Lincoln, NE). The cell density in the medium was determined as DCW because a linear correlation was observed between DCW and optical density measured at OD750 using a UV mini spectrophotometer (Shimadzu, Kyoto, Japan).
2.2. Construction of recombinant strains
The rbcL terminator and downstream regions of slr0168 were amplified from Synechocystis 6803 genomic DNA by PCR using the primer set 5′-CCTCTAGAGTCGACCTGCAGGTTACAGTTTTGGCAATTAC-3′/5′-GCCAGCCCCAACACCTGACGCGTTTCCCCACTTAGATAAAAAATCC-3′ and 5′-TCTAAGTGGGGAAACGCGTCAGGTGTTGGGGCTGGC-3′/5′-TGATTACGCCAAGCTTCTAAGTCAGCGTAAATCTGACAATG-3′, respectively, and then integrated into the PstI and HindIII sites of pBluescript II SK(+) (Agilent Technologies, Palo Alto, CA) using an In-Fusion HD Cloning Kit (Takara Bio, Shiga, Japan) to yield pBluescript-TrbcL-slr0168. A kanamycin-resistance cassette and rbcL promoter were amplified from pCRII-TOPO (Invitrogen, Carlsbad, CA) and Synechocystis 6803 genomic DNA by PCR using the primer set 5′-CGGGCCCCCCCTCGAGCCGGAATTGCCAGCTGGGGC-3′/5′-TGGACTTTCTAATTAGAGCGGCCGCTCAGAAGAACTCGTCAAGA-3′ and 5′-TCTTGACGAGTTCTTCTGAGCGGCCGCTCTAATTAGAAAGTCCA-3′/5′-CCGGGGATCCTCTAGACATATGGGTCAGTCCTCCAT-3′, respectively. The amplified fragments were integrated into the XhoI and XbaI sites of pBluescript-TrbcL-slr0168 using an In-Fusion HD Cloning Kit to yield pBluescript-Kmr-PrbcL-TrbcL-slr0168. The upstream region of slr0168 was amplified from Synechocystis 6803 genomic DNA by PCR using the primer set 5′-TATAGGGCGAATTGGGTACCATGACTATTCAATACACCCCCCTAG-3′/5′-TACCGTCGACCTCGAGCACCAGACCAAAGCCGGGAATTTC-3′ and then integrated into the KpnI and XhoI sites of pBluescript-Kmr-PrbcL-TrbcL-slr0168 using an In-Fusion HD Cloning Kit to yield pBluescript-slr0168-Kmr-PrbcL-TrbcL-slr0168. The NdeI site (CATATG) of pUC19 (Takara Bio) was replaced with CACATG by digesting AatII and EcoRI and inserting the synthetic DNA. Following the digestion of pBluescript-slr0168-Kmr-PrbcL-TrbcL-slr0168 with KpnI and HindIII, the fragment containing slr0168 was integrated into the KpnI/HindIII site of the modified pUC19 vector to yield pSKrbcL-slr0168. The trc promoter was amplified from pTrcHis vector (Invitrogen) by PCR using the primer set 5′-TTCTTCTGAGCGGCCGCCGACTGCACGGTGCACCAAT-3′/5′- TCGACTCTAGACATATGGGTCTGTTTCCTGTGTGAA-3′ and then integrated into the NotI and NdeI site of pSKrbcL-slr0168 to yield pSKtrc-slr0168. The ppc (sll0920) gene encoding phosphoenolpyruvate (PEP) carboxylase was amplified from Synechocystis 6803 genomic DNA by PCR using the primer set 5′-AGGAAACAGACCCATATGAACTTGGCAGTTCCTGC-3′/5′-AACCTGCAGGTCGACTCAACCAGTATTACGCA-3′. The resulting fragment was integrated into NdeI/SalI digested pSKtrc-slr0168 using an In-Fusion HD Cloning Kit to yield pSKtrc-slr0168/sll0920. The GT strain was transformed with pSKtrc-slr0168/sll0920 using a previously described method (Osanai et al., 2011) to yield strain Ppc-ox. Also, the plasmid pSKtrc-slr0168 was introduced into the GT strain to yield the vector control strain. Colonies resistant to 50 μg mL−1 kanamycin were selected, and isolation of a single colony was repeated to achieve complete segregation. The chromosomal integration of ppc and the plasmid-derived sequence was confirmed by PCR using the specific primers 5′-ATGGCACCGATGCGGAATCCCAACAGATTGCCTTTGAC-3′ and 5′-CACGTTGGGTCCCAAGTTTGTGCTGTGGCTGATGCCAT-3′.
2.3. Batch fermentation under anaerobic conditions
After cultivation under phototrophic conditions for 3 days, the cells were collected by filtration using 1-μm pore size polytetrafluoroethylene (PTFE) filter disks (Omnipore; Millipore, Billerica, MA) and transferred into 50 mM Hepes-KOH (pH 7.8). All fermentations were performed at 30 °C under anaerobic conditions with an agitation speed of 170 rpm in 30 mL closed bottles containing 10 mL cell suspension. The initial cell concentration was adjusted to 5 g DCW L−1 (OD750 of 20). The effects of adding NaHCO3 on dark anoxic cultivation of Synechocystis 6803 were investigated using 100 mM Hepes-KOH (pH 7.8) to stabilize the pH of the medium. The concentrations of organic acids in the fermentation medium were analyzed using a high performance liquid chromatography (HPLC) system (Shimadzu) equipped with an Aminex HPX-87H column (300 mm×7.8 mm; Bio-Rad, Hercules, CA) and an RID-10A refractive index detector (Shimadzu). The HPLC system was operated at 50 °C using 5 mM H2SO4 as the mobile phase at a flow rate of 0.6 mL min−1.
2.4. Analysis of intracellular metabolites
Samples for intracellular metabolite analysis were prepared as described previously (Hasunuma et al., 2014). Cyanobacterial cells equivalent to 5 mg DCW were collected from fermentation vessels and immediately filtered using 1-μm pore size PTFE filter disks (Omnipore). After washing the filter disks with 20 mM ammonium carbonate pre-chilled to 4 °C, cells retained on the filters were immediately placed into 2 mL pre-cooled (−30 °C) methanol containing 37.38 μM l-methionine sulfone and 37.38 μM piperazine-1,4-bis(2-ethanesulfonic acid) as internal standards for mass analysis. Cells were suspended by vortexing, then 0.5 mL of the cell suspension was mixed with 0.2 mL of pre-cooled (4 °C) water and 0.5 mL of chloroform at 4 °C. After vortexing for 30 s, the aqueous and organic layers were separated by centrifugation at 14,000xg for 5 min at 4 °C. The aqueous layer (500 μL) was filtered through a Millipore 5 kDa cut-off membrane to remove solubilized proteins. Water was evaporated from the aqueous-layer extracts under vacuum using a FreeZone 2.5 Plus freeze dryer system (Labconco, Kansas City, MO) and the dried metabolites were dissolved in 20 μL of Milli-Q water. The metabolites were analyzed using a capillary electrophoresis-mass spectrometry (CE-MS) system (CE, Agilent G7100; MS, Agilent G6224AA LC/MSD TOF; Agilent Technologies) controlled by MassHunter Workstation Data Acquisition software (Agilent Technologies), as described previously (Hasunuma et al., 2013).
2.5. Global gene expression analysis
Total RNA was obtained after 3, 6, 24, and 48 h of fermentation using a Total RNA Isolation Mini Kit (Agilent Technologies) according to the manufacturer's protocol. RNA concentration and quality were measured using a NanoDrop ND-1000 spectrophotometer (NanoDrop, Wilmington, DE) and an Agilent 2100 Bioanalyzer (Agilent Technologies), respectively. cDNA was reverse-transcribed and labeled with cyanine 3-CTP using a Low-Input Quick Amp Labeling Kit (Agilent Technologies) for hybridization in 8×15k microarrays designed by the author. Hybridization was performed at 65 °C for 17 h and the arrays were scanned using an Agilent Single-Color DNA Microarray Scanner (Agilent Technologies). Gene expression levels were normalized per chip. GeneSpring GX ver. 11.5.1 software (Agilent Technologies) was used to analyze the fold-change in expression data. Each biological sample subjected to microarray analysis was analyzed in triplicate.
2.6. Analysis of intracellular glycogen
Synechocystis cells were harvested from fermentation vessels using PTFE filter disks, as described above. Cells on the filters were washed with 20 mM ammonium carbonate, then immediately frozen in liquid nitrogen and freeze-dried using a freeze dryer (Labconco). Glycogen was extracted as described previously (Izumi et al., 2013). Briefly, glycogen was extracted from 5 mg dry weight cells with 100 μL aqueous KOH (30%, w/v) by incubation in a 90 °C heat block for 90 min, followed by placement on ice. Ethanol (300 μL) pre-chilled to 4 °C was added to the cooled extracts and the extracts were incubated on ice for 1 h. After centrifugation at 7500xg for 5 min at 4 °C, the resulting pellet was washed three times with cold ethanol, then dried for 30 min at 80 °C. Reconstitution in 100 μL water and centrifugation at 14,000xg for 5 min provided the glycogen extract as the supernatant. The glycogen concentration was determined by measuring the glucose released from glycogen by enzymatic hydrolysis. Glycogen extract (50 μL) was mixed with 40 μL of 800 mM sodium acetate buffer (adjusted to pH 4.9 with acetate) and 10 μL of 400 U mL−1 glucoamylase (TOYOBO, Shiga, Japan), then incubated with 200 rpm agitation at 50 °C for 2 h. The enzyme was then denatured at 95 °C for 20 min and the mixture was centrifuged at 14,000xg for 5 min at 4 °C. The supernatant was applied to an HPLC system to determine the amount of glucose hydrolyzed from glycogen. The HPLC system was equipped with an Aminex HPX-87 h column and an RID-10A refractive index detector, as described above.
2.7. Enzyme assay
Glycogen phosphorylase (GP) activity was measured according to a previous method (Andersen and Westergaard, 2002) with minor modifications. Synechocystis cells were cultivated for 72 h under phototrophic conditions followed by 24 h dark anoxic cultivation. The culture (100 μL) was centrifuged at 6000xg for 10 min at 4 °C and the supernatant was discarded. The cells were washed with extraction buffer (18 mM KH2PO4, 27 mM Na2HPO4, 15 mM MgCl2, and 100 μM EDTA, pH 8.0) and suspended in 3 mL of extraction buffer, then disrupted by sonication. Unbroken cells and cell debris were removed by centrifugation (20,000xg for 20 min at 4 °C) and the supernatant was used as a protein extract for the enzyme assay. A 1.5 mL aliquot of reaction mixture contained 100 μL of the protein extract and 1.4 mL of a solution comprising 18 mM KH2PO4, 27 mM Na2HPO4, 15 mM MgCl2, 100 μM EDTA, 340 μM NADP+, 4 μM glucose 1, 6-bisphosphate, 6 units mL−1 glucose 6-phosphate dehydrogenase, 0.8 units mL−1 phosphoglucomutase and 2 mg mL−1 glycogen (from oyster, Nacalai Tesque, Inc., Kyoto, Japan). The same mixture but without the glycogen was used as a control. Enzyme activities were determined by measuring the production of NADPH in the reaction mixtures as a change in absorbance at 340 nm. PEP carboxylase activity was measured according to a previous method (Chen et al., 2002). Synechocystis cells were cultivated for 24 h under phototrophic conditions. A 1.5 mL aliquot of reaction mixture contained 50 μL of the protein extract and 1.45 mL of a solution comprising 50 mM Tris–HCl (pH 7.5), 15 mM MgSO4, 10 mM KHCO3, 100 μM NADH, 1 unit mL−1 malate dehydrogenase, and 5 mM PEP. The same mixture but without the PEP was used as a control. Enzyme activities were determined by measuring the oxidation of NADH in the reaction mixtures as a change in absorbance at 340 nm. The reactions were performed directly in cuvettes. Protein concentrations were determined with the BCA method as described previously (Hasunuma et al., 2013). The data are presented as averages of three independent assays.
2.8. 13C-labeling experiment
In-vivo 13C-labeling was performed to analyze the turnover of metabolites in cyanobacteria. Anaerobic fermentation was initiated by suspending cells in 50 mM Hepes-KOH (pH 7.8) containing 2 g L−1 [U-13C] glucose or 100 mM 13C-bicarbonate. After labeling for 0–6 h, the cells were collected by filtration and processed as described for metabolite analysis. Extracted metabolites were analyzed using CE-MS. Mass spectral peaks of biological origin were identified manually by searching for mass shifts between the 12C- and 13C-mass spectra. The 13C fraction of the metabolites was calculated as described previously (Hasunuma et al., 2010). The relative isotopomer abundance (mi) for each metabolite incorporating i13C atoms was calculated as follows:
where Mi represents the isotopomer abundance for each metabolite incorporating i13C atoms. The 13C fraction of metabolites possessing n carbon atoms is calculated as follows:
3. Results
3.1. Organic acid production during autofermentation by Synechocystis cells
Synechocystis sp. PCC6803 cells were cultivated phototrophically in modified BG11 medium containing 5 mM NaNO3 or NH4Cl as the sole nitrogen source for 3 days, then the cells were transferred to 50 mM Hepes-KOH buffer (pH 7.8) to initiate fermentation under anaerobic conditions. NaNO3 is the typical nitrogen source used in the cultivation of Synechocystis but a recent study (Osanai et al., 2015) used NH4Cl instead; therefore, the two nitrogen sources were compared in this study. During the phototrophic cultivation, no succinate was detected in the medium.
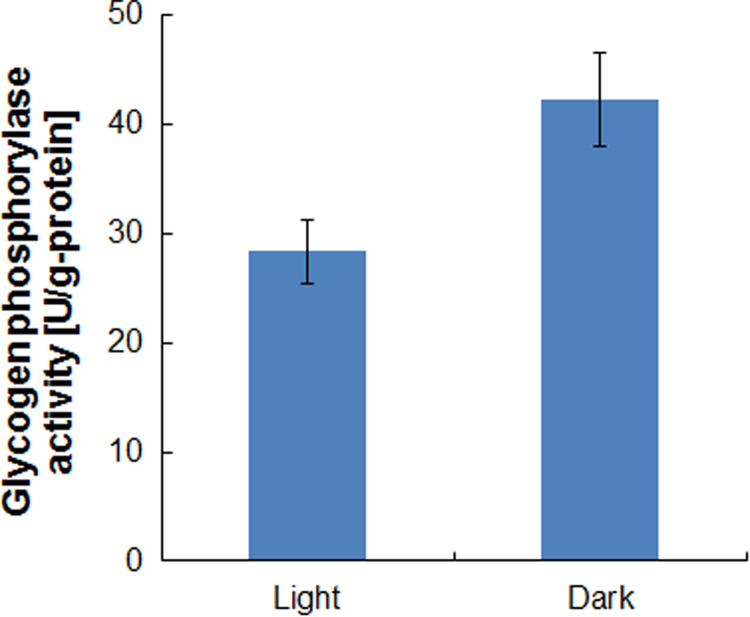
As shown in Fig. 1, under dark anoxic conditions, significant concentrations of the organic acids acetate, 2-ketoglutarate, lactate, malate and succinate were secreted into the fermentation medium, irrespective of the nitrogen source used. The concentration of acetate in the fermentation medium increased with time, to more than 400 mg L−1 after 96 h fermentation, whereas the concentration of lactate plateaued at app. 60 mg L−1 after 24 h. Succinate production was initiated after 6 h of fermentation and increased with time to 100 mg L−1 after 96 h when NH4Cl was used as the nitrogen source. After 72 h fermentation, fumarate concentration was 5 mg L−1 (data not shown). The concentrations of citrate, iso-citrate, fumarate and oxaloacetate in the fermentation medium were lower than 1 mg L−1 (data not shown). NH4Cl supported higher organic-acid production than did NaNO3. The intracellular glycogen content decreased from 0.44 g-glycogen g-DCW−1 when the cells were grown in medium containing NaNO3 to 0.37 g-glycogen g-DCW−1 when the cells were cultivated in the presence of NH4Cl. Since the DCW decreased from 5.2 g L−1 to 3.7 g L−1 during 96-h fermentation, the glycogen in the fermentation vessel decreased from 2.3 g L−1 to 1.3 g L−1 (Fig. 1). The activity of glycogen phosphorylase (GP), which is involved in glycogen hydrolysis by cyanobacteria, was increased 1.5-fold by changing from light to dark anoxic cultivation conditions (Fig. 2).
Fig. 1.
Time-course of the production of organic acids secreted into the medium and glycogen utilization during dark anoxic fermentation by Synechocystis 6803 phototrophically cultivated in the presence of 5 mM NaNO3 (open symbols) or 5 mM NH4Cl (closed symbols) as the sole nitrogen source. Each data point represents the average (±SD) of three independent experiments
Fig. 2.
Glycogen phosphorylase activity under light and dark anoxic conditions. The activity was measured after 72 h phototrophic cultivation and 24 h dark anoxic cultivation. Each data point represents the average (±SD) of three independent experiments.
3.2. Time-course transcriptome analysis during autofermentation
Time-course transcriptome analysis of Synechocystis cells grown in the presence of 5 mM NH4Cl under anaerobic conditions was conducted using DNA microarrays. Switching to dark anoxic conditions resulted in a more than 2-fold increase in the expression level of 10 genes (Table 1) and a more than 2-fold decrease in the expression level of 43 genes (Table 2). The examination of genes involved in glycolysis and the tricarboxylic acid (TCA) cycle showed that the expression of nifJ (sll0741), which encodes pyruvate:ferredoxin oxidoreductase (PFO), was 2.53-fold upregulated after 3 h of cultivation under dark anoxic conditions (Table 1), and genes involved in redox reactions and in hydrogen production, such as ndhD2 (slr1291) and isiB (sll0248), were also upregulated under these conditions. On the other hand, the following genes were all downregulated: cbbA (sll0018), which encodes fructose 1,6-bisphosphate (FBP) aldolase; gap1 (slr0884), which encodes glyceraldehyde 3-phosphate (GAP) dehydrogenase involved in glycolysis; rbcS (slr0012) and rbcX (slr0011), involved in the activity of ribulose 1,5-bisphosphate carboxylase/oxygenase (Rubisco); acnB (slr0665), involved in the TCA cycle; ppc (sll0920), involved in anaplerosis; and glnN (slr0288), glnA (slr1756), gad (sll1641), gltB (sll1502), argG (slr0585) and cysK (slr1842), involved in amino acid metabolism (Table 2). Genes involved in light harvesting and in the photosystem, such as psbC (sll0851), psbB (slr0906), psaJ (sml0008), psaF (sll0819), psbD2 (slr0927), psaE (ssr2831), psaA (slr1834) and apcA (slr2067), psaL (slr1655), ctaB (sll1899), psaB (slr1835), psaC (ssl0563), psbU (sll1194), apcC (ssr3383), petD (slr0343), showed decreased expression.
Table 1.
Genes up-regulated under dark anoxic conditionsa.
| ID | Gene | Fold changeb |
Description | ||
|---|---|---|---|---|---|
| 3 h | 6 h | 24 h | |||
| slr1291 | ndhD2 | 7.53 | 6.71 | 7.41 | NAD(P)H-quinone oxidoreductase subunit 4 |
| slr0518 | abfB | 5.67 | 5.24 | 6.10 | arabinofuranosidase |
| sll0741 | nifJ | 2.53 | 1.32 | 1.27 | pyruvate oxidoreductase |
| sll1894 | ribA | 2.39 | 1.72 | 1.34 | riboflavin biosynthesis protein RibA |
| sll0248 | isiB | 2.16 | 2.08 | 2.48 | flavodoxin FldA |
| sll0070 | purU | 1.67 | 1.73 | 2.53 | formyltetrahydrofolate deformylase |
| slr0905 | bchE | 1.21 | 1.13 | 2.20 | Mg-protoporphyrin IX monomethyl ester oxidative cyclase 66 kD subunit |
| sll1688 | thrC | 1.16 | 1.67 | 2.65 | threonine synthase |
| slr1165 | sat | 1.13 | 1.14 | 2.43 | sulfate adenylyltransferase |
| sll0938 | sll0938 | 1.01 | −1.06 | 2.38 | aspartate transaminase |
Genes annotated in cyanobase (http://genome.microbedb. jp/cyanobase/Synechocystis) were selected.
Fold change is the ratio of expression at 0 h to 3, 6 and 24 h. According to t-test, values were significantly different (P<0.05).
Table 2.
Genes down-regulated under dark anoxic conditionsa.
| ID | Gene | Fold changeb |
Description | ||
|---|---|---|---|---|---|
| 3 h | 6 h | 24 h | |||
| sll0851 | psbC | −8.66 | −11.93 | −13.62 | photosystem II CP43 protein |
| slr0906 | psbB | −6.06 | −6.88 | −12.70 | photosystem II CP47 protein |
| sml0008 | psaJ | −6.05 | −7.10 | −5.85 | photosystem I reaction center subunit IX |
| slr1643 | petH | −5.83 | −7.03 | −8.46 | ferredoxin-NADP oxidoreductase |
| slr0012 | rbcS | −5.71 | −7.59 | −6.85 | ribulose bisphosphate carboxylase small subunit |
| sll0819 | psaF | −5.62 | −6.72 | −4.78 | photosystem I subunit III |
| sll1566 | ggpS | −4.57 | −5.09 | −3.32 | glucosylglycerolphosphate synthase |
| sll0018 | cbbA | −4.47 | −4.43 | −6.46 | fructose 1,6-bisphosphate aldolase |
| sll1535 | rfbP | −4.25 | −3.62 | −2.93 | galactosyl-1-phosphate transferase |
| sll1577 | cpcB | −3.93 | −4.00 | −7.66 | phycocyanin β subunit |
| sll1085 | glpD | −3.84 | −4.03 | −3.18 | glycerol 3-phosphate dehydrogenase |
| slr0288 | glnN | −3.79 | −4.11 | −3.75 | glutamate-ammonia ligase |
| slr0927 | psbD2 | −3.58 | −4.24 | −5.83 | photosystem II D2 protein |
| ssr2831 | psaE | −3.51 | −3.60 | −4.51 | photosystem I reaction center subunit IV |
| slr0054 | dgkA | −3.30 | −4.01 | −5.11 | diacylglycerol kinase |
| slr1756 | glnA | −3.10 | −3.16 | −3.18 | glutamate-ammonia ligase |
| slr1834 | psaA | −3.04 | −5.36 | −9.34 | photosystem I P700 chlorophyll a apoprotein A1 |
| slr2067 | apcA | −2.99 | −3.07 | −5.93 | allophycocyanin α chain |
| slr1655 | psaL | −2.85 | −2.57 | −3.20 | photosystem I reaction center protein subunit XI |
| sll1899 | ctaB | −2.83 | −2.78 | −2.04 | protoheme IX farnesyltransferase |
| slr0884 | gap1 | −2.81 | −3.16 | −2.89 | glyceraldehyde 3-phosphate dehydrogenase |
| slr1835 | psaB | −2.73 | −4.92 | −3.25 | photosystem I P700 chlorophyll a apoprotein A2 |
| sll1641 | gad | −2.71 | −2.46 | −2.93 | glutamate decarboxylase |
| ssl0563 | psaC | −2.64 | −2.85 | −2.89 | photosystem I subunit VII |
| sll1194 | psbU | −2.61 | −2.78 | −3.88 | photosystem II complex extrinsic protein precursor U |
| sll1184 | ho1 | −2.52 | −2.28 | −2.18 | heme oxygenase |
| slr0261 | ndhH | −2.48 | −2.52 | −2.28 | NAD(P)H-quinone oxidoreductase subunit H |
| sll0080 | argC | −2.47 | −2.48 | −2.61 | N-acetyl-gamma-glutamyl-phosphate reductase |
| slr1022 | argD | −2.45 | −2.78 | −3.44 | acetylornithine aminotransferase |
| ssr3383 | apcC | −2.44 | −2.45 | −1.66 | phycobilisome LC linker polypeptide |
| sll0920 | ppc | −2.38 | −2.87 | −2.47 | phosphoenolpyruvate carboxylase |
| slr0665 | acnB | −2.33 | −2.77 | −2.16 | bifunctional aconitate hydratase 2/2-methylisocitrate dehydratase |
| slr0011 | rbcX | −2.28 | −2.62 | −3.95 | possible RubisCO chaperonin |
| sll1502 | gltB | −2.08 | −2.41 | −2.69 | ferredoxin-dependent glutamate synthase |
| slr0343 | petD | −1.85 | −1.83 | −2.77 | cytochrome B6-f complex subunit IV |
| slr0585 | argG | −1.78 | −1.93 | −2.64 | argininosuccinate synthase |
| sll0166 | hemD | −1.76 | −2.23 | −1.26 | uroporphyrin-III synthase |
| sll1893 | hisF | −1.74 | −2.15 | 1.30 | imidazole glycerol phosphate synthase subunit HisF |
| slr1830 | phbC | −1.49 | −1.75 | −1.45 | poly(3-hydroxyalkanoate) synthase |
| slr1842 | cysK | −1.35 | −1.12 | −1.44 | cysteine synthase |
| slr1239 | pntA | −1.29 | −1.81 | −4.34 | NAD(P) transhydrogenase subunit alpha |
| slr1072 | yefA | −1.19 | −1.08 | −2.43 | GDP-d-mannose dehydratase |
| sll0550 | flv3 | 1.04 | −1.08 | −2.05 | flavoprotein |
| slr1064 | rfbU | 1.16 | 1.14 | −2.86 | mannosyltransferase B |
Genes annotated in cyanobase (http://genome.microbedb. jp/cyanobase/Synechocystis) were selected.
Fold change is the ratio of expression at 0 h to 3, 6 and 24 h. According to t-test, values were significantly different (P<0.05).
3.3. Time-course metabolome analysis during anaerobic cultivation
The intracellular metabolome in Synechocystis was measured at 0, 3, 6, 24, 48, 72 and 96 h after the initiation of anaerobic cultivation (Fig. 3). Thirty four hydrophilic metabolites, including amino acids, cofactors, organic acids, sugar nucleotides and sugar phosphates, were analyzed by CE-MS to investigate the metabolic profile of the cells during anaerobic cultivation.
Fig. 3.
Time-course changes of intracellular metabolite pool sizes during dark anoxic fermentation by Synechocystis 6803 phototrophically cultivated in the presence of 5 mM NaNO3 (open symbols) or 5 mM NH4Cl (closed symbols) as the sole nitrogen source. Two-fold upregulated and downregulated genes are shown in red and blue characters, respectively. A reaction catalyzed by ACL is shown with dashed lines. Abbreviations: FBP, fructose 1,6-bisphosphate; F6P, fructose 6-phosphate; GABA, γ-aminobutyrate; GAP, glyceraldehyde 3-phosphate; G1P, glucose 1-phosphate; G6P, glucose 6-phosphate; PEP, phosphoenolpyruvate; 2PGA, 2-phosphoglycerate; 3PGA, 3-phosphoglycerate; R5P, ribose 5-phosphate; Ru5P, ribulose 5-phosphate; RuBP, ribulose 1,5-bisphosphate; S7P, sedoheptulose 7-phosphate; SBP, sedoheptulose 1,7-bisphosphate; DCW, dry cell weight. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
The concentrations of many metabolites, such as ADP-glucose, glucose 6-phosphate (G6P), ribulose 1,5-bisphosphate (RuBP), ribulose 5-phosphate (Ru5P), 3-phosphoglycerate (3PGA), 2-phosphoglycerate (2PGA), PEP, acetyl-CoA, citrate, cis-aconitate, iso-citrate and alanine, transiently increased until 6 h of anaerobic cultivation, and then decreased. In contrast, intracellular malate, fumarate and succinate increased until 24 h, then gradually decreased until 72 h and then stabilized.
Glucose 1-phosphate (G1P), sedoheptulose 7-phosphate (S7P) and aspartate increased in concentration with time. FBP was present at significantly higher concentration than its breakdown products, dihydroxyacetone phosphate (DHAP) and GAP. The concentration of ATP increased up to 24 h of anaerobic cultivation, then decreased with time. Cells grown in the presence of NH4Cl had a higher content of organic acids than cells grown in NaNO3, although the concentrations of glycolysis-related metabolites were the same.
3.4. Metabolite turnover analysis under anaerobic conditions
No carbon source was supplied to cells growing under dark anoxic conditions, so glycogen was apparently converted to organic acids via glycolysis. However, to date there has been no report of the direct observation of glycogen and glucose catabolism.
During metabolism under steady state conditions, metabolites are replaced with newly synthesized compounds at a constant rate and the total amount of each metabolite remains unchanged. Therefore, the determination of time-course changes in the metabolome is insufficient to account for intracellular carbon flow. However, since the Synechocystis GT strain used in this study can utilize glucose as a carbon source, 13C-glucose was added to the medium as a carbon source for the anaerobic cultivation of Synechocystis, and carbon flow during dark anaerobic cultivation was observed by tracer experiments.
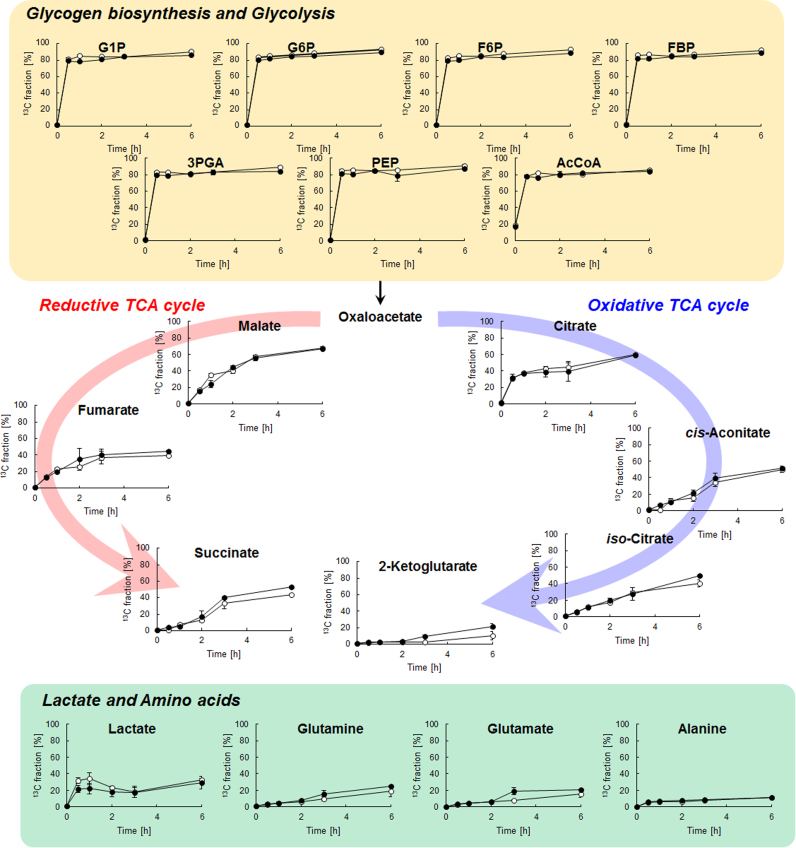
13C was incorporated into metabolites in Synechocystis cells following initiation of dark anoxic cultivation (Fig. 4). The 13C fraction, defined as the ratio of 13C to the total carbon in each metabolite, was calculated from the mass isotopomer distributions. Glucose is phosphorylated to G6P by hexokinase, then metabolized through glycolysis and the pentose phosphate pathway. The 13C fraction of glycolysis metabolites (G6P, F6P, FBP, 3PGA and PEP) and acetyl-CoA reached a maximum of more than 80% after 30 min of labeling, whereas organic acids showed slower 13C incorporation. In particular, 2-ketoglutarate showed the lowest 13C fraction among the metabolites involved in the cyanobacterial TCA cycle, suggesting that succinate is mainly formed from fumarate via the reductive TCA cycle. Lactate and amino acids such as alanine, glutamate and glutamine were labeled by 13C, although their 13C fractions were lower than that of their common precursor, PEP. These data clearly indicated that glucose is catabolized to organic acids and amino acids under anaerobic conditions. Note that the 13C fraction of pyruvate could not be evaluated because of its low abundance in cells.
Fig. 4.
Time-course changes in the metabolite 13C fraction following the addition of 13C-glucose to cultures of Synechocystis sp. PCC6803 phototrophically cultivated in the presence of 5 mM NaNO3 (open symbols) or 5 mM NH4Cl (closed symbols) as the sole nitrogen source. Each data point represents the average (±SD) of three independent experiments.
As shown in Fig. S1, glucose added to the medium was consumed after 24 h and glycogen consumption was repressed during glucose consumption. The amount of succinate produced during the first 48 h was increased from 57 mg L−1 to 74 mg L−1 by the addition of 2 g L−1 glucose.
3.5. Improvement of succinate production based on multiple-omic analyses
The results of dynamic metabolic profiling, shown in Fig. 4, revealed that the 13C fraction of citrate, malate, fumarate and succinate increased more slowly than that of PEP and acetyl-CoA, suggesting that the generation of organic acids involved in the TCA cycle is a rate-limiting step for succinate production. As previously described (Hasunuma et al., 2010), the initial slope of the 13C-fraction versus time curve indicates the turnover rate of the metabolite. Intracellular oxaloacetate could not be detected because of its low abundance, but malate showed a lower turnover rate than citrate (Fig. 4), suggesting that the anaplerotic pathway from PEP to oxaloacetate limits succinate production.
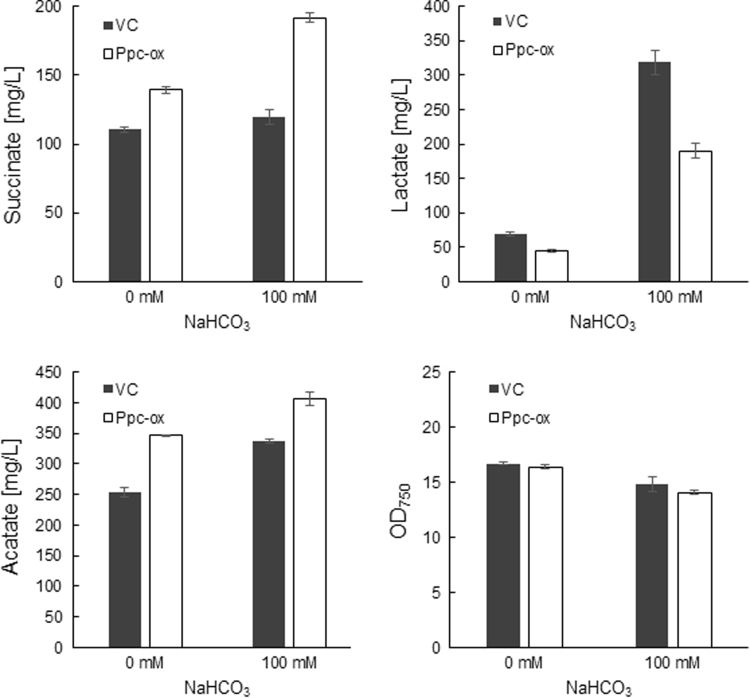
PEP carboxylase converts PEP to oxaloacetate and is encoded by ppc (sll0920). The expression of ppc in Synechocystis 6803 was enhanced by linking ppc to the trc promoter, then integrating into the slr0168 loci through homologous recombination. The chromosomal integration of ppc was confirmed by genomic PCR (Fig. S2). Ppc-ox demonstrated 3-fold higher PEP carboxylase activity than its vector control strain (VC) (Fig. S2). After 72 h fermentation, the recombinant Synechocystis strain, Ppc-ox, exhibited significantly higher production of succinate (140 mg L−1) than VC (111 mg L−1) (Fig. 5). Acetate production was also enhanced by overexpression of sll0920, whereas Ppc-ox showed lower lactate production than VC. Biomass monitored via OD750 was similar between Ppc-ox and VC.
Fig. 5.
Organic acid production in Ppc-ox and its vector control strain in the absence or presence of 100 mM NaHCO3. Organic acids and OD750 were measured after 72 h fermentation. Values represent the average (±SD) of three independent experiments.
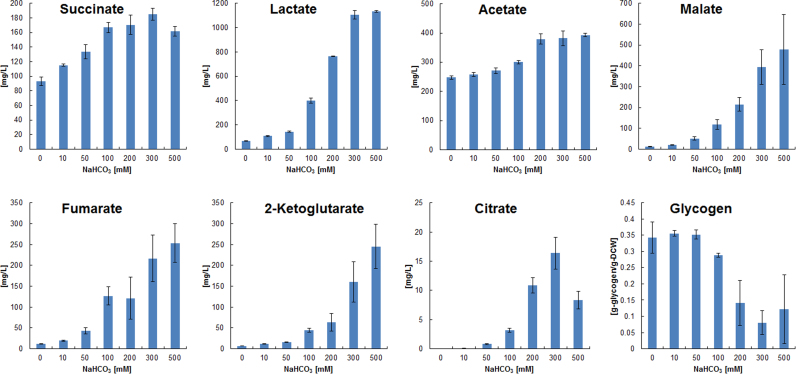
NaHCO3 was added to the medium at the start of anaerobic cultivation of GT strain in order to further improve succinate production. As shown in Fig. 6, succinate production increased as the concentration of NaHCO3 increased and reached 185 mg L−1 after 72 h fermentation in the presence of 300 mM NaHCO3. Lactate and acetate were as high as 1132 mg L−1 and 393 mg L−1, respectively, in the presence of 500 mM NaHCO3, and NaHCO3 addition significantly increased the production of other organic acids such as citrate, fumarate, 2-ketoglutarate and malate that were below 6 mg L−1 in the absence of NaHCO3. Furthermore, in the presence of 100 mM NaHCO3, succinate production was improved from 120 mg L−1 to 192 mg L−1 by ppc overexpression (Fig. 5). The addition of 13C-bicarbonate (NaH13CO3) to the medium resulted in the incorporation of 13C in organic acids such as malate, fumarate and succinate (Fig. 7), demonstrating that bicarbonate is a carbon source for succinate production. Citrate and iso-citrate were labeled with 13C, and the 13C fraction of 2-ketoglutarate was 9.9% after 24 h. Alanine and lactate, which can be produced from pyruvate, were slightly labeled with 13C. In contrast, PEP, acetyl-CoA, and metabolites involved in glycolysis such as F6P and 3PGA were not enriched in 13C.
Fig. 6.
Effect of NaHCO3 addition on organic acid production by Synechocystis sp. PCC6803 GT strain and its effect on glycogen utilization. Organic acids and glycogen were measured after 72 h fermentation. Values represent the average (± SD) of three independent experiments.
Fig. 7.
Time-course changes in the metabolite 13C fraction following the addition of 13C-bicarbonate. Each data point represents the average (±SD) of three independent experiments.
4. Discussion
Time-course analyses of the metabolome, transcriptome and metabolic turnover revealed dynamic changes in the metabolism of the cyanobacterium Synechocystis sp. PCC6803 cultivated under dark anoxic conditions, allowing identification of the rate-limiting steps in sugar catabolism. In addition, rational metabolic engineering based on these metabolic analyses leads to improved succinate production.
ATP is generated for cell maintenance during auto-fermentative metabolism of glycogen (Kumaraswamy et al., 2013, McNeely et al., 2010), but catabolism has never been observed. In the present study, dynamic metabolic profiling clearly indicated that succinate is biosynthesized by sugar catabolism via glycolysis, the pentose phosphate pathway, the anapletotic pathway, and the reductive TCA cycle, since the 12C atoms in sugar (hexose, pentose and triose) phosphates, organic acids and amino acids were replaced by 13C derived from 13C-glucose during anoxic cultivation (Fig. 4). Succinic acid can be produced via three routes in prokaryotic cells: the reductive and oxidative pathways of the TCA cycle, and the glyoxylate shunt (Choi et al., 2015). However, the cyanobacterial TCA cycle is branched into oxidative and reductive routes (Knoop et al., 2013). The genes involved in the glyoxylate shunt of Synechocystis 6803 have not been isolated (Knoop et al., 2013). Recent studies found that the cyanobacterial TCA cycle can be completed via the γ-aminobutyric acid (GABA) shunt constituting a pathway from 2-ketoglutarate, via glutamate, GABA and succinate semialdehyde, to succinate (Xiong et al., 2014) or via 2-ketoglutarate decarboxylase and succinic semialdehyde dehydrogenase (Zhang et al., 2011). However, as shown in Fig. 4, 53% of the 12C was replaced with 13C in succinic acid but only 22% was replaced in 2-ketoglutarate, indicating that succinic acid was produced via the reductive TCA cycle under dark anoxic conditions.
Synechocystis 6803 biosynthesizes glycogen from CO2 assimilated during periods of light exposure (Smith, 1983) and can be hydrolyzed to glucose 1-phosphate by GP encoded by glgP with support from isoamylase encoded by glgX, (Ball and Morell, 2003, Fu and Xu, 2006). In the present study, GP activity was higher under dark anoxic conditions than under light conditions (Fig. 2). Our metabolic analyses elucidated the biological route for succinic acid production from glycogen under dark anoxic conditions, as illustrated in Fig. 3.
Carbon distribution and redox balance during autofermentation have been estimated (McNeely et al., 2010, McNeely et al., 2014) but the in vivo kinetics of these processes have not previously been investigated. Direct measurement of metabolic turnover using the described in vivo 13C-labeling assay demonstrated that there is a rate-limiting step between PEP and oxaloacetate under dark anoxic conditions and that overexpression of PEP carboxylase and the addition of bicarbonate improved succinate production (Fig. 5). In vivo labeling with 13C-bicarbonate clearly showed carbon incorporation into succinate via the anaplerotic pathway and the reductive TCA cycle (Fig. 7). However, the initial slope of succinate was lower than that of fumarate, indicating that the conversion of fumarate to succinate is also a rate-limiting step in succinate biosynthesis. 13C fraction of succinate was lower than 20% after 24 h fermentation, which should be due to glycogen catabolism.
Time-course metabolome analysis indicated other potential rate-limiting steps in succinic acid production. As shown in Fig. 3, G1P increased with time in cells grown with NH4Cl and reached 11.9 μmol g-DCW−1 after 96 h of fermentation, whereas the level of G6P formed from G1P by phosphoglucomutase remained essentially constant (0.3–0.6 μmol g-DCW−1). This raises the possibility that phosphoglucomutase is a rate-limiting step in glycogen catabolism. The FBP level increased from 0.05 to 6.18 μmol g-DCW−1 after 3 h of fermentation and then remained constant, while there were only trace levels of DHAP and GAP, suggesting that FBP aldolase is a rate-limiting step in sugar catabolism. Glycogen catabolism might be improved by increasing the concentrations of phosphoglucomutase and FBP aldolase. Interestingly, ATP levels increased from 0 h to 24 h, then decreased. The catabolism of glycogen can generate ATP during autofermentation. ATP might be used by ATP-citrate lyase (ACL), which converts citrate to acetyl-CoA and oxaloacetate in the reductive TCA cycle (Fig. 3), although genes encoding ALC have not been isolated from Synechocystis 6803.
As shown in Fig. 1, no secretion of succinate was detected from 0 h to 6 h, possibly due to slow 13C incorporation into intracellular succinate between 0 and 2 h (Fig. 4). Intracellular pool size analysis (Fig. 3) showed that maximum levels of acetyl-CoA, citrate, cis-aconitate and iso-citrate were obtained after 6 h, while malate, fumarate and succinate increased until 24 h. The expression of acnB was downregulated after 3 h of fermentation (Table 2). These data suggest that the major carbon flow in the TCA cycle changes from the oxidative to the reductive route during fermentation.
Global gene expression analysis showed that genes involved in light harvesting, photosystems, photosynthetic electron transport and Rubisco were downregulated under dark anoxic conditions (Table 2), likely due to the switching of cyanobacterial metabolism from photoautotrophism to carbohydrate catabolism. The expression of nifJ encoding PFO (Schmitz et al., 2001) increased under anoxic conditions. PFO catalyzes the oxidative cleavage of pyruvate and coenzyme A to acetyl-CoA and CO2, with the simultaneous reduction of ferredoxin or flavodoxin. Under dark anoxic conditions, the expression of nifJ might be enhanced to supply reductants such as reduced ferredoxin and flavodoxin, and NADH for organic acid production. It was previously shown that overexpression of NAD+-dependent GAP dehydrogenase in Synechococcus sp. PCC7002 increased glycogen catabolic capacity, with a concomitant increase in terminal fermentation products, likely due to an increased NADH/NAD+ ratio (Kumaraswamy et al., 2013). The removal of nitrate from the culture medium enhances auto-fermentative hydrogen production in Synechococcus sp. PCC7002 (McNeely et al., 2014). Since nitrate is reduced to NH4+ by nitrate reductase, reductants required for nitrate assimilation are consumed following nitrate removal. In the present study, Synechocystis 6803 cultivated in the presence of NH4Cl showed higher organic acid production than when cultivated in the presence of NaNO3 (Fig. 1), perhaps due to the high accumulation of reductants in cells phototrophically cultivated in the presence of NH4Cl.
Carbon capture and utilization has recently attracted increasing attention as a route to developing an environmentally benign society (von der Assen et al., 2014). The amount of succinate produced by cyanobacteria is lower than that of several strains of rumen bacteria, fungi and metabolically-engineered microorganisms, but they require no carbon source other than CO2. Increased levels of organic acids were observed following the addition of HCO3-, which is in equilibrium with CO2 (pKa=6.35) (Fig. 5), suggesting that the addition of CO2 to the culture can improve succinic acid production via the anaplerotic pathway. Phototrophic cyanobacteria therefore hold great promise for the effective utilization of CO2.
Acknowledgements
The authors thank Ms. Chikako Aoki for technical assistance. This work was supported by ALCA from the Japan Science and Technology Agency, the Ministry of Education, Culture, Sports, Science, and Technology (MEXT), Japan.
Footnotes
Supplementary data associated with this article can be found in the online version at http://dx.doi.org/10.1016/j.meteno.2016.04.003.
Appendix A. Supplementary material
Supplementary material
.
References
- Aikawa S., Joseph A., Yamada R., Izumi Y., Yamagishi T., Matsuda F., Kawai H., Chang J.S., Hasunuma T., Kondo A. Direct conversion of Spirulina to ethanol without pretreatment or enzymatic hydrolysis process. Energy Environ. Sci. 2013;6:1844–1849. [Google Scholar]
- Anfelt J., Kaczmarzyk D., Shabestary K., Renberg B., Rockberg J., Nielsen J., Uhlén M., Hudson E.P. Genetic and nutrient modulation of acetyl-CoA levels in Synechocystis for n-butanol production. Microb. Cell Fact. 2015;14:167. doi: 10.1186/s12934-015-0355-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aikawa S., Nishida A., Ho S.-H., Chang J.S., Hasunuma T., Kondo A. Glycogen production for biofuels by the euryhaline cyanobacteria Synechococcus sp strain PCC 7002 from an oceanic environment. Biotechnol. Biofuels. 2014;7:88. doi: 10.1186/1754-6834-7-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akhtar J., Idris A., Abd Aziz R. Recent advances in production of succinic acid from lignocellulosic biomass. Appl. Microbiol. Biotechnol. 2014;98:987–1000. doi: 10.1007/s00253-013-5319-6. [DOI] [PubMed] [Google Scholar]
- Andersen B., Westergaard N. The effect of glucose on the potency of two distinct glycogen phosphorylase inhibitors. Biochem. J. 2002;367:443–450. doi: 10.1042/BJ20020153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ball S.G., Morell M.K. From bacterial glycogen to starch: understanding the biogenesis of the plant starch granule. Annu. Rev. Plant Biol. 2003;54:207–233. doi: 10.1146/annurev.arplant.54.031902.134927. [DOI] [PubMed] [Google Scholar]
- Beauprez J.J., De Mey M., Soetaert W.K. Microbial succinic acid production: natural versus metabolic engineered producers. Process Biochem. 2010;45:1103–1114. [Google Scholar]
- Bozell J.J., Peterson G.R. Technology development for the production of biobased products from biorefinery carbohydrates – the US Department of Energy's “Top 10” revisited. Green Chem. 2010;12:539–554. [Google Scholar]
- Camañes G., Scalschi L., Vicedo B., González-Bosch C., García-Agustín P. An untargeted global metabolomic analysis reveals the biochemical changes underlying basal resistance and priming in Solanum lycopersicum and identifies 1-methyltryptophan as a metabolite involved in plant responses to Botrytis cinerea and Pseudomonas syringae. Plant J. 2015;84:125–139. doi: 10.1111/tpj.12964. [DOI] [PubMed] [Google Scholar]
- Chen L.M., Omiya T., Hata S., Izui K. Molecular characterization of a phosphoenolpyruvate carboxylase from a thermophilic cyanobacterium, Synechococcus vulcanus with unusual allosteric properties. Plant Cell Physiol. 2002;43:159–169. doi: 10.1093/pcp/pcf019. [DOI] [PubMed] [Google Scholar]
- Chen Y., Nielsen J. Advances in metabolic pathway and strain engineering paving the way for sustainable production of chemical building blocks. Curr. Opin. Biotechnol. 2013;24:965–972. doi: 10.1016/j.copbio.2013.03.008. [DOI] [PubMed] [Google Scholar]
- Choi S., Song C.W., Shin J.H., Lee S.Y. Biorefineries for the production of top building block chemicals and their derivatives. Metab. Eng. 2015;28:223–239. doi: 10.1016/j.ymben.2014.12.007. [DOI] [PubMed] [Google Scholar]
- Cukalovic A., Stevens C.V. Feasibility of production methods for succinic acid derivatives: a marriage of renewable resources and chemical technology. Biofuels Bioprod. Bioref. 2008;2:505–529. [Google Scholar]
- Delhomme C., Weuster-Botz D., Kühn F.E. Succinic acid from renewable resources as a C4 building-block chemical – a review of the catalytic possibilities in aqueous media. Green. Chem. 2009;11:13–26. [Google Scholar]
- Diamond S., Jun D., Rubin B.E., Golden S.S. The circadian oscillator in Synechococcus elongatus controls metabolite partitioning during diurnal growth. Proc. Natl. Acad. Sci. USA. 2015;112:E1916–E1925. doi: 10.1073/pnas.1504576112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dismukes G.C., Carrieri D., Bennette N., Ananyev G.M., Posewitz M.C. Aquatic phototrophs: efficient alternatives to land-based crops for biofuels. Curr. Opin. Biotechnol. 2008;19:235–240. doi: 10.1016/j.copbio.2008.05.007. [DOI] [PubMed] [Google Scholar]
- Fu J., Xu X. The functional divergence of two glgP homologues in Synechocystis sp. PCC6803. FEMS Microbiol. Lett. 2006;260:201–209. doi: 10.1111/j.1574-6968.2006.00312.x. [DOI] [PubMed] [Google Scholar]
- Hasunuma T., Harada K., Miyazawa S., Kondo A., Fukusaki E., Miyake C. Metabolic turnover analysis by a combination of in vivo13C-Labeling from 13CO2 and metabolic profiling with CE-MS/MS reveals rate-limiting steps of the C3 photosynthetic pathway in Nicotiana tabacum leaves. J. Exp. Bot. 2010;61:1041–1051. doi: 10.1093/jxb/erp374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasunuma T., Kikuyama F., Matsuda M., Aikawa S., Izumi Y., Kondo A. Dynamic metabolic profiling of cyanobacterial glycogen biosynthesis under conditions of nitrate depletion. J. Exp. Bot. 2013;64:2943–2954. doi: 10.1093/jxb/ert134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasunuma T., Matsuda M., Senga Y., Aikawa S., Toyoshima M., Shimakawa G., Miyake C., Kondo A. Overexpression of flv3 improves photosynthesis in the cyanobacterium Synechocystis sp. PCC6803 by enhancement of alternative electron flow. Biotechnol. Biofuels. 2014;7:493. doi: 10.1186/s13068-014-0183-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumi Y., Aikawa S., Matsuda F., Hasunuma T., Kondo A. Aqueous sizeexclusion chromatographic method for the quantification of cyanobacterial native glycogen. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2013;930:90–97. doi: 10.1016/j.jchromb.2013.04.037. [DOI] [PubMed] [Google Scholar]
- Kaneko T., Sato S., Kotani H., Tanaka A., Asamizu E., Nakamura Y., Miyajima N., Hirosawa M., Sugiura M., Sasamoto S., Kimura T., Hosouchi T., Matsuno A., Muraki A., Nakazaki N., Naruo K., Okumura S., Shimpo S., Takeuchi C., Wada T., Watanabe A., Yamada M., Yasuda M., Tabata S. Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC6803. II. Sequence determination of the entire genome and assignment of potential protein-coding region. DNA Res. 1996;3:109–136. doi: 10.1093/dnares/3.3.109. [DOI] [PubMed] [Google Scholar]
- Knoop H., Gründel M., Zilliges Y., Lehmann R., Hoffmann S., Lockau W., Steuer R. Flux balance analysis of cyanobacterial metabolism: the metabolic network of Synechocystis sp. PCC 6893. PLoS Comput. Biol. 2013;9:e1003081. doi: 10.1371/journal.pcbi.1003081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumaraswamy G.K., Guerra T., Qian X., Zhang S., Bryant D.A., Dismukes G.C. Reprogramming the glycolytic pathway for increased hydrogen production in cyanobacteria: metabolic engineering of NAD+ -dependent GAPDH. Energy Environ. Sci. 2013;6:3722–3731. [Google Scholar]
- Lee J., Sim S.J., Bott M., Um Y., Oh M.K., Woo H.M. Succinate production from CO2-grown microalgal biomass as carbon source using engineered Corynebacterium glutamicum through consolidated bioprocessing. Sci. Rep. 2014;4:5819. doi: 10.1038/srep05819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Link H., Kochanowski K., Sauer U. Systematic identification of allosteric protein-metabolite interactions that control enzyme activity in vivo. Nat. Biotechnol. 2013;31:357–361. doi: 10.1038/nbt.2489. [DOI] [PubMed] [Google Scholar]
- McNeely K., Xu Y., Bennette N., Bryant D.A., Dismukes G.C. Redirecting reductant flux into hydrogen production via metabolic engineering of fermentative carbon metabolism in a cyanobacterium. Appl. Environ. Microbiol. 2010;76:5032–5038. doi: 10.1128/AEM.00862-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNeely K., Kumaraswamy G.K., Guerra T., Bennette N., Ananyev G., Dismukes G.C. Metabolic switching of central carbon metabolism in response to nitrate: application to autofermentative hydrogen production in cyanobacteria. J. Biotechnol. 2014;182–183:83–91. doi: 10.1016/j.jbiotec.2014.04.004. [DOI] [PubMed] [Google Scholar]
- Osanai T., Oikawa A., Azuma M., Tanaka K., Saito K., Hirai M.Y., Ikeuchi M. Genetic engineering of group 2 σ factor SigE widely activates expression of sugar catabolic genes in Synechocystis species PCC 6803. J. Biol. Chem. 2011;286:30962–30971. doi: 10.1074/jbc.M111.231183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osanai T., Shirai T., Iijima H., Nakaya Y., Okamoto M., Kondo A., Hirai M.Y. Genetic manipulation of a metabolic enzyme and a transcriptional regulator increasing succinate excretion from unicellular cyanobacterium. Front. Microbiol. 2015;6:1064. doi: 10.3389/fmicb.2015.01064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partensky F., Hess W.R., Vaulot D. Prochlorococcus, a marine photosynthetic prokaryote of global significance. Microbiol. Mol. Biol. Rev. 1999;63:106–127. doi: 10.1128/mmbr.63.1.106-127.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rippka R., Deruelles J., Waterbury J.B., Herdman M., Stanier R.Y. Generic assignments, strains histories and properties of pure culture of cyanobacteria. J. Gen. Microbiol. 1979;111:1–61. [Google Scholar]
- Savakis P.E., Angermayr S.A., Hellingwerf K.J. Synthesis of 2,3-butanediol by Synechocystis sp. PCC6803 via heterologous expression of a catabolic pathway from lactic acid- and enterobacteria. Metab. Eng. 2013;20:121–130. doi: 10.1016/j.ymben.2013.09.008. [DOI] [PubMed] [Google Scholar]
- Schmitz O., Gurke J., Bothe H. Molecular evidence for the aerobic expression of nifJ, encoding pyruvate:ferredoxin oxidoreductase, in cyanobacteria. FEMS Microbiol. Lett. 2001;195:97–102. doi: 10.1111/j.1574-6968.2001.tb10504.x. [DOI] [PubMed] [Google Scholar]
- Smith A.J. Modes of cyanobacterial carbon metabolism. Ann. Microbiol. 1983;134B:93–113. doi: 10.1016/s0769-2609(83)80099-4. [DOI] [PubMed] [Google Scholar]
- Tsuge Y., Tateno T., Sasaki K., Hasunuma T., Tanaka T., Kondo A. Direct production of organic acids from starch by cell surface-engineered Corynebacterium glutamicum in anaerobic conditions. AMB Express. 2013;3:72. doi: 10.1186/2191-0855-3-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varman A.M., Xiao Y., Pakrasi H.B., Tang Y.J. Metabolic engineering of Synechocystis sp. strain PCC 6803 for isobutanol production. Appl. Environ. Microbiol. 2013;79:908–914. doi: 10.1128/AEM.02827-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von der Assen N., Voll P., Peters M., Bardow A. Life cycle assessment of CO2 capture and utilization: a tutorial review. Chem. Soc. Rev. 2014;43:7982–7994. doi: 10.1039/c3cs60373c. [DOI] [PubMed] [Google Scholar]
- Wang Y., Sun T., Gao X., Shi M., Wu L., Chen L., Zhang W. Biosynthesis of platform chemical 3-hydroxypropionic acid (3-HP) directly from CO2 in cyanobacterium Synechocystis sp. PCC 6803. Metab. Eng. 2016;34:60–70. doi: 10.1016/j.ymben.2015.10.008. [DOI] [PubMed] [Google Scholar]
- Wijffels R.H., Kruse O., Hellingwerf K.J. Potential of industrial biotechnology with cyanobacteria and eukaryotic microalgae. Curr. Opin. Biotechnol. 2013;24:405–413. doi: 10.1016/j.copbio.2013.04.004. [DOI] [PubMed] [Google Scholar]
- Williams J.G.K. Construction of specific mutations in photosystem II photosynthetic reaction center by genetic engineering methods in Synechocystis 6803. Methods Enzymol. 1988;167:766–778. [Google Scholar]
- Xiong W., Brune D., Vermaas W.F.J. The γ-aminobutyric acid shunt contributes to closing the tricarboxylic acid cycle in Synechocystis sp. PCC 6803. Mol. Microbiol. 2014;93:786–796. doi: 10.1111/mmi.12699. [DOI] [PubMed] [Google Scholar]
- Zhang S., Bryant D.A. The tricarboxylic acid cycle in cyanobacteria. Science. 2011;334:1551–1553. doi: 10.1126/science.1210858. [DOI] [PubMed] [Google Scholar]
- Zhou J., Zhang H., Zhang Y., Li Y., Ma Y. Designing and creating a modularized synthetic pathway in cyanobacterium Synechocystis enables production of acetone from carbon dioxide. Metab. Eng. 2012;14:394–400. doi: 10.1016/j.ymben.2012.03.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material