Abstract
2-Pyrrolidone is a valuable bulk chemical with myriad applications as a solvent, polymer precursor and active pharmaceutical intermediate. A novel 2-pyrrolidone synthase, ORF27, from Streptomyces aizunensis was identified to catalyze the ring closing dehydration of γ-aminobutyrate. ORF27's tendency to aggregate was resolved by expression at low temperature and fusion to the maltose binding protein (MBP). Recombinant Escherichia coli was metabolically engineered for the production of 2-pyrrolidone from glutamate by expressing both the genes encoding GadB, a glutamate decarboxylase, and ORF27. Incorporation of a GadB mutant lacking H465 and T466, GadB_ΔHT, improved the efficiency of one-pot 2-pyrrolidone biosynthesis in vivo. When the recombinant E. coli strain expressing the E. coli GadB_ΔHT mutant and the ORF27-MBP fusion was cultured in ZYM-5052 medium containing 9 g/L of l-glutamate, 7.7 g/L of l-glutamate was converted to 1.1 g/L of 2-pyrrolidone within 31 h, achieving 25% molar yield from the consumed substrate.
Keywords: 2-Pyrrolidone, E. coli, Glutamate, Metabolic engineering, Biosynthesis
Graphical abstract
Highlights
-
•
ORF27 from Streptomyces aizunensis catalyzes formation of 2-pyrrolidone from γ-aminobutyrate.
-
•
Recombinant Escherichia coli with GadB and ORF27 produces 2-pyrrolidone from glutamate.
-
•
Engineered strain capable of producing 1.1 g/L of 2-pyrrolidone from 9 g/L of glutamate within 31 h.
1. Background
2-Pyrrolidone was identified by the US Department of Energy as an important C4 “Top Value-Added Chemical from Biomass” that can potentially be derived from glutamate (Werpy, 2004). 2-Pyrrolidone is currently used as precursor for the production of N-vinylpyrrolidone, a solvent for animal injection, a building block for active pharmaceutical ingredients, optical co-solvent for water-based ink formulation, process solvent for membrane filters and a copolymer for floor polish (BASF, 2015). 2-Pyrrolidone can also be used in ring-opening polymerization to produce nylon-4, a fiber material with better thermal stability and the highest hydrophilicity in the nylon family of materials (Park, 2013). With a variety of applications, 2-pyrrolidone continues to be a product of huge commercial interest.
Current industrial production of 2-pyrrolidone involves the dehydrogenation of 1,4-butanediol to form γ–butyrolactone, followed by reacting aqueous γ-butyrolactone with ammonia (Fig. 1A) (Harreus et al., 2011). By using low cost glutamate as starting material, as well as avoiding harsh reaction conditions, biological production of 2-pyrrolidone offers the potential for a cheaper and more environmentally friendly synthesis route. Therefore, we propose a two-step enzymatic process for 2-pyrrolidone biosynthesis from glutamate: (1) decarboxylation of glutamate to form γ-aminobutyrate (GABA), and (2) enzymatic ring closing of GABA into 2-pyrrolidone (Fig. 1B).
Fig. 1.
Routes for production of 2-pyrrolidone. (A) BASF petrochemical route for 2-pyrrolidone production. (B) Microbial 2-pyrrolidone biosynthetic route.
While the first enzymatic step is known (Ma, 2012, Park, 2013, Shi, 2013, Takahashi, 2012, Vo, 2012), the second step has not been demonstrated under any fermentation conditions (Stavila, 2013). We employed a targeted strategy to identify appropriate enzyme candidates for the GABA activation step by conducting retro-biosynthetic analysis of polyketides. Here we report the discovery in Streptomyces aizunensis of ORF27, an auxiliary enzyme in the linearmycin A biosynthetic cluster that performs the GABA activation step to form 2-pyrrolidone under mild fermentation conditions. Coupling this 2-pyrrolidone synthase with glutamate decarboxylase, which forms GABA from glutamate, we achieved the first demonstration of the full 2-pyrrolidone biosynthetic pathway in Escherichia coli.
2. Materials and methods
2.1. Enzyme scouting by retro-biosynthetic analysis of PKS natural products
It was hypothesized that an enzyme employing a GABA activation mechanism on GABA's acid group followed by spontaneous irreversible intramolecular cyclization could be utilized as a 2-pyrrolidone synthase. However, there has been no documentation of AMP-activating enzymes using GABA as a substrate. Type I polyketide synthases (PKSs) are mega-synthases whose enzymatic domain organization predictably correlate with their natural product's chemical structure – usually referred to as the colinearity rule (Du, 2001, Du, 2010, Dutta, 2014, Khosla, 2009, Li, 2009, Menzella, 2005, Tran, 2010, Weissman, 2008, Wong, 2010, Yadav, 2009). PKSs are responsible for biosynthesis of the characteristic polyketide aglycone backbone, and auxiliary enzymes are involved in starter unit activation and/or post aglycone modification (Kalaitzis, 2009, Li, 2005, Llewellyn, 2007, Moorea, 2002, Ogasawara, 2004, Shinohara, 2011, Simunovic, 2006). The close positioning of PKS genes with auxiliary enzymes in the same gene cluster makes it relatively easy to identify the substrates upon which these auxiliary enzymes act. PKS databases were explored to identify polyketides with positively charged, amine-containing starter units and auxiliary enzymes clustered near the PKS (see Table S1 for lists of PKS databases). Desertomycin A/B, Eco-0501, linearmycin A were selected for further retrobiosynthetic analysis (Fig. S1A). From the arrangement of the polyketide synthase genes, 4-aminobutyrate-like starter unit was predicted to be loaded onto the ACP0 domain (Fig. S1B). Since polyketide starter unit loading requires the substrate to be activated by an ATP dependent mechanism, the linearmycin A biosynthetic gene cluster was further analyzed for auxiliary enzymes with this function. Two ORFs in the gene cluster, ORF27 and ORF36, were predicted to be AMP-dependent synthetases (Fig. S1B). The hypothesized candidates were reverse transcribed according to the optimized E. coli codon usage using DNA 2.0's algorithm and synthesized.
2.2. Bacterial strains and chemicals
E. coli strain DH10B [F− mcrA Δ(mrr-hsdRMS-mcrBC) φ80dlacZΔM15 ΔlacX74 recA1 endA1 araD139 Δ(ara, leu)7697 galU galK λ− rpsL (Strr) nupG] was used for all molecular biology manipulations. DH10B or BL21 Star (DE3) [F− ompT hsdSB (rB− mB−) gal dcm rne131 (DE3)] were used as hosts for production of 2-pyrrolidone. All the strains and plasmids utilized in this study are listed in Table 1. For high-density shake flask cultures, Studier's autoinduction ZYM-5052 medium was prepared according to the published protocol (Studier, 2005). Chloramphenicol (25 μg/ml), kanamycin (20 μg/ml) and ampicillin (100 μg/ml) were added where desired to provide selective pressure for plasmid maintenance. During 2-pyrrolidone production, the following antibiotic concentration was used: chloramphenicol (6.25 μg/ml), kanamycin (5 μg/ml) and ampicillin (25 μg/ml). 2-Pyrrolidone, glutamic acid, GABA, and ATP were purchased from Sigma-Aldrich (St. Louis, Missouri).
Table 1.
E. coli strains, plasmids and oligonucleotides used.
| Name | Relevant genotype | References |
|---|---|---|
| Strains | ||
| DH10B | F− mcrA (crmrr-hsdRMS-mcrBC) (r-hsdRMS-mcrBC) and oligonucleotide139 Δ(ara, leu)7697 galU galK alrpsL nupG | Life technologies (Carlsbad, CA) |
| BL21 Star (DE3) | F− ompT hsdSB (rB− mB−) gal dcm rne131 (DE3) | Life technologies (Carlsbad, CA) |
| JW2637-4 | F–, Δ(araD-araB)567, ΔlacZ4787(:rrnB-3), λ–, ΔgabT743:kan, rph-1, Δ(rhaD-rhaB)568, hsdR514 | CGSC #11775 |
| JW0036-1 | F–, Δ(araD-araB)567, ΔlacZ4787(:rrnB-3), λ–, ΔCaiC750:kan, rph-1, Δ(rhaD-rhaB)568, hsdR514 | CGSC #8352 |
| Plasmids | ||
| pDNA2.0-ORF27 | pUC ori, KanR, ORF27 with Biobrick overhang | DNA 2.0 (Menlo Park, CA) |
| pBbE2C- RFP | ColE ori, CmR, tetR, RFP | (2011) |
| pBbE2C-ORF27 | ColE ori, CmR, tetR, ORF27 | This study |
| pBbE2C-CaiC | ColE ori, CmR, tetR, CaiC | This study |
| pBbS2C-ORF27 | SC101 ori, CmR, tetR, ORF27 | This study |
| pET28b | pBR322 ori, KanR, lacI, MCS after PT7 | EMD Millipore (Billerica, MA) |
| pET28a-MBP | pBR322 ori, KanR, lacI, MCS after MBP fusion driven by PT7 | EMD Millipore (Billerica, MA) |
| pET28a-MBP-ORF27 | pET28a-MBP with ORF27 inserted into NdeI and XhoI | This study |
| pBbE2C-MBP-ORF27 | ColE ori, CmR, tetR, MBP-ORF27 | This study |
| pBbA5a-GadB | p15A ori, AmpR,Placuv5, GadB | This study |
| pBbA5a-GadB_ΔHT | p15A ori, AmpR, Placuv5, GadB_ΔHT | This study |
| pBbA5a-GadB_Δ1-14 | p15A ori, AmpR, Placuv5, GadB_ Δ1-14 | This study |
| pBbA5a-GadB_Δ1-14, ΔHT | p15A ori, AmpR, Placuv5, GadB_ Δ1-14, ΔHT | This study |
| pBbA7a-RFP | p15A ori, AmpR,PT7, RFP | T.S. Lee (2011) |
| pBbA7a-GadB | p15A ori, AmpR,PT7, GadB | This study |
| pBbA7a-GadB_ΔHT | p15A ori, AmpR,PT7, GadB_ΔHT | This study |
| pBbA7a-GadB_Δ1-14 | p15A ori, AmpR,PT7, GadB_ Δ1-14 | This study |
| pBbA7a-GadB_Δ1-14_ΔHT | p15A ori, AmpR,PT7, GadB_ Δ1-14_ΔHT | This study |
| Oligonucleotides | 5′→ 3′ Sequence, restriction site underlied, synthesized by Integrated DNA technologies, Inc (Coralville, IA) | Target gene |
| JZ_MBP-ORF27_f | GCGCGgaattcaaaagatcttttaagaaggagatatacatatgggcagcagccatcatca | MBP-ORF27 |
| JZ_MBP-ORF27_r | GCGCGctcgagtttggatccTCATTCTGCCGCCATACGGG | MBP-ORF27 |
| JZ_GadB_f | gcgcgCATATGgataagaagcaagtaacg | GadB, GadB_ΔHT, |
| JZ_GadB_r | gcgcgGGATCCTTAtcaggtatgtttaaagctgtt | GadB, GadB_Δ1-14 |
| JZ_GadBΔ1-14_f | gcgcgCATATGGGTTCACGTTTTGGTGCGA | GadB_Δ1-14, GadB_Δ1-14_ΔHT |
| JZ_GadBΔHT_r | gcgcgGGATCCTTAtcatttaaagctgttctgttggg | GadB_Δ1-14_ΔHT GadB_ΔHT |
| JZ_CaiC_f | GCGCgaattcaaaagatcttttaagaaggagatatacatATGGATAGAGGTGCAATGGAT | CaiC |
| JZ_CaiC_r | GCGCG ctcgagtttggatccTTATTTCAGATTCTTTCTAATTATTTTCCCC | CaiC |
| Strains | Plasmids | Host |
| JZ-171 | pBbE2C-ORF27 | JW2637-4 |
| JZ-172 | pBbE2C-RFP | JW2637-4 |
| JZ-298 | pBbS2C-ORF27 | JW2637-4 |
| JZ-299 | pBbE2C-MBP-ORF27 | JW2637-4 |
| JZ-336 | pBbE2C-MBP-ORF27+pBbA5a-GadB WT | BL21 Star (DE3) |
| JZ-338 | pBbE2C-MBP-ORF27+pBbA5a-GadB_ΔHT | BL21 Star (DE3) |
| JZ-339 | pBbE2C-MBP-ORF27+pBbA5a-GadB_Δ1-14 | BL21 Star (DE3) |
| JZ-340 | pBbE2C-MBP-ORF27+pBbA5a-GadB_Δ1-14, ΔHT | BL21 Star (DE3) |
| JZ-342 | pET28a-MBP-ORF27+pBbA7a-GadB WT | BL21 Star (DE3) |
| JZ-344 | pET28a-MBP-ORF27+pBbA7a-GadB_ΔHT | BL21 Star (DE3) |
| JZ-345 | pET28a-MBP-ORF27+pBbA7a-GadB_Δ1-14 | BL21 Star (DE3) |
| JZ-346 | pET28a-MBP-ORF27+pBbA7a-GadB_Δ1-14, ΔHT | BL21 Star (DE3) |
| JZ-348 | pBbE2C-MBP-ORF27+pBbA7a-GadB WT | BL21 Star (DE3) |
| JZ-350 | pBbE2C-MBP-ORF27+pBbA7a-GadB_ΔHT | BL21 Star (DE3) |
| JZ-351 | pBbE2C-MBP-ORF27+pBbA7a-GadB_Δ1-14 | BL21 Star (DE3) |
| JZ-352 | pBbE2C-MBP-ORF27+pBbA7a-GadB_Δ1-14, ΔHT | BL21 Star (DE3) |
| JZ-370 | pBbE2C-CaiC | JW2637-4 |
| JZ-386 | pET28b-N-CaiC+pBbA7a-GadB_ΔHT | BL21 Star (DE3) |
| JZ-393 | pBbE2C-RFP | JW0036-1 |
2.3. Plasmid construction
Genes encoding ORF27 (GenBank: AAX98201.1) and ORF36 (GenBank: AAX98210.1) were recoded using E. coli codon usage with biobrick overhangs 5′-gaattcaaaAGATCTAGGAGGCAT-3′ on the 5′ end and 5′-TAAGGATCCAAACTCGAG-3′ on the 3′ end. DNA 2.0 (Menlo Park, CA) cloned the genes into plasmid vectors creating pDNA2.0-ORF27 and pDNA2.0-ORF36, respectively. The genes encoding wild-type GadB and the variant GadB_ΔHT, were amplified from E. coli MG1655 genomic DNA using the primers specified. GadB_ΔHT lacks histidine 465 and threonine 466 of E. coli GadB. The construction of each plasmid is described in Table 2 and Table S2.
Table 2.
Plasmid construction.
| Constructed plasmid | Backbone source (restriction site) | Gene source (direct digestion or PCR) | PCR primers |
|---|---|---|---|
| pBbE2C-ORF27 | pBbE2C-RFP (BglII, XhoI) | pDNA2.0-ORF27 (BglII, XhoI) | N/A |
| pBbS2C-ORF27 | pBbS2C-RFP (BglII, XhoI) | pDNA2.0-ORF27 (BglII, XhoI) | N/A |
| pET28b-N-CaiC | pET28b (NdeI, XhoI) | pBbE2C-CaiC (NdeI, XhoI) | N/A |
| pBbE2C-ORF27 | pBbE2C-RFP (BglII, XhoI) | pDNA2.0-ORF27 (BglII, XhoI) | N/A |
| pBbE2C-CaiC | pBbE2C-RFP (EcoRI, XhoI) | E. coli MG1655 gDNA PCR | JZ_CaiC_f, JZ_CaiC_r |
| pET28a-MBP-ORF27 | pET28a-MBP (NdeI, XhoI) | pDNA2.0-ORF27 (NdeI, XhoI) | N/A |
| pBbE2C-MBP-ORF27 | pBbE2C-RFP (BglII, XhoI) | pET28a-MBP-ORF27 PCR | JZ_MBP-ORF27_f, JZ_MBP-ORF27_r |
| pBbA5a-GadB | pBbA5a-RFP (NdeI, BamHI) | E. coli MG1655 gDNA PCR | JZ_GadB_f, JZ_GadB_r |
| pBbA5a-GadB_ΔHT | pBbA5a-RFP (NdeI, BamHI) | E. coli MG1655 gDNA PCR | JZ_GadB_f, JZ_GadB_ΔHT _r |
| pBbA5a-GadB_Δ1-14 | pBbA5a-RFP (NdeI, BamHI) | E. coli MG1655 gDNA PCR | JZ_GadBΔ1-14_f, JZ_GadB_r |
| pBbA5a-GadB_Δ1-14, ΔHT | pBbA5a-RFP (NdeI, BamHI) | E. coli MG1655 gDNA PCR | JZ_GadBΔ1-14_f, JZ_GadB_ΔHT _r |
| pBbA7a-GadB | pBbA7a-RFP (NdeI, BamHI) | pBbA5a-GadB (NdeI, BamHI) | N/A |
| pBbA7a-GadB_ΔHT | pBbA7a-RFP (NdeI, BamHI) | pBbA5a-GadB_ΔHT(NdeI, BamHI) | N/A |
| pBbA7a-GadB_Δ1-14 | pBbA7a-RFP (NdeI, BamHI) | pBbA5a-GadB_Δ1-14 (NdeI, BamHI) | N/A |
| pBbA7a-GadB_Δ1-14_ΔHT | pBbA7a-RFP (NdeI, BamHI) | pBbA5a-GadB_Δ1-14, ΔHT(NdeI, BamHI) | N/A |
2.4. 2-Pyrrolidone, GABA and glutamate product assays
2.4.1. Liquid chromatography method for 2-pyrrolidone, GABA and glutamate separation
Liquid chromatography (LC) separation of 2-pyrrolidone was conducted at 55 °C with an Inertsil ODS-3 reverse-phase C18 column (250 mm length, 2.1 mm internal diameter, 3 μM particle size; GL Sciences) using a 1100 series high-performance LC system (Agilent Technologies). The mobile phase was composed of 0.1% formic acid in H2O (solvent A) and 0.1% formic acid in MeOH (solvent B). Butyrolactam was separated with the following gradient: 40–60% B for 4.5 min, 60–100% B for 0.5 min, 100–40% B for 0.5 min, held at 10% B for 8.5 min. A flow rate of 0.18 mL/min was used throughout.
2.4.2. Mass spectrometry analysis of 2-pyrrolidone, GABA and glutamate
The LC system was coupled to an Agilent Technologies LC-MSD SL electrospray ionization mass spectrometer (ESI MS). Nitrogen gas was used as both the nebulizing and drying gas to facilitate the production of gas-phase ions. The drying and nebulizing gases were set to 10 L/min and 20 psig, respectively, and a drying gas temperature of 300 °C was used throughout. ESI was conducted in the positive-ion mode with a capillary voltage of 4 kV. Mass measurements were carried out in the selected ion monitoring (SIM) mode (2-pyrrolidone, m/z 86; GABA, m/z 104; glutamate, m/z 148) for the detection of [M+H]+ ions. Data acquisition and processing were performed using ChemStation (Agilent technologies).
2.5. 2-Pyrrolidone production titer determination
600 μL of culture was cooled on ice and centrifuged at 18,000g for 5 min at 4 °C. 250 μL of the supernatant was mixed with 250 μL methanol to a final concentration of 50% (v/v), and the mixed solution filtered through 10 K Amicon Ultra-0.5 mL Centrifugal Filters (Millipore) by centrifuging at 20,000×g for 15 min. The filtered solution was diluted into the respective linear range of detection for 2-pyrrolidone and analyzed by LC–MS in the SIM mode described above.
2.6. Culture conditions
2.6.1. Inducible 2-pyrrolidone production from GABA
E. coli strains (JZ-298, JZ-171, JZ-299, JZ-370, and JZ-393) harboring plasmids containing genes encoding the proposed GABA activating enzyme (ORF27, MBP-ORF27 or CaiC) were inoculated into 25 mL LB medium with chloramphenicol (25 μg/ml) and grown at 37 °C. E. coli expressing RFP was utilized as a negative control (JZ-172). When the OD600 reached around 0.5, the culture was cooled to various temperatures (18–37 °C). 50 ng/mL of anhydrotetracycline (aTc) was added for protein production and GABA was supplied to a final concentration of 0–10 mM. The 2-pyrrolidone titer was analyzed 24 h after induction.
2.6.2. Inducible 2-pyrrolidone production from glutamate
E. coli strains (JZ-336, JZ-338–JZ-340, JZ-342, JZ-344–JZ-346, JZ-348, JZ-350–JZ-352, and JZ-386) harboring plasmids containing genes encoding both glutamate decarboxylase and a GABA-activating enzyme were inoculated into 25 mL LB medium containing various concentrations of glutamate (0–9 g/L) with appropriate antibiotics and grown at 37 °C. When the OD600 reached around 0.6, the culture was cooled to 25 °C. IPTG and aTet were added to a final concentration of 500 μM and 50 ng/mL, respectively to induce protein expression. The pH was titrated by adding 0.6 N HCl solution, and the culture was placed in a 25 °C incubator. 2-Pyrrolidone titer was analyzed at 24 h after induction.
2.6.3. Autoinducible 2-pyrrolidone production from glutamate
E. coli strains (JZ-344 and JZ-386) harboring plasmids containing genes encoding both glutamate decarboxylase and a GABA-activating enzyme were inoculated into 10 mL of LB or LB plus 5 g/L glutamate overnight. On day 2, the overnight culture was inoculated 1:100 (v/v) into 25 mL Studier's autoinduction ZYM-5052 medium with various concentrations of glutamate (0–9 g/L) and appropriate antibiotics (Studier, 2005). The culture was incubated at 37 °C. When the OD600 reached around 0.6, the culture was cooled to 25 °C. The pH was titrated to 5.25 by adding 0.6 N HCl solution. The culture was then placed at 25 °C, and 2-pyrrolidone titer was analyzed at 24 h later.
3. Results
3.1. ORF27 and E. coli's native CaiC catalyze 2-pyrrolidone formation in vivo
E. coli JW2637-4 from the KEIO knockout library contains a knockout of gabT, which encodes a GABA transaminase involved in GABA catabolism (Tomoya Baba et al., 2006). This host was initially used to confirm production of 2-pyrrolidone in vivo. ORF36 overexpression did not result in 2-pyrrolidone production (data not shown). E. coli JZ-171 (ORF27) and JZ-172 (RFP negative control) were grown in LB medium containing 0, 1, and 10 mM GABA. At 1 mM GABA, 2-pyrrolidone was observed only in the presence of ORF27 (Fig. 2A). Surprisingly, at 10 mM GABA, slight 2-pyrrolidone production was observed in the RFP control strain, indicating that nonspecific activation of GABA by a native E. coli enzyme also contributes to 2-pyrrolidone formation. To determine which native E. coli enzymes were responsible for the activity, genes encoding various AMP activating enzymes, such as acs, prpE and caiC, were cloned and overexpressed. Overexpression of one of these enzymes, CaiC (strain JZ-370), a previously described betaine-CoA ligase (AMP activating enzyme family), led to 2-pyrrolidone formation (Bernal, 2008). E. coli JW0036-1, the KEIO collection caiC deletion mutant, was transformed with pBbE2C-RFP, creating strain E. coli JZ-393, which overexpresses RFP; this strain was no longer able to produce 2-pyrrolidone when fed 10 mM GABA (Fig. 2B).
Fig. 2.
2-Pyrrolidone production in vivo. (A) At 1 mM GABA, 2-pyrrolidone was observed in the strain where the ORF27 gene (JZ-171) is expressed. RFP expression strain (JZ-172) serves as negative control. (B) At 10 mM GABA concentration, 2-pyrrolidone production is still observed in JZ-172 due to background CaiC expression, when caiC was knocked out (JZ-393), 2-pyrrolidone is no long produced.
3.2. Increased ORF27 functional expression improves GABA to 2-pyrrolidone conversion in vivo
ORF27 had the tendency to aggregate and become insoluble one day after induction depending on the temperature (Fig. S2). By balancing reaction rate and enzyme functional expression, the optimal temperature was determined to be 25 °C for ORF27 to maximize 2-pyrrolidone titer from GABA. An MBP-ORF27 fusion increased solubility of ORF27 and resulted in a 1.5× to 2× improvement in overall titer. ORF27 was expressed from a low copy number plasmid harboring the SC101 origin of replication (pBbS2C-ORF27). However, use of this plasmid decreased the titer, presumably due to lower expression of the enzyme (Fig. S3). Chaperone coexpression was tested as another strategy to improve solubility (related strains listed in Table S2). This strategy, however, also resulted in decreased titer (Fig. S4).
3.3. Inducible 2-pyrrolidone production from glutamate
3.3.1. pH profile experiment
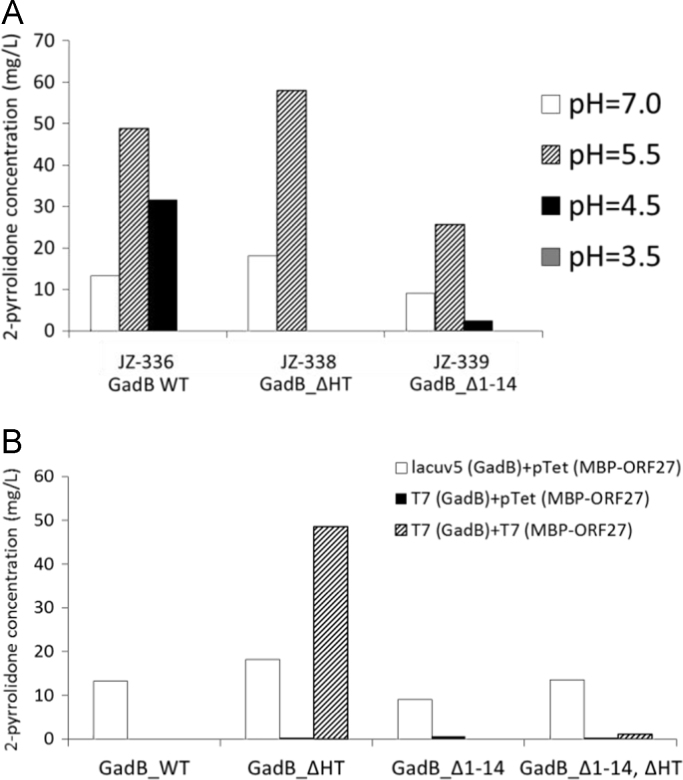
A two-step 2-pyrrolidone biosynthetic route from glutamate consists of the E. coli glutamate decarboxylase GadB and MBP-ORF27 as the 2-pyrrolidone synthase. E. coli's GadB has a pH optimum of 3.8 and its catalytic activity significantly decreases when the pH rises above 5.0 (Pennacchietti, 2009). In GadB, H465 plays an important role in blocking the enzyme active site at pHs near neutral by forming aldamine between its distal nitrogen on the imidazole ring with GadB's PLP-L276 schiff base; GadB mutants, such as H465A or GadB_ΔHT (lacking H465 and T466), are able to maintain relatively high activity at more alkaline pH (Pennacchietti, 2009). As shown in the extracellular pH profile for 2-pyrrolidone biosynthesis strains carrying either wild type or one of two GadB mutants (JZ-336, JZ-338, and JZ-339), pH ~5.5 resulted in the highest 2-pyrrolidone titer for both GadB wild type and variants (Fig. 3A). Sensitivity analysis showed that strains carrying GadB ΔHT robustly gave optimum yield within the pH range of 5.0 – 5.5, and declined when the exogenous pH drifted above 6.0 (Fig. S5).
Fig. 3.
(A) GadB mutants and the effect of extracellular pH on 2-pyrrolidone titer when feeding 10 mM glutamic acid. (B) The effect of promoter strength on 2-pyrrolidone titer when feeding 10 mM glutamic acid at an extracellular pH of 7.0.
3.3.2. Promoter engineering
To increase the flux through the pathway, various inducible promoters (lacUV5, Tet, T7) were tested (JZ-336, JZ-338–JZ-340, JZ-342, JZ-344–JZ-346, JZ-348, and JZ-350–JZ-352). Production of 2-pyrrolidone was highest when strong promoters were used, so the T7 promoter was chosen to drive expression of GadB ΔHT and MBP-ORF27 for optimal 2-pyrrolidone production (Fig. 3B).
3.4. Autoinducible 2-pyrrolidone production from glutamate
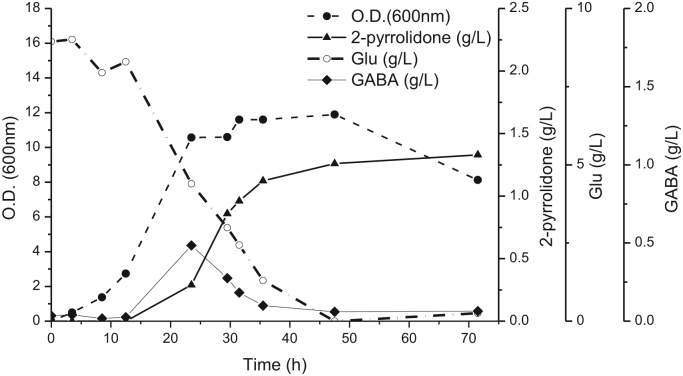
Studier's ZYM-5052 is a high density culture medium that enables autoinduction of protein expression in E. coli (Studier, 2005). Strain JZ-344, grown overnight in LB medium, was tested through 2-stage autoinducible 2-pyrrolidone production from glutamate: stage I, cell growth to OD600~0.6 at 37 °C; stage II, pH titration to 5.25 and incubation at 25 °C for 2-pyrrolidone production. Production in medium with 9 g/L and 0 g/L of glutamate showed that over 98% of 2-pyrrolidone was converted from glutamate supplied in the medium (Fig. S6). Overall, 1.1 g/L of 2-pyrrolidone was produced by converting 7.7 g/L of l-glutamate within 31 h, achieving 25% molar yield from consumed substrate (Fig. 4). The formation of GABA from glutamate likely occurred during 12–40 h, since during this time, the pH increased from 5.36 to 8.07 (Fig. S7). There was an approximate 12-hour time delay between extracellular GABA accumulation and glutamate consumption, presumably due to delayed autoinduction of enzyme expression. GABA accumulated around 12–30 h, yet was consumed quickly. This could result from transcriptional activation of GABA catabolic pathway in E. coli. GABA was rapidly transformed into 2-pyrrolidone when MBP-ORF27 was solubly expressed in day 1. 2-Pyrrolidone formation slowed down, presumably due to inactivation at approximately 48 h (Fig. 4). The OD600 of the culture decreased from 12 to 8, presumably due to either cell clumping or lysis.
Fig. 4.
E. coli strain #344 Production 2-pyrrolidone from glutamic acid (closed triangle) and OD600 of E. coli growth (closed circle). Glutamic acid feed consumption (open circle), GABA intermediate accumulation (closed diamond).
3.5. E. coli CaiC
E. coli CaiC was also tested (Bernal, 2008). Although CaiC has better thermostability and outperformed ORF27 in 2-pyrrolidone biosynthesis during GABA feeding, expression of E. coli CaiC under glutamate feeding conditions resulted in only 18.7% of the 2-pyrrolidone titer of that achieved when engineered ORF27 fused with MBP was used (data not shown).
4. Discussion
Through years of engineering, the petrochemical industry created over 4000 bulk chemicals (from ICIS). Currently, around 200 products are made via microbial fermentation (excluding food and beverages), including amino acids, bioactive compounds, etc. With advances in synthetic biology, it is now possible to engineer microbes to produce chemicals traditionally made via petrochemical processes at lower cost.
An important step towards building molecules of increasing complexity is to discover enzymes for substrate activation, such as C–C bond and C–N bond formation (Dougherty, 2009). Here we successfully demonstrated an efficient methodology to scout for enzymes from the secondary metabolite linearmycin A biosynthetic gene cluster that activate γ-aminobutyrate. The huge diversity of secondary metabolites and the functional groups embedded in these molecules made them a rich source of gene candidates (Medema, 2011, Yadav, 2009). Because polyketide biosynthesis follows the co-linearity rule and is highly predictable in terms of its biosynthetic pathway, polyketide biosynthetic clusters are convenient sources of substrate activating enzymes.
Although the dehydration of γ-aminobutyrate to form 2-pyrrolidone is thermodynamically favorable, due to its high activation barrier, a significant temperature (>200 °C) is required for the reaction to proceed even in the presence of Al2O3 catalyst. This reaction requires several days to complete, and the harsh conditions also lead to off pathway reactions, producing oligomers or cyclic GABA dimer or trimer (Stavila, 2013). The discovery of S. aizunensis ORF27 enables the first demonstration of 2-pyrrolidone production from γ-aminobutyrate at mild fermentation conditions.
S. aizunensis ORF27 was prone to aggregation when heterologously overexpressed in E. coli. Utilizing the MBP fusion alleviated ORF27 aggregation and maintained enzymatic activity for prolonged 2-pyrrolidone production, thus leading to an increased final titer. However ORF27 still suffers from a tendency to aggregate. For prolonged 2-pyrrolidone biosynthesis to be sustained during a production process, it would be desirable to evolve ORF27 to be more soluble and stable. In addition, use of a glutamate or GABA overproduction host, such as Corynebacterium glutamicum, would be desirable for production of 2-pyrrolidone from glucose or another carbon source (Choi, 2015, Lothar, 2005).
ORF27 and CaiC exhibited different comparative 2-pyrrolidone formation behavior. During 10 mM GABA feeding, CaiC had better performance, while during direct production from glutamate, ORF27 performed better. ORF27 might have a low Km for GABA, therefore high intracellular GABA production during glutamate feeding experiment result in much higher 2-pyrrolidone formation for ORF27, although more enzymology work needs to be conducted.
5. Conclusion
In this study, we utilized retro-biosynthetic analysis of polyketide natural products as a targeted method to prospect for novel γ-aminobutyrate activating enzymes for performing unprecedented reactions such as GABA conversion to 2-pyrrolidone. E. coli's native CaiC, a betaine-CoA ligase, was also discovered to be able to catalyze 2-pyrrolidone formation. Protein modification, such as MBP fusion, increased the activity of expressed ORF27. Metabolic engineering and process optimization collectively improved 2-pyrrolidone titer from glutamate. 1.1 g/L of 2-pyrrolidone was produced from 7.7 g/L of glutamate, representing a 25% yield from consumed substrate.
Acknowledgments
This work was funded by the Joint BioEnergy Institute (http://www.jbei.org/), which is supported by the US Department of Energy, Office of Science, Office of Biological and Environmental Research, through Contract DE-AC02-05CH11231 between Lawrence Berkeley National Laboratory and the US Department of Energy, and by the Synthetic Biology Engineering Research Center (SynBERC) through National Science Foundation Grant NSF EEC 0540879.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.meteno.2015.11.001.
Appendix A. Supplementary material
Supplementary material
References
- BASF, 2015. 2-Pyrrolidone, vol. 2015
- Bernal V. Role of betaine:CoA ligase (CaiC) in the activation of betaines and the transfer of coenzyme A in Escherichia coli. J. Appl. Microbiol. 2008;105:42–50. doi: 10.1111/j.1365-2672.2008.03740.x. [DOI] [PubMed] [Google Scholar]
- Choi J.W. Enhanced production of gamma-aminobutyrate (GABA) in recombinant Corynebacterium glutamicum by expressing glutamate decarboxylase active in expanded pH range. Microb. Cell. Factories. 2015;14:21. doi: 10.1186/s12934-015-0205-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dougherty M.J. Directed evolution: new parts and optimized function. Curr. Opin. Biotechnol. 2009;20:486–491. doi: 10.1016/j.copbio.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du L. Hybrid peptide-polyketide natural products: biosynthesis and prospects toward engineering novel molecules. Metab. Eng. 2001;3:78–95. doi: 10.1006/mben.2000.0171. [DOI] [PubMed] [Google Scholar]
- Du L. PKS and NRPS release mechanisms. Nat. Prod. Rep. 2010;27:255–278. doi: 10.1039/b912037h. [DOI] [PubMed] [Google Scholar]
- Dutta S. Structure of a modular polyketide synthase. Nature. 2014;510:512–517. doi: 10.1038/nature13423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harreus R.B., Eichler J.-O., Feuerhake R., Jakel C., Mahn U., Pinkos R., Vogelsang R. Wiley-VCH Verlag GmbH & Co. KGaA; Weinheim: 2011. 2-Pyrrolidone. Ullmann's Encyclopedia of Industrial Chemistry. [Google Scholar]
- Kalaitzis J.A. In vitro biosynthesis of unnatural enterocin and wailupemycin polyketides. J. Nat. Prod. 2009;72:469–472. doi: 10.1021/np800598t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khosla C. Revisiting the modularity of modular polyketide synthases. Curr. Opin. Chem. Biol. 2009;13:135–143. doi: 10.1016/j.cbpa.2008.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, T. S., Krupa, R. A., Zhang, F., Hajimorad, M., Holtz, W. J., Prasad, N., Lee, S. K., Keasling, J. D., 2011. BglBrick vectors and datasheets: A synthetic biology platform for gene expression. Journal of biological engineering. 5, 12. [DOI] [PMC free article] [PubMed]
- Li Y. Biosynthesis of the unique amino acid side chain of butirosin: possible protective-group chemistry in an acyl carrier protein-mediated pathway. Chem. Biol. 2005;12:665–675. doi: 10.1016/j.chembiol.2005.04.010. [DOI] [PubMed] [Google Scholar]
- Li C. Analysis of the indanomycin biosynthetic gene cluster from Streptomyces antibioticus NRRL 8167. Chembiochem Eur. J. Chem. Biol. 2009;10:1064–1072. doi: 10.1002/cbic.200800822. [DOI] [PubMed] [Google Scholar]
- Llewellyn N.M. Biosynthesis of butirosin: transfer and deprotection of the unique amino acid side chain. Chem. Biol. 2007;14:379–386. doi: 10.1016/j.chembiol.2007.02.005. [DOI] [PubMed] [Google Scholar]
- Lothar E. CPC Press, Taylor & Francis Group; Boca Raton, London, New York: 2005. Handbook of Corynebacterium glutamicum. [Google Scholar]
- Ma D. Structure and mechanism of a glutamate-GABA antiporter. Nature. 2012;483:632–636. doi: 10.1038/nature10917. [DOI] [PubMed] [Google Scholar]
- Medema M.H. Synthetic biology in Streptomyces bacteria. Methods Enzymol. 2011;497:485–502. doi: 10.1016/B978-0-12-385075-1.00021-4. [DOI] [PubMed] [Google Scholar]
- Menzella H.G. Combinatorial polyketide biosynthesis by de novo design and rearrangement of modular polyketide synthase genes. Nat. Biotechnol. 2005;23:1171–1176. doi: 10.1038/nbt1128. [DOI] [PubMed] [Google Scholar]
- Moorea B.S. Biosynthesis and attachment of novel bacterial polyketide synthase starter units. Nat. Prod. REp. 2002;19:70–99. doi: 10.1039/b003939j. [DOI] [PubMed] [Google Scholar]
- Ogasawara Y. Cloning, sequencing, and functional analysis of the biosynthetic gene cluster of macrolactam antibiotic vicenistatin in Streptomyces halstedii. Chem. Biol. 2004;11:79–86. doi: 10.1016/j.chembiol.2003.12.010. [DOI] [PubMed] [Google Scholar]
- Park S.J. Synthesis of nylon 4 from gamma-aminobutyrate (GABA) produced by recombinant Escherichia coli. Bioprocess Biosyst. Eng. 2013;36:885–892. doi: 10.1007/s00449-012-0821-2. [DOI] [PubMed] [Google Scholar]
- Pennacchietti E. Mutation of His465 alters the pH-dependent spectroscopic properties of Escherichia coli glutamate decarboxylase and broadens the range of its activity toward more alkaline pH. J. Biol. Chem. 2009;284:31587–31596. doi: 10.1074/jbc.M109.049577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi F. Enhancement of gamma-aminobutyric acid production in recombinant Corynebacterium glutamicum by co-expressing two glutamate decarboxylase genes from Lactobacillus brevis. J. Ind. Microbiol. Biotechnol. 2013;40:1285–1296. doi: 10.1007/s10295-013-1316-0. [DOI] [PubMed] [Google Scholar]
- Shinohara Y. A natural protecting group strategy to carry an amino acid starter unit in the biosynthesis of macrolactam polyketide antibiotics. J. Am. Chem. Soc. 2011;133:18134–18137. doi: 10.1021/ja208927r. [DOI] [PubMed] [Google Scholar]
- Simunovic V. Myxovirescin A biosynthesis is directed by hybrid polyketide synthases/nonribosomal peptide synthetase, 3-hydroxy-3-methylglutaryl-CoA synthases, and trans-acting acyltransferases. Chembiochem Eur. J. Chem. Biol. 2006;7:1206–1220. doi: 10.1002/cbic.200600075. [DOI] [PubMed] [Google Scholar]
- Stavila E. Synthesis of lactams using enzyme-catalyzed aminolysis. Tetrahedron Lett. 2013;54:370–372. [Google Scholar]
- Studier W. Protein production by auto-induction in high-density shaking cultures. Protein Expr. Purif. 2005;41:207–234. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- Takahashi C. Robust production of gamma-amino butyric acid using recombinant Corynebacterium glutamicum expressing glutamate decarboxylase from Escherichia coli. Enzym. Microb. Technol. 2012;51:171–176. doi: 10.1016/j.enzmictec.2012.05.010. [DOI] [PubMed] [Google Scholar]
- Tomoya Baba T.A., Hasegawa Miki, Takai Yuki, Okumura Yoshiko, Baba Miki, Datsenko Kirill A., Tomita Masaru, Wanner Barry L., Mori Hirotada. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2006;2 doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran L. Insights into protein–protein and enzyme–substrate interactions in modular polyketide synthases. Chem. Biol. 2010;17:705–716. doi: 10.1016/j.chembiol.2010.05.017. [DOI] [PubMed] [Google Scholar]
- Vo L. Effects of glutamate decarboxylase and gamma-aminobutyric acid (GABA) transporter on the bioconversion of GABA in engineered Escherichia coli. Bioprocess Biosyst. Eng. 2012;35:645–650. doi: 10.1007/s00449-011-0634-8. [DOI] [PubMed] [Google Scholar]
- Weissman K.J. Protein–protein interactions in multienzyme megasynthetases. Chembiochem Eur. J. Chem. Biol. 2008;9:826–848. doi: 10.1002/cbic.200700751. [DOI] [PubMed] [Google Scholar]
- Werpy, P., 2004. Top Value Added Chemicals From Biomass. Volume I – Results of Screening for Potential Candidates from Sugars and Synthesis Gas
- Wong F.T. Protein–protein recognition between acyltransferases and acyl carrier proteins in multimodular polyketide synthases. Biochemistry. 2010;49:95–102. doi: 10.1021/bi901826g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav G. Towards prediction of metabolic products of polyketide synthases: an in silico analysis. PLoS Comput. Biol. 2009;5:e1000351. doi: 10.1371/journal.pcbi.1000351. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material