Abstract
The potential advantages of hyaluronic acid (HA) production by metabolically-engineered Lactococcus lactis is constrained by the lower molecular weight and yield of HA obtained in these strains, compared to natural producers. Earlier studies have correlated lower HA yield with excessive lactate production in L. lactis cultures (Chauhan et al., 2014). In the present study, a three-fold increase was observed in the amount as well as molecular weight of HA produced by recombinant ldh-mutant L. lactis strains. The diversion from lactate production in the ldh-mutant strains resulted in excess ethanol and acetoin production and higher NAD+/NADH ratio in these cultures. The initial NAD+/NADH ratio showed a positive correlation with HA molecular weight as well as with the HA-precursor ratio (UDP-GlcUA/UDP-GlcNAc). The influence of NAD+/NADH ratio on regulation of the concerned metabolic pathways was assessed by transcriptional analysis of key genes having putative binding sites of the NADH-binding transcriptional factor, Rex.
Keywords: Hyaluronic acid molecular weight, Lactococcus lactis, Lactate dehydrogenase mutant, NAD+/NADH ratio, HA-precursor concentration ratio, Transcriptional regulator Rex
Highlights
-
•
Hyaluronan MW and production is higher in engineered ldh-mutant L. lactis strains.
-
•
Excess ethanol production in ldh-L. lactis cultures increased the NAD+/NADH ratio.
-
•
The initial NAD+/NADH ratio showed a positive correlation with HA molecular weight.
-
•
The NAD+/NADH ratio showed a positive correlation with the HA-precursor ratio.
-
•
Rex-repressor regulates transcription of genes in HA and other metabolic pathways.
1. Introduction
Hyaluronic acid (HA) production by metabolically-engineered recombinant organisms is being studied intensively, due to potential advantages over current commercial sources. Some of the recombinant GRAS organisms used for production of HA include Escherichia coli (Mao et al., 2009, Yu and Stephanopoulos, 2008), Bacillus subtilis (Chien and Lee, 2007a, Jia et al., 2013, Widner et al., 2005), Agrobacterium (Mao and Chen, 2007) and Lactococcus lactis (Badle et al., 2014, Chauhan et al., 2014, Chien and Lee, 2007b, Hmar et al., 2014, Prasad et al., 2010, Sheng et al., 2009). A comparison of various recombinant bacterial strains used for HA production, in terms of HA concentration, yield and molecular weight (based on the standards used) is given in Table 1. One of the main challenges with these systems is to produce high molecular weight HA, comparable to animal sources and native producers such as the pathogen, Streptococcus zooepidemicus.
Table 1.
Comparison of different recombinant bacterial strains producing HA.
| HA produced (g/l) | HA yield (YHA/S) (g/g) | HA molecular weight (MDa) and standards used | Reference |
|---|---|---|---|
| Bacillus subtilis | |||
| – | – | 1.1–1.2 | Widner et al. (2005) |
| 1.8 | 0.18 | – | Chien and Lee (2007a) |
| 6.8 | 0.34 | 4.5 (Polystyrene sodium sulfate) | Jia et al. (2013) |
| E. coli | |||
| 0.16 | – | 0.39–1.6 (HA) | Yu and Stephanopoulos (2008) |
| 3.7 | – | 1.5 (Dextran) | Mao et al. (2009) |
| L. lactis | |||
| 0.65 | 0.065 | – | Chien and Lee (2007b) |
| 0.234 | 0.023 | 2.8 (Pullulan) | Prasad et al. (2010) |
| 0.595 | 0.06 | – | Prasad et al. (2012) |
| 0.89 | 0.007 | 1.79 (PEO) | Chauhan et al. (2014) |
| 0.68 | 0.068 | 4.3(PEO) | Hmar et al. (2014) |
| 0.49 | 0.049 | – | Sheng et al. (2014) |
| 3.03 | 0.101 | 1.40 (HA) | Present study |
| Agrobacterium | |||
| 0.3 | – | 0.7–2 (Dextran) | Mao and Chen (2007) |
In the native producer of HA (S. zooepidemicus) two parallel pathways originate from glucose-6-phosphate and lead to formation of HA-precursors i.e. UDP-glucuronic acid and UDP-N-acetyl glucosamine (Fig. 1). The genes involved in HA synthesis are arranged in the has operon. In S. zooepidemicus, these genes are hasA, hasB, hasC, hasD and hasE. The most commonly used recombinant hosts for HA production do not contain the hyaluronan synthase (hasA) gene, whereas the homologs corresponding to the rest of genes are available. These homologous genes corresponding to hasB, hasC, hasD and hasE are UDP-glucose dehydrogenase (ugd), UDP-glucose pyrophosphorylase (galU), pyrophosphorylase (glmU) and phosphoglucoisomerase (pgi), respectively (Fig. 1). However, many studies have demonstrated that incorporation and expression of heterologous hasA alone results in low HA production. Co-expression of hasA with other has genes enhances HA yield and molecular weight (Chauhan et al., 2014, Prasad et al., 2010, Widner et al., 2005, Yu and Stephanopoulos, 2008). In L. lactis, it was shown that combination of three genes (hasABC and hasABD) was the optimum for higher HA yield (Chauhan et al., 2014, Prasad et al., 2010, Prasad et al., 2012).
Fig. 1.
Biosynthetic pathway of HA in L. lactis. The has genes shown in oval are the heterologous genes from S. zooepidemicus, which can be potentially expressed in L. lactis. The genes in hexagon are the homologous genes in L. lactis (adapted from Prasad et al. (2010)).
L. lactis presents an attractive alternative for HA production due to its well-studied genome and availability of wide range of genetic manipulation tools. However, HA production in this organism is limited by the excess lactic acid production. In L. lactis, nearly 70% of glucose gets fluxed towards lactate pathway (Chong et al., 2005). An inverse correlation was observed between HA yield and lactate yield in recombinant L. lactis strains (Badle et al., 2014, Chauhan et al., 2014) and S. zooepidemicus (Shah et al., 2013). Therefore, the objective of the present study was to investigate HA production in recombinant lactate dehydrogenase (ldh) mutant L. lactis strains, containing heterologous has-genes (hasABC and hasABD) taken from S. zooepidemicus.
In homo-fermentative cultures of lactic acid bacteria, the tight coupling of cofactor utilization and regeneration ensures a stoichiometric relation between glucose consumption and lactic acid production. During fermentative metabolism of glucose, every mole of glucose consumed produces 2 mol of NADH. Due to the absence of respiratory activity, NADH is regenerated to NAD+ primarily through lactate production, thus maintaining redox balance inside the cell (Cocaign-Bousquet et al., 1996). In the absence of ldh genes, carbon gets re-distributed from lactate production to other pathways for production of ethanol, acetoin, acetate, formate etc. (Bongers et al., 2003, Lopez De Felipe et al., 1997, Mehmeti et al., 2011). The emergence of other rescue pathways to replenish pools of NAD+ changes overall NAD+ and NADH levels and their ratio. The change in NAD+/NADH ratio has significant effects on glycolytic flux (Garrigues et al., 1997), as NAD+/NADH ratio affects GAPDH activity (Payot et al., 1998). The other key intermediate of glycolysis, fructose 1,6-bisphosphate, is also influenced by NAD+ levels (Neves et al., 1999). In HA synthesis, two equivalents of NAD+ are involved in the production of the limiting precursor UDP-Glucuronic acid (UDP-GlcUA). Therefore, we investigated the effect of changes in NAD+/NADH ratio on precursor levels and HA synthesis.
When the redox balance is disturbed, the cell rearranges its carbon distribution to attain redox homeostasis. This happens through up-regulation or down-regulation of genes coding for enzymes involved in cofactor-utilizing reactions. Many studies have shown that the redox-sensing regulator Rex is sensitive to NAD+/NADH ratio (Brekasis and Paget, 2003) and regulates the expression of genes coding for NAD+ or NADH-utilizing enzymes (Gyan et al., 2006, Pagels et al., 2010). Rex is composed of two structural domains: an N-terminal domain that interacts with DNA and a C-terminal domain that binds with NADH (Du and Pene, 1999, Sickmier et al., 2005, Wang et al., 2008). Its DNA-binding domain is strongly influenced by NADH level. Increased NADH level stabilizes its two DNA-binding domains, preventing the binding of Rex to its regulatory site in the upstream region of the gene (Gyan et al., 2006, Pagels et al., 2010, Sickmier et al., 2005).
Transcriptional regulation by Rex has been found in many organisms like B. subtilis (Gyan et al., 2006), Staphylococcus aureus (Pagels et al., 2010), Enterococcus faecalis (Mehmeti et al., 2011). Rex is found to be universally conserved between Streptococcaceae and Lactobacillaceae lineages (Ravcheev et al., 2013). The present study also sought to examine the possible role of Rex in regulating the expression of genes involved in NAD+ regeneration in recombinant L. lactis.
2. Materials and methods
2.1. Bacterial strains and plasmids
The strains used in this study were L. lactis NZ9000 and L. lactis NZ9020. Both the strains are based on the nisin-inducible (NICE) expression system. L. lactis NZ9000 was constructed from L. lactis MG1363 by integrating nisRK genes in its chromosome (Kuipers et al., 1998). L. lactis NZ9020 is a lactate dehydrogenase (ldh) mutant strain, in which two out of three ldh genes have been knocked out (Bongers et al., 2003). Both L. lactis NZ9000 and L. lactis NZ9020 were procured from NIZO (Netherlands). The plasmids used were pSJR3 (hasABC gene combination) and pSJR6 (hasABD gene combination), constructed by Prasad et al., 2010, Prasad et al., 2012. L. lactis SJR3 and L. lactis SJR6 were respectively constructed by transformation of L. lactis NZ9000 with pSJR3 and pSJR6 (Prasad et al., 2010, Prasad et al., 2012). In the present study, L. lactis MKG3 and L. lactis MKG6 were obtained by transformation of L. lactis NZ9020 with pSJR3 and pSJR6, respectively.
2.2. Culture media
The media used for agar plates, seed culture and bioreactor studies had the following components (g/l): Glucose (varying concentration), Yeast extract (5.0), brain heart infusion (5.0), KH2PO4 (0.5), K2HPO4 (1.5), MgSO4·7H2O (0.5), ascorbic acid (0.5). All the media components were procured from HiMedia (India). Glucose concentration in agar media and seed culture was 5 g/l. Initial glucose concentration varied from 10–80 g/l for bioreactor studies. During the investigations on the effect of acetate addition on HA production, different initial concentrations (5–15 g/l) of sodium acetate were used.
All the strains used in the study were stored at −80 °C as glycerol stocks and cultured for 24 h on agar plates at 30 °C. Colonies from agar plates were cultured in 10 ml tubes for 12–14 h. These tubes were then used to inoculate 100 ml of seed culture media, kept in static condition at 30 °C. The selection marker used for L. lactis SJR3 and L. lactis SJR6 strains was chloramphenicol (10 μg/ml). For L. lactis MKG3 and L. lactis MKG6, the selection markers were chloramphenicol (10 μg/ml) and tetracycline (2 μg/ml).
All the batch bioreactor experiments were carried out in a 2.4 l bioreactor (Bioengineering, Switzerland) with working volume of 1.2 l. The process conditions were: unaerated conditions, impeller speed 200 rpm, pH 7, 30 °C, 8% inoculum. HA production was induced by 2 ng/ml nisin (Sigma-Aldrich, USA) when the culture reached OD600=0.6.
2.3. Analytical methods
Samples were treated with 0.1% SDS to remove HA capsule from the cell surface and then centrifuged at 10,000 rpm to pellet HA-free biomass. The pellets were dissolved in 0.9% NaCl and the optical density was measured at 600 nm. The dry cell weight (DCW) concentration was measured and correlated to optical density using the correlation: 1 OD600=0.45 gDCW/l. The supernatant was used to estimate glucose, HA and other metabolites. Glucose was estimated by glucose-oxidase–peroxidase (GOD–POD) method (Merck Pvt. Ltd., India). HA was quantified by modified carbazole assay (Bitter and Muir, 1962).
HA molecular weight was estimated using PolySep-GFC-P 6000 size-exclusion column (300×7.8 mm2), in a HPLC fitted with RID. Mobile phase used was 0.2 N NaNO3 at 0.6 ml/min. HA standards were procured from Calbiochem (0.6 MDa) and Lifecore Biomedical (1.5 MDa and 1.8 MDa) and were used for estimation of molecular weight of HA. Although, many other studies have used polymer standards other than HA (Chauhan et al., 2014, Im et al., 2009, Lai et al., 2012, Liu et al., 2008a), it has been recently observed in our laboratory that it leads to overestimation of HA molecular weight (Supplementary data, Table S1).
Lactate, acetate, formate, ethanol and acetoin were estimated by HPLC (Shimadzu, Japan) at 50 °C using a 300×7.8 mm2 column (Phenomenex Rezex). The eluent used was 5 mM H2SO4 at 0.6 ml/min. Lactate, formate and acetate were detected by diode-array detector at 210 nm. Ethanol and acetoin were detected by refractive index detector (RID). The NAD+/NADH ratio was estimated using a quantification kit (Sigma-Aldrich, USA; Cat no.: MAK037). For extraction of NAD+ and NADH, 1 ml of extraction buffer was added to each sample and sonicated using QSONICA sonicator at 60 Hz for 5 min with pulse time of 10 s ON and 2 s OFF. Rest of the protocol was followed as described in the manual supplied with the kit.
Intracellular precursors of HA (UDP-GlcUA and UDP-GlcNAc) were extracted using method described by Ramos et al. (2001). Their estimation was done using Ion-pair reverse phase HPLC. The column used was Luna 5u C18(2) 100 Å, 250×4.6 mm2 Phenomenex column. For elution, Buffer A (100 mM potassium phosphate buffer, pH 6.4, supplemented with 8 mM tetrabutylammonium hydrogen sulfate as ion-pair reagent) and Buffer B (70% Buffer A with 30% acetonitrile) were used. The elution gradient was as follows: 100% Buffer A for 13 min; 0–77% linear gradient of Buffer B for 22 min; 77–100% gradient of Buffer B for 1 min; and 100% Buffer B for 14 min (Nakajima et al., 2010). The flow rate was maintained at 1 ml/min and the metabolites were detected using diode array detector at 254 nm.
2.4. Bioinformatic analysis of Rex binding sites in L. lactis MG1363
The manually-curated database of Rex regulon is available on RegPrecise database (http://regprecise.lbl.gov) for L. lactis subsp. cremoris SK11, which has 99% identity with L. lactis subsp. cremoris MG1363. From the given sequences of Rex-binding sites, a DNA motif was constructed to screen the genome of L. lactis subsp. cremoris MG1363 using Regulatory Sequence Analysis Tool (http://prokaryotes.rsat.eu/). Under the ‘pattern matching’ option, the ‘genome-scale-DNA-pattern’ was selected. The query pattern WWGWDRWWNHNDWHAHVW was searched in L. lactis subsp. cremoris MG1363 uid58837. The various parameters used were: feature type: CDS; sequence type: upstream from −500 to −1; overlap with upstream ORFs was allowed; sequence label: gene identifier; search strands: both strands; overlapping matches were prevented; threshold on match counts: 1; substitutions: 0.
2.5. Real-time quantification PCR
The expression level of adhe, adr and ugd was measured, based on protocols developed by Hmar et al. (2014). It consisted of initial denaturation at 95 °C for 15 min, followed by 40-cycle sequence of denaturation (15 s, 94 °C), annealing (57 °C, 30 s), and extension (72 °C, 30 s). The sequence of primers used for the genes is shown in Supplementary data (Table S2). The samples were collected at different time points (4, 6, 10, and 16 h) and normalized to their biomass concentrations. The collection times reflected critical stages of the culture, such as early, mid and late-exponential phases of growth and time periods corresponding to highest glucose uptake rate.
3. Results and discussion
3.1. Effect of ldh mutation on HA production and molecular weight
The L. lactis NZ9000 and L. lactis NZ9020 (ldh mutant) strains were transformed with recombinant plasmids containing the heterologous hasABC (L. lactis SJR3; L. lactis MKG3) and hasABD (L. lactis SJR6; L. lactis MKG6) gene combinations. Previous studies in our laboratory have established that these gene combinations give maximum HA production in recombinant L. lactis cultures (Chauhan et al., 2014, Prasad et al., 2010, Prasad et al., 2012). The four recombinant strains were compared under similar conditions for the concentration and molecular weight of HA produced by each strain in batch bioreactor cultures. It was observed that HA production was nearly three-fold higher in L. lactis MKG cultures as compared to L. lactis SJR cultures. The lowest HA concentration produced was 0.92 g/l by L. lactis SJR3 and the highest HA concentration produced was 3.03 g/l by L. lactis MKG6 (Fig. 2A). This result was anticipated due to the inverse correlation between HA yield and lactate yield (Badle et al., 2014, Chauhan et al., 2014).
Fig. 2.
Comparison of different recombinant strains in terms of (A) HA concentration and relative HA molecular weight; (B) acetoin, ethanol and lactate concentrations; (C) formate and acetate concentrations. The data are presented as the mean±SD from three replicates. L. lactis MKG3 and L. lactis MKG6 were statistically compared to L. lactis SJR3 and L. lactis SJR6, respectively. ** Indicates p<0.01 and *** indicates p< 0.001.
Our investigation threw up another interesting and unexpected result. The highest molecular weight of HA was produced by L. lactis MKG6 strain (~1.4 MDa), which was about three-fold higher (Fig. 2A) as compared to recombinant L. lactis SJR3 strain (0.47 MDa). Fig. 2A has shown only relative molecular weight increase for HA, since the values reported for absolute molecular weight in literature are dependent on the polymer standards used for calibration and estimating HA molecular weight. As can be observed from Table S1 (Supplementary data), the molecular weight of HA estimated using PEO-standards is nearly twice that estimated using HA-standards. Therefore, it is not appropriate to compare our data with the HA molecular weight reported in literature using non-HA polymer standards (Table 1). This study focuses on factors leading to molecular weight enhancement; therefore, relative molecular weight of HA is reported. The relative HA molecular weight is the molecular weight of HA estimated for each sample normalized to the molecular weight estimated for HA produced by L. lactis SJR3 grown at 30 g/l initial glucose in a batch bioreactor.
Very few studies have reported simultaneous increase in HA production as well as molecular weight. The present study showed significant enhancement in HA molecular weight along with increased HA production due to an ldh mutation in L. lactis strains. 3.2, 3.4 highlight the possible factors responsible for the increase in molecular weight of HA produced by recombinant L. lactis MKG strains.
As expected, lactate accumulation was low in case of L. lactis MKG cultures (Fig. 2B). The carbon overflow in L. lactis MKG3 and L. lactis MKG6 cultures was re-distributed towards formation of metabolites such as ethanol, acetoin, acetate and formate. Much higher concentrations of these metabolites were observed in L. lactis MKG cultures, as compared to L. lactis SJR cultures (Fig. 2B and C).
In general, the recombinant L. lactis MKG strains grew slower than the SJR strains (Supplementary data, Fig. S1). Many studies have shown an increase in HA molecular weight with decrease in specific growth rate (Badle et al., 2014, Chong et al., 2005, Jagannath and Ramachandran, 2010, Shah et al., 2013). In our study, the increase in molecular weight of HA produced by L. lactis MKG strains was much more drastic for the relatively small reduction in specific growth rate compared in L. lactis SJR strains (Supplementary data, Fig. S1).
Earlier studies in our laboratory have shown that initial glucose concentration affects yield and molecular weight of HA and an optimum value of 30–40 g/l has been reported for L. lactis SJR cultures (Chauhan et al., 2014). Therefore, experiments were conducted by varying initial glucose from 10–80 g/l for L. lactis MKG strains (Supplementary data, Tables S3 and S4). Unlike the inverse correlation between lactate yield and HA yield reported for recombinant L. lactis SJR cultures (Badle et al., 2014, Chauhan et al., 2014), a positive correlation was observed between ethanol and HA yields across various glucose concentrations in recombinant L. lactis MKG cultures (Fig. 3A). Similarly, a positive correlation was also observed between acetoin and HA yields (Fig. 3B).
Fig. 3.
Batch bioreactor cultures for L. lactis MKG6 (♦) and L. lactis MKG3 (■) across various initial glucose concentrations, showing (A) yield of HA per C-mol glucose consumed (YHA/S) versus C-mol yield of ethanol on glucose (YEthanol/S); (B) C-mol yield of HA on glucose (YHA/S) versus C-mol yield of acetoin on glucose consumed (YAcetoin/S).
3.2. Effect of NAD+/NADH ratio on HA production and molecular weight
In L. lactis MKG strains, the carbon redistribution towards ethanol and acetoin due to ldh deletion can potentially generate a higher amount of NAD+, as compared to lactate-producing L. lactis SJR strains. While each mole of lactate produced generates one mole of NAD+, each mole of acetoin and ethanol produced generates one and two moles of NAD+, respectively. Therefore, higher yield of ethanol and acetoin will give a higher yield of NAD+ as compared to lactate yield (Supplementary data, Table S5). An MFA study by Gao et al. (2006) showed that HA production is inhibited under high NADH flux. Subsequent studies showed that HA synthesis was found to be facilitated by oxidative environment inside the cell i.e. with availability of higher NAD+ (Liu et al., 2008b). However, as shown in Section 3.5, it is not NAD+ per se, but the NAD+/NADH ratio which is critical for the transcriptional regulation of many genes coding for NADH-utilizing enzymes (Brekasis and Paget, 2003). Such transcriptional regulation can affect the metabolic fluxes through the HA-precursor biosynthesis pathway and other competing pathways. In S. zooepdemicus, enhanced NAD+/NADH ratio was shown to increase HA yield indirectly (Zhang et al., 2006). Therefore, production and molecular weight of HA were analysed as a function of NAD+/NADH ratios in L. lactis MKG and L. lactis SJR cultures.
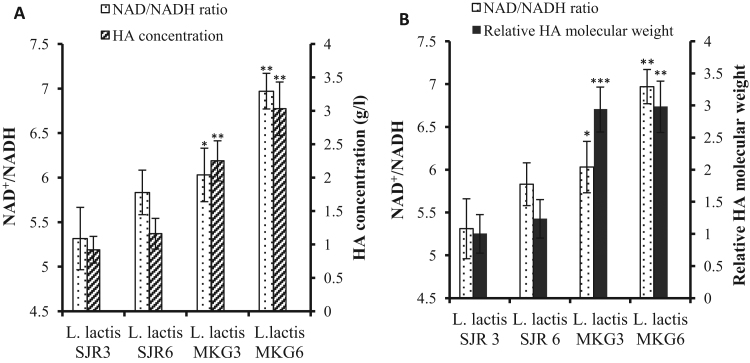
The initial NAD+/NADH ratio showed strong positive correlation with HA produced. The lowest initial NAD+/NADH ratio obtained was 5.31 in L. lactis SJR3 and the highest was 6.97 in L. lactis MKG6. This ratio was in general higher for L. lactis MKG strains as compared to L. lactis SJR strains and correlated well with the higher amount of HA produced by the former (Fig. 4A). It has been shown with other NAD+-dependent products that even a small increase in initial NAD+/NADH ratio, from 4.0 to 6.7 in a recombinant E. coli strain, results in a drastic eight-fold increase in yield of the NAD+-dependent product (Zhou et al., 2013).
Fig. 4.
Effect of initial NAD+/NADH ratio on (A) HA concentration; (B) Relative HA molecular weight for L. lactis SJR3, L. lactis SJR6, L. lactis MKG3 and L. lactis MKG6. The data are presented as the mean±SD from three replicates. L. lactis MKG3 and L. lactis MKG6 were statistically compared to L. lactis SJR3 and L. lactis SJR6, respectively. *p<0.05, **p<0.01 and ***p<0.001.
In our studies, a positive correlation was also found between the initial NAD+/NADH ratio and HA molecular weight obtained in L. lactis MKG and L. lactis SJR cultures. The highest relative molecular weight for HA (2.98) and NAD+/NADH ratio (6.97) were found in L. lactis MKG6 whereas the lowest relative molecular weight for HA (1.0) and NAD+/NADH ratio (5.31) were found in L. lactis SJR3 (Fig. 4B).
3.3. Effect of NAD+/NADH ratio on HA precursor ratio
It has been suggested that the polymerization activity of HA synthase is affected by competition between precursors for each other’s binding site (Chen et al., 2009, Tlapak-Simmons et al., 2004). This implies that chain termination could occur when one of the precursors is at a much higher concentration as compared to the other. It has been observed that a balance in concentration between the two HA-precursors is required for production of high molecular weight HA (Badle et al., 2014, Chen et al., 2009, Chong et al., 2005, Hmar et al., 2014 ). The cellular content of UDP-GlcNAc is found to be three to seven times higher than UDP-GlcUA in eukaryotic cells (Tammi et al., 2011). Vigetti et al. (2006) have shown that over-expressing genes for UDP-GlcUA enhanced HA production. Badle et al. (2014) have shown positive correlations between UDP-GlcUA/UDP-GlcNAc ratio and HA molecular weight. Hmar et al. (2014) showed that the higher molecular weight HA produced by recombinant L. lactis strains (with genome-integrated has genes) correlated with the UDP-GlcUA/UDP-GlcNAc ratio being close to unity in these cultures. Thus, the limiting precursor in HA synthesis seems to be UDP-GlcUA, whose synthesis requires two equivalents of NAD+.
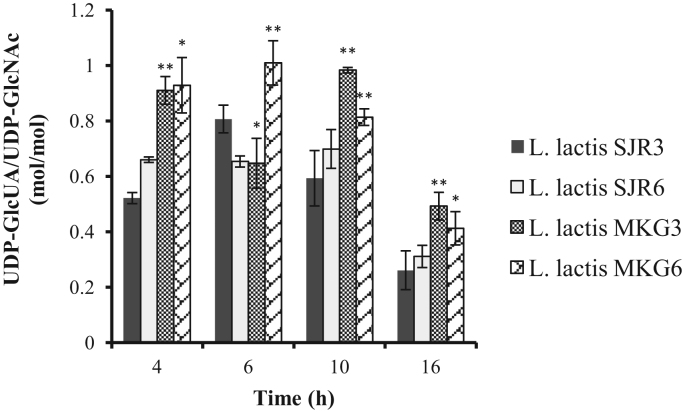
In the present study, the intracellular concentrations of UDP-GlcUA and UDP-GlcNAc were measured during the exponential and stationary phase of L. lactis MKG and L. lactis SJR cultures. The range of values obtained was comparable to that in S. zooepidemicus (Marcellin et al., 2009). The intracellular concentration of UDP-GlcNAc was found to be higher than UDP-GlcUA across all the strains (Fig. 5). On an average, the concentration of UDP-GlcUA was found to be higher in L. lactis MKG strains as compared to L. lactis SJR strains, the highest being in L. lactis MKG6. The ratio of UDP-GlcUA/UDP-GlcNAc was calculated for L. lactis MKG and L. lactis SJR strains (Fig. 6). During the initial and mid log phase, the balance of concentrations for UDP-GlcUA and UDP-GlcNAc was less skewed for L. lactis MKG6 as compared to the rest three strains.
Fig. 5.
The concentration of the UDP-sugars, UDP-GlcUA and UDP-GlcNAc, during various stages of growth. (A) L. lactis MKG6; (B) L. lactis MKG3; (C) L. lactis SJR6; (D) L. lactis SJR3. The data are presented as the mean±SD from three replicates. The concentration of UDP-GlcUA in L. lactis MKG6 and L. lactis MKG3 was statistically compared to L. lactis SJR6 and L. lactis SJR3. *p<0.05 and **p<0.01.
Fig. 6.
Ratio of UDP-GlcUA/UDP-GlcNAc for L. lactis MKG6, L. lactis MKG3, L. lactis SJR6 and L. lactis SJR3. The data are presented as the mean±SD from three replicates. The ratio of UDP-GlcUA/UDP-GlcNAc in L. lactis MKG6 and L. lactis MKG3 was statistically compared to L. lactis SJR6 and L. lactis SJR3. *p<0.05 and **p<0.01.
Since the cofactor NAD+ is involved in production of UDP-GlcUA, the effect of NAD+/NADH ratio on UDP-GlcUA/UDP-GlcNAc ratio was studied in L. lactis MKG and L. lactis SJR cultures. When measured with different recombinant L. lactis strains and across different time points, it was observed that NAD+/NADH ratio showed a strong positive correlation with UDP-GlcUA/UDP-GlcNAc ratio (Fig. 7). With increase in NAD+/NADH ratio, the UDP-GlcUA/UDP-GlcNAc ratio was found to approach unity, which is favorable for production of higher molecular weight HA. Badle et al. (2014) have shown that this precursor ratio correlates strongly with molecular weight of HA produced by S. zooepidemicus as well as recombinant L. lactis strains. This shows that higher NAD+/NADH ratio helps in producing higher molecular weight HA. To further validate these observations, a process strategy was adopted to increase NAD+/NADH ratio inside the cell.
Fig. 7.
Batch bioreactor studies showing the effect of NAD+/NADH ratio on ratio of UDP-GlcUA/UDP-GlcNAc. The strains used were L. lactis MKG6 (♦), L. lactis MKG3 (▀), L. lactis SJR6 (▲) and L. lactis SJR3 (●). Each data point corresponds to the mean value obtained from three repeats.
3.4. Effect of acetate addition on redox balance and HA molecular weight
The NAD+/NADH ratio can be manipulated through process strategies (Liu and Chen, 2011; San et al., 2002) or genetic strategies (Berríos-Rivera et al., 2004, Liu and Chen, 2011, San et al., 2002, Wang et al., 2013). Hols et al. (1999) have shown that acetate addition enhanced ethanol formation in ldh-mutant L. lactis strains. We adopted the same strategy to enhance ethanol production, which in turn was expected to increase NAD+/NADH ratio, and thereby HA production and molecular weight. The strain L. lactis MKG6, which produced HA at the highest concentration and molecular weight, was chosen for the experiments.
Acetate was added at different concentrations (5, 10 and 15 g/l) at the beginning of the bioreactor culture, keeping the initial glucose concentration at 30 g/l. Interestingly, with 5 g/l acetate addition there was 25% increase in HA molecular weight (Table 2). The yields of acetoin and ethanol also increased from 0.43 C-mol/C-mol each to 0.52 C-mol/C-mol and 0.47 C-mol/C-mol, respectively. The initial NAD+/NADH ratio showed an increase from 6.97 to 8.46.
Table 2.
Effect of acetate addition on HA production and molecular weight in L. lactis MKG6.
| Without acetate | 5 g/l Acetate | 10 g/l Acetate | 15 g/l Acetate | |
|---|---|---|---|---|
| Biomass (g/l) | 3.36±0.16 | 6.83±0.10 | 7.99±0.23 | 7.2±0.05 |
| HA (g/l) | 3.03±0.4 | 2.5±0.21 | 1.76±0.11 | 1.6±0.06 |
| Relative HA molecular weight | 3.52±0.2 | 4.4±0.14 | 3.5±0.15 | 3.6±0.11 |
| Lactic acid (g/l) | 0.6±0.16 | 0.9±0.12 | 0.83±0.11 | 0.92±0.12 |
| Formate (g/l) | 4.1±0.16 | 7.09±0.11 | 6.2±0.23 | 6.0±0.26 |
| Acetoin (g/l) | 9.28±0.4 | 11.8±0.07 | 10.1±0.03 | 10.8±0.2 |
| Ethanol (g/l) | 9.86±0.32 | 11.31±0.38 | 6.96±0.23 | 8.59±0.14 |
| Acetic acid (g/l) | 1.23±0.05 | 3.5±0.24 | 6.52±0.14 | 10.2±0.07 |
| Yacetoin/S (C-mol/C-mol) | 0.43 | 0.52 | 0.44 | 0.45 |
| Yethanol/S (C-mol/C-mol) | 0.43 | 0.47 | 0.29 | 0.33 |
| NAD+/NADH ratio | 6.97±0.3 | 8.46±0.17 | 6.1±0.09 | 5.99±0.21 |
The yields of acetoin and ethanol as well as initial NAD+/NADH ratio decreased with increased acetate addition (>5 g/l), with a concomitant decrease in HA molecular weight. This experiment validated the hypothesis that increase in initial NAD+/NADH ratio contributes towards higher HA molecular weight. However, the HA concentration inexplicably dropped from 3.03 g/l to 2.5 g/l at even at higher initial NAD+/NADH ratio (obtained at acetate addition of 5 g/l). The HA concentration decreased further with increased acetate addition.
3.5. Transcriptional regulation of genes involving cofactor utilization
During the fermentation at different growth phases, NAD+/NADH ratio showed dynamic variation (Supplementary data, Fig. S2). This change in level of NAD+/NADH ratio is a part of dynamic homeostasis between different redox states. It is mediated inside the cell through redox sensors and transcriptional regulators like Rex (Mehmeti et al., 2011).
The deletion of ldh genes in L. lactis led to re-distribution of fluxes, resulting in increased production of ethanol and acetoin, as well as shift in NAD+/NADH ratios. The fluxes for HA synthesis as well as competing pathways may be regulated by transcriptional regulation of critical genes in these pathways.
Screening of L. lactis subsp. cremoris MG1363 showed 11 genes in central carbon metabolism that could contain Rex-binding sites. Of these, the genes for bifunctional acetaldehyde-CoA/alcohol dehydrogenase (adhe), acetoin (diacetyl) reductase (adr) and UDP-glucose dehydrogense (ugd) were selected for transcriptional analysis in L. lactis MKG6. The mRNA level of these genes were measured relative to the host strain L. lactis NZ9020. The relative mRNA level was correlated with the NAD+/NADH ratio, measured at selected time-points in the recombinant L. lactis MKG6 cultures. For Rex-regulated genes, a high NAD+/NADH ratio would lead to binding the Rex repressor and low mRNA levels. Conversely, a low NAD+/NADH ratio would de-repress the gene and lead to high mRNA levels. The relative mRNA levels for adhe, adr and ugd showed an inverse correlation with NAD+/NADH ratio (Fig. 8). This indicated that the expression of these genes is probably regulated by the global regulator Rex, whose binding activity is controlled by NAD+/NADH ratio.
Fig. 8.
Relative mRNA level of adhe, adr and ugd genes in L. lactis MKG6 strain.
While the Rex-regulation understandably acts as a feedback regulation mechanism for adhe and adr, it becomes a feed-forward repression mechanism for ugd. At high NAD+/NADH ratio, the production of UDP-GlcUA is controlled at transcriptional level. Prasad et al. (2012) have found the hasB mRNA level to be several-fold lower than other has genes in S. zooepidemicus, and similar results were obtained for ugd in L. lactis cultures, suggesting its tight regulation at transcriptional level. Not surprisingly, several studies have shown that co-expression of hasB (ugd) along with hasA leads to multifold increase in HA production (Chauhan et al., 2014, Chien and Lee, 2007a, Chien and Lee, 2007b, Prasad et al., 2010, Widner et al., 2005).
When NAD+/NADH ratio decreases, the transcriptional regulation of ugd is relaxed, but the lowered thermodynamic driving force leads to lower precursor production. The equilibrium constant for the reaction converting UDP-glucose and 2 NAD+ to UDP-GlcUA and 2 NADH, has a 2nd order dependence on NAD+/NADH ratio. The equilibrium constant changes sharply with variation in NAD+/NADH ratio. Therefore, even with over-expression of ugd gene, HA production will not be thermodynamically favored unless the NAD+/NADH ratio is high. Our study confirms this hypothesis while comparing HA production by the L. lactis NZ9000 and L. lactis NZ9020 (ldh-mutant) strains containing the same combination of has genes.
In addition to the transcriptional regulation, Ugd experiences cofactor and product inhibition under certain conditions. In E. coli, Mainprize et al. (2013) have shown substrate inhibition by NAD+ at concentrations beyond 1 mM. They suggested that at high concentration of NAD+, multiple NAD+ molecules fit into the cofactor binding pocket in non-productive geometries, which decreases Ugd activity. In Klebsiella pneumonia, the mechanism of allosteric regulation of Ugd was proposed (Chen et al., 2011). When UDP-GlcUA accumulates (Ki=283 µM), it inactivates Ugd either by competing for substrate binding site or by binding to the allosteric site. The heterologous expression of the ugd gene leads to high enzyme levels, reducing inhibition by the co-factor or the product. Due to the multiple regulations, over-expression of ugd as well as high NAD+/NADH ratio is required for high production of UDP-GlcUA and obtaining high molecular weight HA.
4. Conclusions
This study has highlighted the importance of cofactor distribution and availability in metabolic processes for enhancing the yield and productivity of cofactor-dependent products. The pathways for HA synthesis involve different cofactors including NAD+, UTP and Acetyl CoA. Therefore, metabolic engineering for HA production provides a rich paradigm for studying optimal cofactor distribution between cellular needs and product synthesis. This study demonstrated how the change in redox balance (viz. the NAD+/NADH ratio) impacts production and molecular weight of HA in pathway-engineered L. lactis strains. Although, the shift in redox balance occurred primarily due to the ldh-mutation, which inactivated lactate synthesis, we have shown that suitable process strategies (viz. acetate addition) can also be adopted to achieve this objective. Through the screening of Rex-binding sites and correlation of gene-expression with NAD+/NADH ratio, we have indirectly shown the possibility of transcriptional regulation of several critical genes by Rex. Although, this study did not investigate direct mechanistic evidence for this hypothesis, it is nevertheless an interesting observation worth further investigation. Cofactor utilization is tightly regulated within a cell, often at multiple levels and this issue has wider implications for metabolic engineering of cofactor-dependent products.
Acknowledgments
We acknowledge the Ministry of Human Resource Development (Government of India) for providing financial aid to Mandeep Kaur and the Department of Biotechnology (Government of India) for the funding under Project no. BT/IN/Finland/30/GJ/2013. We would also like to acknowledge Dr. Shashi Bala Prasad for providing the recombinant plasmids and NIZO (Netherlands) for the Lactococcus lactis strains.
Footnotes
Supplementary data associated with this article can be found in the online version at http://dx.doi.org/10.1016/j. meteno.2016.01.003.
Appendix A. Supplementary material
Supplementary material
Supplementary material
References
- Badle S.S., Jayaraman G., Ramachandran K.B. Ratio of intracellular precursors concentration and their flux influences hyaluronic acid molecular weight in Streptococcus zooepidemicus and recombinant Lactococcus lactis. Bioresour. Technol. 2014;163:222–227. doi: 10.1016/j.biortech.2014.04.027. [DOI] [PubMed] [Google Scholar]
- Berríos-Rivera S.J., Sánchez A.M., Bennett G.N., San K.Y. Effect of different levels of NADH availability on metabolite distribution in Escherichia coli fermentation in minimal and complex media. Appl. Microbiol. Biotechnol. 2004;65:426–432. doi: 10.1007/s00253-004-1609-3. [DOI] [PubMed] [Google Scholar]
- Bitter T., Muir H.M. A modified uronic acid carbazole reaction. Anal. Biochem. 1962;4:330–334. doi: 10.1016/0003-2697(62)90095-7. [DOI] [PubMed] [Google Scholar]
- Bongers R.S., Hoefnagel M.H.N., Starrenburg M.J.C., Siemerink M.A.J., Arends J.G.A., Hugenholtz J., Kleerebezem M. IS981-Mediated adaptive evolution recovers lactate production by ldhB transcription activation in a lactate dehydrogenase-deficient strain of Lactococcus lactis. J. Bacteriol. 2003;185:4499–4507. doi: 10.1128/JB.185.15.4499-4507.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brekasis D., Paget M.S. A novel sensor of NADH/NAD redox poise in Streptomyces coelicolor A3(2) EMBO J. 2003;22:4856–4865. doi: 10.1093/emboj/cdg453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauhan A.S., Badle S.S., Ramachandran K.B., Jayaraman G. The P170 expression system enhances hyaluronan molecular weight and production in metabolically-engineered Lactococcus lactis. Biochem. Eng. J. 2014;90:73–78. [Google Scholar]
- Chen W.Y., Marcellin E., Hung J., Nielsen L.K. Hyaluronan molecular weight is controlled by UDP-N-acetylglucosamine concentration in Streptococcus zooepidemicus. J. Biol. Chem. 2009;284:18007–18014. doi: 10.1074/jbc.M109.011999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y.Y., Ko T.P., Lin C.H., Chen W.H., Wang A.H.J. Conformational change upon product binding to Klebsiella pneumoniae UDP-glucose dehydrogenase: a possible inhibition mechanism for the key enzyme in polymyxin resistance. J. Struct. Biol. 2011;175:300–310. doi: 10.1016/j.jsb.2011.04.010. [DOI] [PubMed] [Google Scholar]
- Chien L.J., Lee C.K. Enhanced hyaluronic acid production in Bacillus subtilis by coexpressing bacterial hemoglobin. Biotechnol. Prog. 2007;23:1017–1022. doi: 10.1021/bp070036w. [DOI] [PubMed] [Google Scholar]
- Chien L.J., Lee C.K. Hyaluronic acid production by recombinant Lactococcus lactis. Appl. Microbiol. Biotechnol. 2007;77:339–346. doi: 10.1007/s00253-007-1153-z. [DOI] [PubMed] [Google Scholar]
- Chong B.F., Blank L.M., Mclaughlin R., Nielsen L.K. Microbial hyaluronic acid production. Appl. Microbiol. Biotechnol. 2005;66:341–351. doi: 10.1007/s00253-004-1774-4. [DOI] [PubMed] [Google Scholar]
- Cocaign-Bousquet M., Garrigues C., Loubiere P., Lindley N.D. Physiology of pyruvate metabolism in Lactococcus lactis. Antonie Van Leeuwenhoek. 1996;70:253–267. doi: 10.1007/BF00395936. [DOI] [PubMed] [Google Scholar]
- Du X., Pene J.J. Identification, cloning and expression of p25, an AT-rich DNA-binding protein from the extreme thermophile, Thermus aquaticus YT-1. Nucleic Acids Res. 1999;27:1690–1697. doi: 10.1093/nar/27.7.1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao H.J., Du G.C., Chen J. Analysis of metabolic fluxes for hyaluronic acid (HA) production by Streptococcus zooepidemicus. World J. Microbiol. Biotechnol. 2006;22:399–408. [Google Scholar]
- Garrigues C., Loubiere P., Lindley N.D., Cocaign-Bousquet M. Control of the shift from homolactic acid to mixed-acid fermentation in Lactococcus lactis: predominant role of the NADH/NAD ratio. J. Bacteriol. 1997;179:5282–5287. doi: 10.1128/jb.179.17.5282-5287.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyan S., Shiohira Y., Sato I., Takeuchi M., Sato T. Regulatory loop between redox sensing of the NADH/NAD(+) ratio by Rex (YdiH) and oxidation of NADH by NADH dehydrogenase Ndh in Bacillus subtilis. J. Bacteriol. 2006;188:7062–7071. doi: 10.1128/JB.00601-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hmar R.V., Prasad S.B., Jayaraman G., Ramachandran K.B. Chromosomal integration of hyaluronic acid synthesis (has) genes enhances the molecular weight of hyaluronan produced in Lactococcus lactis. Biotechnol. J. 2014;9:1554–1564. doi: 10.1002/biot.201400215. [DOI] [PubMed] [Google Scholar]
- Hols P., Ramos A., Hugenholtz J., Delcour J., de Vos W.M., Santos H., Kleerebezem M. Acetate utilization in Lactococcus lactis deficient in lactate dehydrogenase : a rescue pathway for maintaining redox balance. J. Bacteriol. 1999;181:5521–5526. doi: 10.1128/jb.181.17.5521-5526.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Im J.H., Song J.M., Kang J.H., Kang D.J. Optimization of medium components for high-molecular-weight hyaluronic acid production by Streptococcus sp. ID9102 via a statistical approach. J. Ind. Microbiol. Biotechnol. 2009;36:1337–1344. doi: 10.1007/s10295-009-0618-8. [DOI] [PubMed] [Google Scholar]
- Jagannath S., Ramachandran K.B. Influence of competing metabolic processes on the molecular weight of hyaluronic acid synthesized by Streptococcus zooepidemicus. Biochem. Eng. J. 2010;48:148–158. [Google Scholar]
- Jia Y., Zhu J., Chen X., Tang D., Su D., Yao W., Gao X. Metabolic engineering of Bacillus subtilis for the efficient biosynthesis of uniform hyaluronic acid with controlled molecular weights. Bioresour. Technol. 2013;132:427–431. doi: 10.1016/j.biortech.2012.12.150. [DOI] [PubMed] [Google Scholar]
- Kuipers O.P., de Ruyter P.G.G.A., Kleerebezem M., de Vos W.M. Quorum sensing-controlled gene expression in lactic acid bacteria. J. Biotechnol. 1998;64:15–21. [Google Scholar]
- Lai Z.W., Rahim R.A., Ariff A.B., Mohamad R. Biosynthesis of high molecular weight hyaluronic acid by Streptococcus zooepidemicus using oxygen vector and optimum impeller tip speed. J. Biosci. Bioeng. 2012;114:286–291. doi: 10.1016/j.jbiosc.2012.04.011. [DOI] [PubMed] [Google Scholar]
- Liu L., Chen J. Cofactor Engineering Enhances the Physiological Function of an Industrial Strain. In: Carpi A., editor. Progress in molecular and environmental bioengineering – from analysis and modeling to technology applications. Intech; Croatia: 2011. pp. 427–444. ISBN 978-953-307-268-5. [Google Scholar]
- Liu L., Du G., Chen J., Wang M., Sun J. Influence of hyaluronidase addition on the production of hyaluronic acid by batch culture of Streptococcus zooepidemicus. Food Chem. 2008;110:923–926. doi: 10.1016/j.foodchem.2008.02.082. [DOI] [PubMed] [Google Scholar]
- Liu L., Wang M., Du G., Chen J. Enhanced hyaluronic acid production of Streptococcus zooepidemicus by an intermittent alkaline-stress strategy. Lett. Appl. Microbiol. 2008;46:383–388. doi: 10.1111/j.1472-765X.2008.02325.x. [DOI] [PubMed] [Google Scholar]
- Lopez De Felipe F., Starrenburg M.J.C., Hugenholtz J. The role of NADH-oxidation in acetoin and diacetyl production from glucose in Lactococcus lactis subsp. lactis MG1363. FEMS Microbiol. Lett. 1997;156:15–19. [Google Scholar]
- Mainprize I.L., Bean J.D., Bouwman C., Kimber M.S., Whitfield C. The UDP-glucose dehydrogenase of Escherichia coli K-12 displays substrate inhibition by NAD that is relieved by nucleotide triphosphates. J. Biol. Chem. 2013;288:23064–23074. doi: 10.1074/jbc.M113.486613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Z., Chen R.R. Recombinant synthesis of hyaluronan by Agrobacterium sp. Biotechnol. Prog. 2007;23:1038–1042. doi: 10.1021/bp070113n. [DOI] [PubMed] [Google Scholar]
- Mao Z., Shin H.D., Chen R. A recombinant E. coli bioprocess for hyaluronan synthesis. Appl. Microbiol. Biotechnol. 2009;84:63–69. doi: 10.1007/s00253-009-1963-2. [DOI] [PubMed] [Google Scholar]
- Marcellin E., Nielsen L.K., Abeydeera P., Kromer J.O. Quantitative analysis of intracellular sugar phosphates and sugar nucleotides in encapsulated streptococci using HPAEC-PAD. Biotechnol. J. 2009;4:58–63. doi: 10.1002/biot.200800197. [DOI] [PubMed] [Google Scholar]
- Mehmeti I., Jönsson M., Fergestad E.M., Mathiesen G., Nes I.F., Holo H. Transcriptome, proteome, and metabolite analyses of a lactate dehydrogenase-negative mutant of Enterococcus faecalis V583. Appl. Environ. Microbiol. 2011;77:2406–2413. doi: 10.1128/AEM.02485-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima K., Kitazume S., Angata T., Fujinawa R., Ohtsubo K., Miyoshi E., Taniguchi N. Simultaneous determination of nucleotide sugars with ion-pair reversed-phase HPLC. Glycobiology. 2010;20:865–871. doi: 10.1093/glycob/cwq044. [DOI] [PubMed] [Google Scholar]
- Neves A.R., Ramos A., Nunes M.C., Kleerebezem M., Hugenholtz J., De Vos W.M., Almeida J., Santos H. In vivo nuclear magnetic resonance studies of glycolytic kinetics in Lactococcus lactis. Biotechnol. Bioeng. 1999;64:200–212. doi: 10.1002/(sici)1097-0290(19990720)64:2<200::aid-bit9>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- Pagels M., Fuchs S., Pané-Farré J., Kohler C., Menschner L., Hecker M., McNamarra P.J., Bauer M.C., Von Wachenfeldt C., Liebeke M., Lalk M., Sander G., Von Eiff C., Proctor R.A., Engelmann S. Redox sensing by a Rex-family repressor is involved in the regulation of anaerobic gene expression in Staphylococcus aureus. Mol. Microbiol. 2010;76:1142–1161. doi: 10.1111/j.1365-2958.2010.07105.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payot S., Guedon E., Cailliez C., Gelhaye E., Petitdemange H. Metabolism of cellobiose by Clostridium cellulolyticum growing in continuous culture: evidence for decreased NADH reoxidation as a factor limiting growth. Microbiology. 1998;144:375–384. doi: 10.1099/00221287-144-2-375. [DOI] [PubMed] [Google Scholar]
- Prasad S.B., Jayaraman G., Ramachandran K.B. Hyaluronic acid production is enhanced by the additional co-expression of UDP-glucose pyrophosphorylase in Lactococcus lactis. Appl. Microbiol. Biotechnol. 2010;86:273–283. doi: 10.1007/s00253-009-2293-0. [DOI] [PubMed] [Google Scholar]
- Prasad S.B., Ramachandran K.B., Jayaraman G. Transcription analysis of hyaluronan biosynthesis genes in Streptococcus zooepidemicus and metabolically engineered Lactococcus lactis. Appl. Microbiol. Biotechnol. 2012;94:1593–1607. doi: 10.1007/s00253-012-3944-0. [DOI] [PubMed] [Google Scholar]
- Ramos A., Boels I.C., de Vos M.W., Santos H. Relationship between glycolysis and exopolysaccharide biosynthesis in Lactococcus lactis. Appl. Environ. Microbiol. 2001;67:33–41. doi: 10.1128/AEM.67.1.33-41.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravcheev D.A., Best A.A., Sernova N.V., Kazanov M.D., Novichkov P.S., Rodionov D.A. Genomic reconstruction of transcriptional regulatory networks in lactic acid bacteria. BMC Genom. 2013;14:1–14. doi: 10.1186/1471-2164-14-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- San K.Y., Bennett G.N., Berríos-Rivera S.J., Vadali R.V., Yang Y.T., Horton E., Rudolph F.B., Sariyar B., Blackwood K. Metabolic engineering through cofactor manipulation and its effects on metabolic flux redistribution in Escherichia coli. Metab. Eng. 2002;4:182–192. doi: 10.1006/mben.2001.0220. [DOI] [PubMed] [Google Scholar]
- Shah M.V., Badle S.S., Ramachandran K.B. Hyaluronic acid production and molecular weight improvement by redirection of carbon flux towards its biosynthesis pathway. Biochem. Eng. J. 2013;80:53–60. [Google Scholar]
- Sheng J., Ling P., Wang F. Constructing a recombinant hyaluronic acid biosynthesis operon and producing food-grade hyaluronic acid in Lactococcus lactis. J. Ind. Microbiol. Biotechnol. 2014;42:197–206. doi: 10.1007/s10295-014-1555-8. [DOI] [PubMed] [Google Scholar]
- Sheng J.Z., Ling P.X., Zhu X.Q., Guo X.P., Zhang T.M., He Y.L., Wang F.S. Use of induction promoters to regulate hyaluronan synthase and UDP-glucose-6-dehydrogenase of Streptococcus zooepidemicus expression in Lactococcus lactis: a case study of the regulation mechanism of hyaluronic acid polymer. J. Appl. Microbiol. 2009;107:136–144. doi: 10.1111/j.1365-2672.2009.04185.x. [DOI] [PubMed] [Google Scholar]
- Sickmier E.A., Brekasis D., Paranawithana S., Bonanno J.B., Paget M.S.B., Burley S.K., Kielkopf C.L. X-ray structure of a Rex-family repressor/NADH complex insights into the mechanism of redox sensing. Structure. 2005;13:43–54. doi: 10.1016/j.str.2004.10.012. [DOI] [PubMed] [Google Scholar]
- Tammi R.H., Passi A.G., Rilla K., Karousou. E., Vigetti D., Makkonen K., Tammi M.I. Transcriptional and post-translational regulation of hyaluronan synthesis. FEBS J. 2011;278:1419–1428. doi: 10.1111/j.1742-4658.2011.08070.x. [DOI] [PubMed] [Google Scholar]
- Tlapak-Simmons V.L., Baron C.A., Weigel P.H. Characterization of the purified hyaluronic acid synthase from Streptococcus equisimilis. Biochemistry. 2004;43:9234–9242. doi: 10.1021/bi049468v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigetti D., Ori M., Viola M., Genasetti A., Karousou E., Rizzi M., Pallotti F., Nardi I., Hascall V.C., De Luca G., Passi A. Molecular cloning and characterization of UDP-glucose dehydrogenase from the amphibian Xenopus laevis and its involvement in hyaluronan synthesis. J. Biol. Chem. 2006;281:8254–8263. doi: 10.1074/jbc.M508516200. [DOI] [PubMed] [Google Scholar]
- Wang E., Bauer M.C., Rogstam A., Linse S., Logan D.T., Von Wachenfeldt C. Structure and functional properties of the Bacillus subtilis transcriptional repressor Rex. Mol. Microbiol. 2008;69:466–478. doi: 10.1111/j.1365-2958.2008.06295.x. [DOI] [PubMed] [Google Scholar]
- Wang Y., San K.Y., Bennett George N. Cofactor engineering for advancing chemical biotechnology. Curr. Opin. Biotechnol. 2013;24:994–999. doi: 10.1016/j.copbio.2013.03.022. [DOI] [PubMed] [Google Scholar]
- Widner B., Behr R., Von Dollen S., Tang M., Heu T., Sloma A., Sternberg D., DeAngelis P.L., Weigel P.H., Brown S. Hyaluronic acid production in Bacillus subtilis. Appl. Environ. Microbiol. 2005;71:3747–3752. doi: 10.1128/AEM.71.7.3747-3752.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H., Stephanopoulos G. Metabolic engineering of Escherichia coli for biosynthesis of hyaluronic acid. Metab. Eng. 2008;10:24–32. doi: 10.1016/j.ymben.2007.09.001. [DOI] [PubMed] [Google Scholar]
- Zhang J., Hao N., Chen G.Q. Effect of expressing polyhydroxybutyrate synthesis genes (phbCAB) in Streptococcus zooepidemicus on production of lactic acid and hyaluronic acid. Appl. Microbiol. Biotechnol. 2006;71:222–227. doi: 10.1007/s00253-005-0164-x. [DOI] [PubMed] [Google Scholar]
- Zhou Y.J., Yang W., Wang L., Zhu Z., Zhang S., Zhao Z.K. Engineering NAD+ availability for Escherichia coli whole-cell biocatalysis: a case study for dihydroxyacetone production. Microb. Cell Fact. 2013;12:1–11. doi: 10.1186/1475-2859-12-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Supplementary material