Abstract
Acidithiobacillus ferrooxidans is a gram-negative chemolithoautotrophic γ-proteobacterium. It typically grows at an external pH of 2 using the oxidation of ferrous ions by oxygen, producing ferric ions and water, while fixing carbon dioxide from the environment. A. ferrooxidans is of great interest for biomining and environmental applications, as it can process mineral ores and alleviate the negative environmental consequences derived from the mining processes. In this study, the first genome-scale metabolic reconstruction of A. ferrooxidans ATCC 23270 was generated (iMC507). A total of 587 metabolic and transport/exchange reactions, 507 genes and 573 metabolites organized in over 42 subsystems were incorporated into the model. Based on a new genetic algorithm approach, that integrates flux balance analysis, chemiosmotic theory, and physiological data, the proton translocation stoichiometry for a number of enzymes and maintenance parameters under aerobic chemolithoautotrophic conditions using three different electron donors were estimated. Furthermore, a detailed electron transfer and carbon flux distributions during chemolithoautotrophic growth using ferrous ion, tetrathionate and thiosulfate were determined and reported. Finally, 134 growth-coupled designs were calculated that enables Extracellular Polysaccharide production. iMC507 serves as a knowledgebase for summarizing and categorizing the information currently available for A. ferrooxidans and enables the understanding and engineering of Acidithiobacillus and similar species from a comprehensive model-driven perspective for biomining applications.
Keywords: Biomining, Acidithiobacillus ferrooxidans, Systems biology, Strain design
Highlights
-
•
First genome scale reconstruction metabolic network of Acidithiobacillus ferrooxidans.
-
•
Genome scale based proton translocation stoichiometry estimation.
-
•
Strain designs for Extracellular Polysaccharide growth-coupling production.
1. Introduction
The mining industry is a major force in the world economy, occupying a primary position at the start of the resource supply chain, supporting 14.4% of the world's total economy, while using less than 1% of the global surface area (CIA, 2011). Production patterns are driven by consumption, which continues to rise in middle- to high-income countries, and is reaching unprecedented levels in low-income countries, whose appetite for the world's minerals reflects their rapid development (Fischer-Kowalski and Swilling, 2011). However, extraction and processing are associated with a number of sustainable development challenges, including economic, environmental and social issues. For example, poor waste management practice, one of the most conspicuous features of the global mineral industry, can result in severe and long-term environmental and social consequences. Furthermore, it can also impose costs on mining companies by eroding share value, increasing the risks of temporary or permanent shut down, exposure to compensation, fines and litigation costs, lost future opportunities and increased remediation and monitoring (Franks et al., 2011).
A way to alleviate the negative consequences of mining is through the application of microbial processes, referred generically as “biomining”. They do not require the high amounts of energy used during roasting or smelting and do not produce sulfur dioxide or other environmentally harmful gaseous emissions. Furthermore, mine tailings and wastes produced from physicochemical processes when exposed to rain and air may be biologically leached, producing unwanted acid and metal pollution. Tailings and wastes from biomining operations are less chemically active, and the biological activity they can support is reduced by at least the extent to which they have already been bioleached. From an economical point of view, biomining has a clear advantage in the extraction and recovery of precious and base metals from low-grade ores, where many metals are not economically recoverable by non-biological methods (Rawlings, 2002) (ores of copper, nickel, cobalt, zinc and uranium). At least 20% of the copper produced worldwide today comes from bioleaching (Rawlings and Johnson, 2007).
There are two major microbial mediated processes in biomining. The first is bioleaching, which is a strategy for metal recovery, whose underlying mechanism is the oxidation of metallic and/or sulfuric compounds by either enzymatic or mediated chemical oxidation caused by the catabolism of microorganisms. Depending on the mineral, chemical attack is by a combination of ferric iron and acid (protons), whereas the role of the microorganisms is to generate the ferric iron and acid. The second process is called biooxidation. This strategy applies mainly to the recovery of gold from difficult-to-treat arsenopyrites ores and concentrates. The aim is to use biooxidation to decompose the mineral matrix and expose entrapped gold (Rawlings et al., 2003). These processes are mediated by a consortium of Gram-negative bacteria (Acidithiobacillus, Leptospirillum, Sulfobacillus, Acidimicrobium) and archaeal genus (Ferroplasma, Sulfolobulus and Metallosphaers). There are many factors that affect the microbial composition of ores, such as, the type of mineral to be treated, temperature, and type of reactor used. Industrial applications use both mixed populations (Ishigaki et al., 2005) as well as isolated cultures (Falco et al., 2003, Sand et al., 1992).
Bioleaching processes are extensively used in copper extraction. Typically, these processes can be summarized in three steps. First, copper ores are pulverized and placed in heaps. Second, In order to promote the microbial consortia metabolism for iron and sulfur compound oxidation, the heaps are sprinkled with sulfuric acid. In this step, the microbial consortia oxidizes Fe(II) to Fe(III). And thirdly, the Fe(III) generated from the microbial metabolism is used to oxidize Cu(I) to the more soluble form Cu(II). One of the most important, and by far the best characterized members of the bioleaching microbial consortia is Acidithiobacillus ferrooxidans (Brandl, 2008, Edwards, 1990, Ingledew, 1982, Rawlings, 2002). Formerly known as Thiobacillus ferrooxidans, it is a gram-negative, highly acidophilic, chemolithoautotrophic γ-proteobacterium (Rohwerder et al., 2003). For bioleaching, this organism is particularly important, since it drives Fe(II) oxidation thus allowing the copper solubilization for further recovery by physic-chemical means (Valdes et al., 2008).
Beyond its biomining capabilities, A. ferrooxidans offers exceptional opportunities to study life under extreme conditions. It typically grows at an external pH of 2 or lower using the oxidation of ferrous ions (fe2) by oxygen (o2), producing ferric ions (fe3) and water (h2o), while fixing carbon dioxide (co2) from the environment. It can also obtain energy by the oxidation of Reduced Inorganic Sulfur Compounds (RISCs), hydrogen (Drobner et al., 1990), and formate (Pronk et al., 1991). A. ferrooxidans has the potential to drive respiration by directly transferring electrons from electrodes to the microorganism (Carbajosa et al., 2010, Li et al., 2010). Extracellular polymeric substances (EPS) production in A. ferrooxidans is crucial for the bioleaching process. It has been demonstrated that EPS activation in A. ferrooxidans, significantly increases the pyrite bioleaching capacity (Gehrke et al., 1998). EPS aids the process by mediating the bacterial adhesion to the sulfide mineral surface, and by concentrating ferric ion in the mineral-microorganism interface by complexation with uronic acids or the EPS residues, allowing the oxidative attack on the sulfur to take place (Sand and Gehrke, 2006). Due to the lack of well-developed systems for genetic manipulations, the study and exploration of the molecular biology and physiology of A. ferrooxidans has proven to be deficient. In terms of the behavior of the complete system, different aspects of metabolism, such as, iron oxidation, co2 uptake and fixation, and the anaerobic metabolism of sulfur-coupled iron reduction remain little described. Furthermore, this organism has often proved to be the source of some confusion, because it requires understanding of the consequences of both growing at very acidic external pH and of using a relatively weak reductant (ferrous iron) as the sole source of electrons for respiration (Ferguson and Ingledew, 2008). Several aspects regarding its energetic metabolism remain weakly described in quantitative terms, such as, how it balances the use of iron as both a micronutrient and as a required energy source and how proton-driven membrane transport and energy processes function in face of a proton motive force across the inner membrane that is several orders of magnitude higher, and how the large pH gradient is maintained across the cytoplasmic membrane of A. ferrooxidans (Ferguson and Ingledew, 2008).
Based on the complete genome sequence of A. ferrooxidans (Valdes et al., 2008), several studies have provided insights into its physiological properties, including the most relevant parts of the metabolism (Esparza et al., 2010, Ferguson and Ingledew, 2008, Osorio et al., 2013, Valdes et al., 2008). However, from this knowledge it is not possible to either predict or quantitatively describe the physiological outcome from the annotated sequence alone. Only small scale metabolic models for A. ferrooxidans (Hold et al., 2009, Sepúlveda et al., 2011), have been proven successful on describing the main aspects of its metabolism. However, these models lack the ability to capture the complex physiological characteristics, behavior and metabolic capabilities of the cell as a whole integrated system. In order to overcome this difficulty and fully explore metabolic genotype-phenotype relationships, implementation of constraint-based reconstruction and analysis (COBRA) methods for A. ferrooxidans have been employed. The “cornerstone” of this method is represented by the genome-scale network reconstruction (GENRE) (Thiele and Palsson, 2010) that is built systematically using genome annotation, “omics” data sets and legacy knowledge. Genes, proteins, reactions and metabolites that participate in the metabolism are identified, categorized and systematically interconnected, enabling a mechanistic description of metabolic physiology. A GENRE combined with constraints-based methods can be used to formulate mechanistic predictions of metabolic physiology that can be used in a prospective manner to elucidate new biological knowledge and understanding, as well as design and engineer the cellular metabolism (McCloskey et al., 2013). Several workflows have been implemented to predict metabolic phenotypes, by integrating high-throughput data sets with COBRA methods (Bordbar et al., 2014).
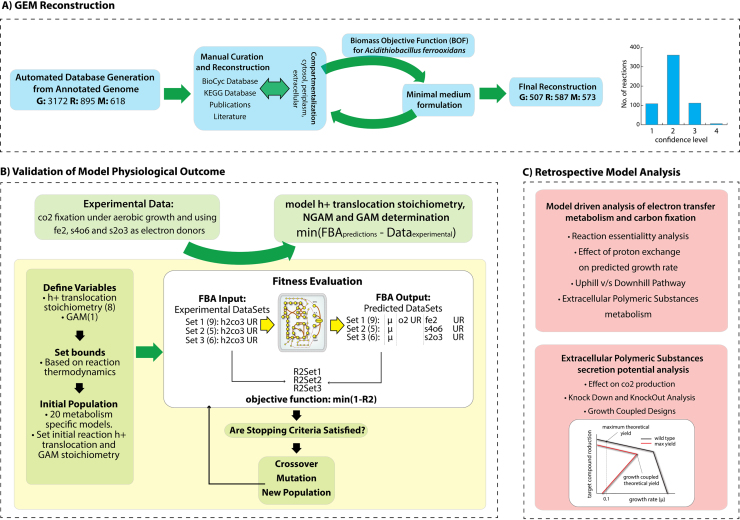
In this work, a GENRE was reconstructed, validated and analyzed to better understand key metabolic capabilities of A. ferrooxidans ATCC 23270 (see Fig. 1). A systematic workflow for the elucidation of proton translocation stoichiometry of key enzymes based on physiological data was implemented under three different aerobic chemolithoautotrophic conditions: using ferrous ion, tetrathionate and thiosulfate as electron donors. The electron transport system and central carbon metabolism was characterized and studied. Furthermore, in order to determine the potential production of extracellular polymeric substances in A. ferrooxidans, a retrospective model-driven analysis was performed.
Fig. 1.
General workflow to generate, validate and further analysis of iMC507: (A) the workflow detailing the iterative model building procedure used to generate iMC507. The reconstruction process was initiated based on the annotated genome generated by PathoLogic. Manual curation and reconstruction was performed by using the Insilico Biotechnology software aided by the KEGG and MetaCyc databases. Publications and literature sources were used to refine the network content, assigning a specific confidence score to each reaction. The reconstructed network in conjunction with the BOF, were used to formulate a minimum medium for the three different metabolism studied. (B) Proton translocation for 8 different membrane reaction and GAM stoichiometry were estimated using a genetic algorithm. Experimental data for growth under three different electron donors and FBA was used to decipher the model parameters that best represent cell behavior. Based on these results the iMC507 network was validated with physiological data for growth under ferrous ion, tetrathionate and thiosulfate. (C) To study the most relevant aspects of the electron transfer metabolism and carbon fixation, a retrospective model analysis was performed by using iMC507. Further analysis on EPS secretion potential for growth-coupled production through knock-outs was performed.
2. Methods
2.1. Network reconstruction process
The reconstruction software Insilico Discovery™, version 3.3 (Insilico Biotechnology AG, Stuttgart, Germany), was used to build and curate the first draft of the A. ferrooxidans genome-scale network. First, the specific Pathway/Genome Database (PGDB) for A. ferrooxidans ATCC 23270, version 14.1, was downloaded from BioCyc (Caspi et al., 2012), where based on the annotated genome sequence, the PathoLogic program (Dale et al., 2010, Karp et al., 2010) automatically infers metabolic pathways, assigns enzymes to reactions they catalyze, and infers transport reactions among others capabilities. The draft genome accounts for 3217 protein-coding genes, of which 64% were assigned a putative function (Valdes et al., 2008). Next, in order to connect the database with the reconstruction platform, four different files were generated by Insilico Biotechnology AG (Stuttgart, Germany) from the original A. ferrooxidans ATCC 23270 PGDB. The generated files specify different kinds of metabolic interaction levels, such as, metabolites, reactions, genes and operons. At the beginning, most gene-protein-reaction (GPR) association assignments were made from the annotated genome and the model was reconstructed on a pathway basis manually. In general, the reconstruction process was implemented to minimize the number of grouped, or lumped, reactions in the network reconstruction. During the manual curation process online database, KEGG (Kanehisa et al., 2006), MetaCyc (Caspi et al., 2012) and BRENDA (Schomburg et al., 2002), were extensively used. The ORFs that encode the proteins included were integrated into the GPRs associations for the reactions in which they participate. GPR associations were also determined directly from biochemical evidence presented in journal publications and reviews. Transport reactions were added to the network from the genome annotation or from physiological data. All reactions included in the model were both elementally and charge balanced. Reaction reversibility was determined from thermodynamic considerations. Confidence scores were assigned for each reaction, based on the available evidence of its presence in the model (Thiele and Palsson, 2010). Biochemically characterized enzymes received a confidence score of 4. If genetic knockout information or physiological evidence was available, a score of 3 was assigned. A score of 2 was assigned to reactions for which indirect evidence or sequence homology information was available. During gap-filling and evaluation of the network functionality some reactions were added with a confidence score of 1. It is worthwhile to mention that throughout the entire manuscript, metabolites and reactions are named after the corresponding model abbreviations (see Supplementary Tables 1 and 2).
2.2. Generation of the Biomass Objective Function (BOF)
In order to simulate a cell that strives to maximize biomass production from available media substrates, a detailed and precise biomass reaction is needed for realistic metabolic network analysis. The BOF is a linear equation consisting of the fractional molar amounts of metabolites that constitute the dry weight content of the cell along with a growth associated maintenance (GAM) reaction to account for non-metabolic growth activity (e.g., energy required for macromolecular synthesis), represented in the BOF as ATP hydrolysis reaction. In addition to the BOF, a non-growth associated maintenance (NGAM) reaction (which is an independent ATP hydrolysis reaction) was used as an energy “drain” on the system during the linear programming calculations, and accounts for non-growth cellular activities (e.g., turgor pressure). The A. ferrooxidans BOF was formulated according to the procedures described elsewhere (Thiele and Palsson, 2010). Biosynthetic macromolecule fractional content was obtained from experimentally reported values when possible, if not, the corresponding fractional content was estimated as reported in iAF1260 (Feist et al., 2007). Experimental values for proteins (60%) (Kuenen, 1979) and inorganic ions (0.3%) (Sublette, 1988) were found in the literature. Assuming a linear behavior between genome size and DNA cell fractional content in different organisms, the corresponding DNA fractional content for A. ferrooxidans was estimated from a correlation specified in Supplementary Table 4. RNA fractional content was estimated according to the genome composition. The remaining glycogen, murein, LPS, phospholipids and soluble pool content was extrapolated from the BOF formulated for Escherichia coli (Feist et al., 2007). This approach has been shown to be a valid assumption when comparing the same gram-negative cell morphology (Feist et al., 2014). It is worthwhile mentioning that from a mathematical perspective, growth rate and unmeasured uptake rate predictions are relatively insensitive to biomass macromolecular weight fraction variations (Feist et al., 2007).
The relative fraction of amino acids was obtained from previous studies (Sublette, 1988). The nucleotide content for DNA and RNA was estimated based on the genome composition. The relative fraction of fatty acids (Mykytczuk et al., 2010), phospholipids (Shively and Benson, 1967), LPS (Mayer et al., 1989) and inorganic ions (Sublette, 1988) was taken from experimental data reported for Acidithiobacillus. EPS production was modeled as a biomass independent reaction, and fractional precursor content was obtained from experimental results (Harneit et al., 2006). The GAM reaction stoichiometry and the NGAM flux were estimated by using a genetic algorithm and previously reported experimental results. A detailed description of the iMC507's biomass reactions is shown in Supplementary Tables 3 and 4.
2.3. Modeling simulations
In order to mathematically represent the reconstructed metabolic network, a stoichiometric matrix (Smxn) was generated, where m is the number of metabolites and n is the number of reactions. Each entry Sij, represents the stoichiometric coefficient for the corresponding ith metabolite in the jth reaction. More details can be found elsewhere (Thiele and Palsson, 2010). Flux Balance Analysis (FBA) (Orth et al., 2010) was utilized for predicting growth and analyzing the reaction flux (vj) through the metabolic network. Based on the stoichiometry matrix, a linear programming (LP) problem is solved by maximizing the BOF reaction (vBOF) under steady-state criteria. Additionally, reactions are constrained by setting an upper (ub) and lower bound (lb), which define the maximum and minimum allowable flux of the reactions. The general LP problem can be represented as:
For reversible reactions and for reactions containing metabolites present in the extracellular space the lb and ub constraints were set to −1000 and 1000, respectively. For irreversible reactions and reactions containing metabolites that are not in the medium, meaning that the metabolite could leave the cell but not enter the system, lb and ub constraints were set to 0 and 1000 respectively. Based on the inorganic electron donor, three different A. ferrooxidans's aerobic chemolithoautotrophic metabolisms were studied. Specifically for ferrous ion, tetrathionate and thiosulfate, by using as a unique carbon source carbonic acid (h2co3). Flux variability analysis (FVA) was used to find the minimum and maximum flux for reactions in the network, while maintaining a predefined flux state of the network. More details about the FVA method can be found elsewhere (Mahadevan and Schilling, 2003). In this work, FVA was used to find the minimum and maximum flux through each reaction while supporting 100% of the maximal growth rate. Linear programming calculations were performed using Insilico Discovery™, version 3.3 (Insilico Biotechnology AG, Stuttgart, Germany) and the MATLAB® version 8.1.0.604 (The MathWorks Inc., Natick, MA) linked to the COBRA Toolbox 2.0 (Schellenberger et al., 2011). The linear programming package GUROBI version 5.5.0 (Gurobi Optimization Inc., Houston, TX) was used as a solver.
2.4. Minimal medium formulation
A synthetic minimal medium was determined based on the biomass composition specifically determined in the BOF. Biomass constituents were grouped in two different sets. The first group corresponds to inorganic ions. By assuming that inorganic ions do not impose a growth restriction, the corresponding exchange reactions were allowed to freely enter and leave the network by setting lb and ub to −1000 and 1000, respectively. For carbon based biomass constituents, a manual gap filling procedure was performed. Biomass components were sequentially added to the BOF individually and further FBA was performed for BOF maximization. If the optimization leads to a positive flux through the biomass reaction, a subsequent component was added to the BOF and simulation was re-ran. For optimizations resulted in no flux through the biomass reaction, the network was updated by adding the needed reactions able to sustain growth. This process was repeated until all biomass constituents were added to the BOF. Simulations were performed under aerobic chemolithoautotrophic conditions (external oxygen reaction exchange lb and ub were set to −1000 and 1000) and using h2co3 as a unique carbon source for the three major electron donors: fe2, tetrathionate (ttton) and thiosulfate (tsul). In order to avoid operation of the rusticyanin complex for tetrathionate and thiosulfate simulations, CYT2 reaction bounds were set to 0. GAM and NGAM were not considered for the gap filling procedure and determination of minimal media. Three different minimal synthetic media were generated according to the specific electron donor related to the corresponding metabolism (Supplementary Table 1). Metabolism specific simulations were performed by using the previously determined media conditions, which define A. ferrooxidans aerobic chemolithoautotrophic growth, using 3 three different electron donors. The metabolisms studied were abbreviated based according to the specific electron donor: specifically for aerobic ferrous ion (FIM), tetrathionate (TTM) and thiosulfate (TSM) metabolism.
2.5. Genetic algorithm for proton translocation stoichiometry estimation and sensitivity analysis
The proton translocation stoichiometry for ATPS5rpp (ATP synthase), NADHI (NADH dehydrogenase, ubiquinone-8), CYTAA32 (cytochrome c oxidase, aa3-type), CYTAA31 (cytochrome c oxidase, aa3type), CYTRED (bc1 complex), CYTBC1 (cytochrome oxidase bc), CYTBO3 (cytochrome oxidase bo3, ubiquinol-8), CYTBD (cytochrome oxidase bd, ubiquinol-8) (see Fig. 3), the GAM reaction stoichiometry and NGAM flux were estimated based on the analysis outlined in Fig. 1B. A genetic algorithm was implemented in order to adjust the model parameters based on previously reported experimental results (Boon, 1996, Eccleston and Kelly, 1978, Hold et al., 2009) using three different electron donors (i.e. fe2, ttton, and tsul) under aerobic chemolithoautotrophic metabolic conditions. In total 20 different experimental points specifying the growth rate, h2co3 uptake rate, o2 uptake rate (only for FIM), and the corresponding final electron donor (fe2, ttton or tsul) were used for simulations (see Supplementary Table 5). For the proton translocation reactions, upper and lower bounds on the number of allowable protons able to cross the periplasmic membrane were estimated. These bounds were estimated based on Mitchell's chemiosmotic theory and also obtained from experimental data (Cox et al., 1979) (see Supplementary Note 1). Bounds on GAM were set according to reasonable reported values regarding similar analysis. Furthermore, to reduce the number of unknown variables, the NGAM was set as 2.5% of the GAM (Feist et al., 2006). In order to initialize the genetic algorithm, first, 20 different metabolism-specific models were generated by setting the corresponding metabolism constraints and the specific experimental h2co3 uptake rate. Second, the corresponding reaction to evaluate, and initial values for proton translocation and GAM reactions stoichiometry were set for each one of the 20 models. Initial parameter lower and upper bounds for genetic algorithm initialization were set randomly inbetween the corresponding minimum lower and maximum upper allowable bound space. Third, genetic algorithm performance variables were defined, such as: crossover function, mutation function and stopping criteria. After models and genetic algorithm initialization, the algorithm started. For each round, FBA was utilized for optimizing growth and quantitative phenotypic evaluation. Fitness was calculated by comparing FBA predictions with previously reported experimental results (Supplementary Table 5), specifically for o2, fe2, ttton and tsul uptake rate and growth rate, for each one of the 20 models. The coefficient of determination (R2) was evaluated for each dataset (R2Set1 (fe2), R2Set2 (ttton) and R2Set3 (tsul)), and the average (R2m) was used for fitness evaluation. The objective function for the genetic algorithm was the minimization of (1-R2m). MATLAB® version 8.1.0.604 (The MathWorks Inc., Natick, MA) genetic algorithm was used for all simulations. Once proton translocation, GAM and NGAM stoichiometry were estimated, a metabolism specific sensitivity analysis for each estimated variable was performed. For each metabolism, the estimated parameters were independently varied from the optimal values. According to the variation, FBA was used to calculate the corresponding R2Set1, R2Set2 and R2Set3 deviation, observed from the FBA predictions and experimental results.
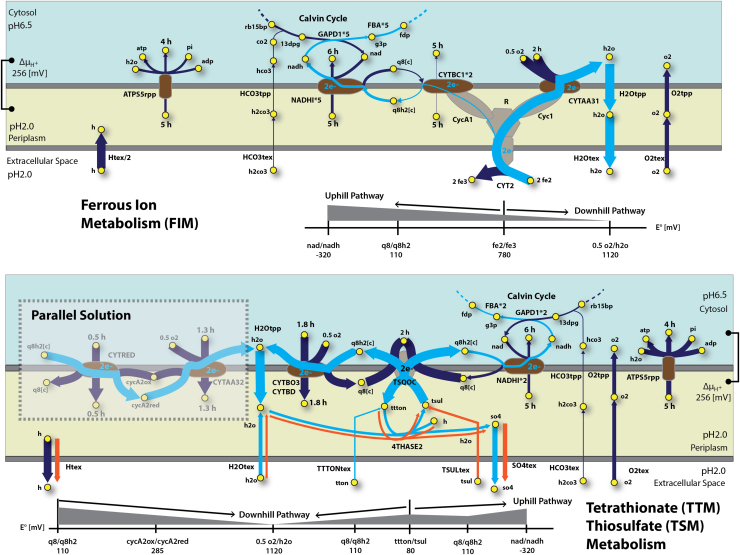
Fig. 3.
Electron transfer flux distribution predicted for ferrous ion, tetrathionate and thiosulfate metabolism: A flux map illustrating the electron transfer metabolism for ferrous ion (FIM), tetrathionate (TTM) and thiosulfate (TSM) metabolism. Reactions are specified in capital letters, while metabolites in lowercase letters and yellow circles. Arrows indicate the direction of enzymatic activity and the arrow thicknesses are proportional to the flux through each reaction (a thicker arrow has a larger flux). Light blue arrows represent the electron flow in the network. Visually undetectable fluxes for being so small were increased by a corresponding factor specified next to the corresponding reaction in the map. The diagram shows the energy-conserving ion translocating reactions, each labeled with the stoichiometry of the translocated ion. Proton translocation stoichiometry predicted reactions are shown in brown. The different colored regions correspond to the tree modeled spaces: cytoplasm (light blue), periplasm (yellow) and the extracellular space (white). For each network the corresponding standard reduction potential (in mV) associated with a specific transformation was specified. The uptake of h2co3 for each simulation was constraint at 2 mmol/g DW/h, and the corresponding electron donor was allowed to freely enter the system. FBA was performed for FIM, TTM and TSM. Due to flux similarities results, TTM and TSM were plotted together. For TTM, reactions, metabolites and fluxes we potted according to the previous description. In the case of TSM, reaction fluxes different from the ones obtain in TTM, were plotted in orange. For TTM and TSM, an additional flux distribution, called as parallel solution, was plotted. This solution was obtained when constraining CYTBO3 and CYTBD ub and lb to 0. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
2.6. Reaction essentiality analysis
Reaction essentiality analysis was performed for FIM, TTM and TSM. The analysis consists of the sequential independent reaction removal from the model, followed by FBA simulations for growth maximization. For each simulation the h2co3 uptake rate was set to 2.34 mmol/g DW/h. Reactions were removed by setting the corresponding lb and ub to 0. Simulations predicting growth rates higher than 0 were defined as non-lethal reaction knock-outs. Lethal knock-outs were clustered in terms of subsystems and further analysis was performed.
2.7. Reaction knock-down simulations
Reaction knock-down simulations were performed only for aerobic chemolithoautotrophic FIM. The h2co3 uptake rate for each simulation was set to 2 mmol/g DW/h. fe2 and o2 were freely allowed to enter and leave the cell. In order to analyze the growth-rate and EPS production due to metabolic co2 production and independent reaction flux constriction, 12 different simulations were calculated for each of the reactions in the model. Those conditions where defined by two parameters. First, each reaction was allowed to proceed by constraining the optimal flux at 0%, 25%, 50%, 75% and 100%. When constraining reaction flux at 0%, all reactions were freely allowed to proceed according to the corresponding directionality. On the contrary, reaction fluxes constrained at 100% mean a knock-out. The others constraining percentages represent the knock-down ratio from the optimum. Additionally, a co2 transport was incorporated and constrained in the same manner. By constraining this transport reaction, metabolically produced co2 was allowed to leave the cell at 0%, 50% and 100%. FBA was used to maximize BOF, and for each reaction and condition, the corresponding growth rate and EPS production rate were saved.
2.8. Reaction knock-out simulations
Based on the genome-scale reconstruction, model-driven growth-coupled designs through reactions knock-outs for EPS production were calculated. These growth-coupled designs could be difficult to achieve and may require multiple knock-outs. A number of algorithms, such as OptKnock (Burgard et al., 2003), OptGene (Patil et al., 2005), RobustKnock (Tepper and Shlomi, 2010), and GDLS (Lun et al., 2009), have been proposed for designing production strains through gene knock-outs. Still, the search for knock-out phenotypes is computationally extensive, since the solution of one or more mixed-integer linear problems (MILP) are involved. This means that the time taken to solve MILPs arising from network reconstructions becomes prohibitive. Instead of formulating a MILP, an exhaustive search over all single, double, and triple knockout mutants was performed. A major benefit of this strategy is that it finds all growth-coupled designs instead of a single mutant returned by most of the MILP based algorithms. Reaction knock-out simulations were performed only for aerobic chemolithoautotrophic ferrous ion metabolism. The h2co3 uptake rate for each simulation was set to 2.34 mmol/g DW/h. fe2 and o2 were freely allowed to enter and leave the cell. Furthermore, a metabolic co2 transport reaction step was added to the model. Single, double and triple knock-outs were simulated. First, in order to decrease the number of simulations, a subset of reactions was determined. Specifically, all non-lethal reactions were taken into account for the simulations. In total 180 reactions were used for this analysis. Second, from this subset of reactions, all possible combination knock-outs for single, double, and triple deletions were determined. Third, for each deletion combination, FBA was used to optimize growth. Finally, in order to obtain EPS production associated to growth, knock-out combinations associated with phenotypes able to grow and simultaneously produced EPS were saved. Knock-out reactions were simulated by setting the ub and lb to 0.
3. Results
The results are presented following the scheme shown in Fig. 1. First, a description on the content included in the genome-scale metabolic reconstruction of A. ferrooxidans and then conversion into a computational model is described. Second, by using a genetic algorithm in conjunction with experimental data, the GENRE was validated and proton translocation stoichiometry for key metabolic reactions was estimated. Third, a retrospective model-driven analysis describing A. ferrooxidans key metabolic capabilities and potential applications for EPS production was studied.
3.1. Acidithiobacillus ferrooxidans ATCC 23270 genome-scale metabolic network reconstruction and unique metabolic capabilities
A genome-scale metabolic reconstruction of A. ferrooxidans ATCC 23270, iMC507, was generated by performing a bottom-up reconstruction approach (Fig. 1A). The final reconstruction captures all major known metabolic pathways and contained 507 genes, 587 metabolic and transport reactions, and 573 nonunique metabolites, which were distributed over 42 subsystems and three different cellular compartments: extracellular, periplasm and cytoplasm. In general, the reconstruction accounts for 16%, 69%, and 92% of the initial automated database genes, reactions, and metabolites respectively. Reactions were subdivided into 13 high-level functional categories based on the major metabolic roles of the cell. The largest number of reactions was involved in the biosynthesis of amino acids, cofactors and prosthetic groups. In total, 79% of the reactions have a GPR association (see Supplementary Fig. 2A). The transport functional category shows the lowest number of GPR associated reactions, only 24%. Another 50% of transport reactions were associated with outer membrane porin transport, no GPR association was assigned. The high number of transport reactions with no gene assignments, points to the fact that further work is needed to characterize the biochemical foundations involved in the transport of molecules in A. ferrooxidans. Almost all genes were specific for each functional category. In the case of metabolites, this is completely the opposite (see Supplementary Fig. 2B-C), showing that almost all metabolites are shared among the different functional categories.
Since A. ferrooxidans has the ability to grow under extremely low pH and on sources of electrons that yield scarcely sufficient energy for ATP synthesis and other endergonic processes vital for the cell (Ferguson and Ingledew, 2008), a detailed characterization of the electron and hydrogen transfer pathways through all three cellular compartments was crucial for enabling the systems analysis of the energetic processes and understanding it's unique capabilities. An extensive effort was made to construct the most reliable model that exceeds previously published work in detail and coverage (Hold et al., 2009). Together with reconstructing the main carbon and precursor metabolism for biomass, the reconstruction process was focused on describing the energy metabolism regarding three different electron donors: ferrous ion (FIM), tetrathionate (TTM) and thiosulfate (TSM) metabolism. All major metabolic subsystems are outlined in Supplementary Fig. 1, specifying the reaction name and the corresponding gene association. Furthermore, a detailed characterization and analysis of the energy metabolism can be found in the Supplementary Note 2.
The number of essential reactions for growth under aerobic FIM, TSM and TTM was determined using FBA (see Section 2). The results of such calculations are presented in Supplementary Fig. 6. These results have to be interpreted with caution, since calculations were based on the assumption that all enzymes are expressed. Transcriptomics or proteomic data would considerably improve the results by constraining the internal fluxes. Approximately 68% of all possible single reaction deletions would be lethal for the organism under aerobic FIM, TTM and TSM. In general, when comparing the percentage of knock-out reactions non-essential for growth, for all studied subsystems, no considerable fluctuations were observed under FIM, TSM, and TTM. The only substantial difference between these three metabolisms on reaction essentiality was observed for oxidative phosphorylation and sulfur metabolism, where in both cases the number of knock-out reactions being non-essential for growth was higher for TSM and TTM compared to FIM. Also a higher percentage of knock-out reactions non-essential for growth are associated to Carbon and Energy metabolism compared to amino acid and related molecules metabolism. This shows how flexible the carbon and energy metabolism and how rigid for amino acid and related molecules metabolism are in A. ferrooxidans due to knock-out perturbations. It is interesting to note in Supplementary Fig. 6 that the majority of the reactions in the sulfur metabolism are non-essential during growth with all electron donors tested. This behavior was expected when using fe2, where the sulfur and iron metabolism can work independently from each other. Nevertheless, when using ttton and tsul as a sole electron source, approximately 80% of the reactions were assigned as non-essential. Most of the reactions assigned to the sulfur metabolism are specifically associated to the RISC metabolism. This metabolism is responsible for oxidizing a range of different sulfur compounds through many different metabolic routes, thus providing the cell with sufficient electrons for growth. Based on the latter, the network shows a flexible behavior when simulating growth by using RISCs as a sole electron source. This allows re-routing the fluxes when reaction deletions associated to the sulfur metabolites are performed, and explains the high percentage of non-essential reactions in the sulfur metabolism. When using ttton and tsul as electron donors, approximately 80% of the reactions in the oxidative phosphorylation where assigned as non-essential for growth. After analyzing the reactions involved in this metabolism, approximately 50% correspond exclusively to the fe2 metabolism. This set of reactions under which RISC conditions become non-essential, explains the high percentage on non-essential reactions in the oxidative phosphorylation for ttton and tsul.
It should be noted that the macromolecular fraction values in the BOF for glycogen, murein, LPS, phospholipids and the soluble pool were extrapolated from iAF1260. These similarities might lead to redundancies when comparing gene essentiality results. These redundancies reflect the similarities in the BOF between both species, which in this case where assumed to be valid specifically for glycogen, murein, LPS and phospholipid metabolism, due to the similar gram-negative cell morphology. On the other hand, reaction essentiality redundancies for reactions contained within the pathways producing soluble pools, should be carefully considered, since no evidence regarding the soluble pool content in A. ferrooxidans was found.
Based on the available experimental evidence, a confidence score was assigned to each reaction in the metabolic network. On completion, the GENRE had an overall average confidence of 2.02. In fact, 19% of all reactions in A. ferrooxidans included in iMC507 have been very well or well-studied, while 58% were fundamentally based on the genome annotation and 18% of all reactions reflects that no evidence is available, but the reaction is required for modeling. Future research efforts should be directed towards this latter group, which is described in Supplementary Fig. 3. Furthermore, in order to increase the accuracy and utility of the model, BOF should be reevaluated in future efforts, since some of the precursors were determined based on previous work non-related to A. ferrooxidans genome-scale models, specifically iAF1260. According to the latter, problems might arise when conditionally essential metabolites are inappropriately included in the BOF, leading to false reaction essentiality and growth-coupled design predictions.
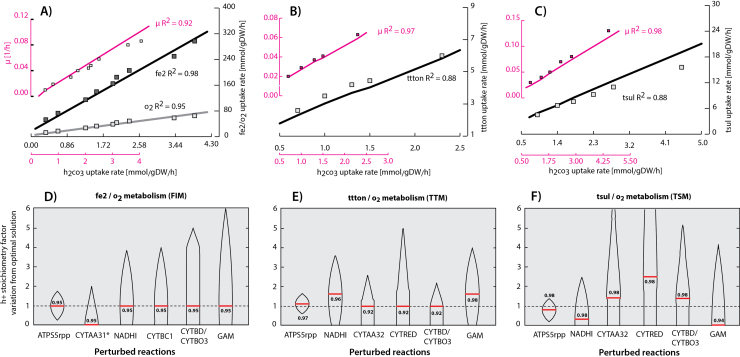
3.2. GENRE proton translocation stoichiometry estimation and network validation
A. ferrooxidans's proton translocation metabolism and transmembrane enzyme configuration, has been studied during many years. Nevertheless, accurate quantitative estimation of the parameters involved in the proton translocation process has not been accurately described. In these work, a model driven systems approach was implemented and used for estimating the previously described parameters (see Section 2). Based on the genetic algorithm results and further analysis (see Supplementary Note 3), proton translocation stoichiometry for ATPS5rpp, CYTAA31, NADHI, CYTBC1, CYTAA32, CYTRED, CYTBO3 and CYTBD were set to 5, 0, 5, 5, 1.3, 0.5, 1.8 and 1.8 [h+/2e−] respectively. GAM stoichiometry was set to 139 and NGAM to 3.48. Based on this solution, for all 7 adjusted curves (Fig. 2A, B and C), an average R2 equal to 0.92 was calculated. Under the simulated conditions, iMC507 exhibited a growth rate, oxygen uptake rate and the corresponding electron donor uptake rate practically identical to the experimental results. This result shows the predictive potential and preciseness of COBRA methods for simulation of cellular outcomes by using additional constraints. In order to analyze the predicted proton translocation and GAM stoichiometry sensitivity on the specific R2Set (1:fe2, 2:ttton and 3:tsul), independent perturbations were imposed for each parameter as a factor from the local optimal solution and a further R2Set was calculated by using FBA. R2Set variations were represented as a violin plot along the y-axis, and the highest calculated value for each studied parameter was reported and represented as red lines. Since CYTAA31 proton translocation stoichiometry was calculated as 0, variations were performed not as a factor from the optimal solution, but instead the direct proton translocation value corresponding to the y-axis was set for overall R2Set sensitivity analysis (Fig. 2D). The most sensitive and constrained reaction in the metabolism is ATPS5rpp. Under FIM (Fig. 2D), when setting the proton translocation and atp consumption stoichiometry in the GAM reaction to 0, in the y-axis, no significant changes in the overall R2 were observed for NADHI, CYTBC1, CYTBD/CYTBO3 and GAM. Furthermore for these reactions, when increasing the parameters by approximately 100% (factor equal to 2), no considerable decrease in the R2 was observed. Specifically for reactions NADHI and CYTBC1, when increasing the number of translocated protons, FBA predictions shows an increase in reaction CYTAA31 flux, thus calculating ferrous ion and oxygen uptake rates greater than the experimental results, displacing the R2Set ratio to 0 (see Fig. 3 for flux visualization). For reaction CYTBD/CYTBO3, proton translocation was modeled leaving the cytoplasm. When the proton translocation stoichiometry is increased, FBA predicts that less oxygen and ferrous ions are needed to balance the cytoplasmic hydrogen in reaction CYTAA31. When GAM is increased, FBA predicted that ATPS5rpp needs to generate more ATP to fulfill the GAM and NGAM demand. Similarly, as the generation of ATP by ATPS5rpp is associated with a net flux of protons inside the cell, CYTAA31 needs to balance the cytoplasmic protons by increasing oxygen and ferrous ion uptake rates, deviating the R2Set ratio. Due to the existence of uphill and downhill pathways, electrons can be diverted for biomass production (through reaction CYTBC1) and for balancing cytoplasmic protons (through reaction CYTAA31). For TTM and TSM, the same kind of behavior regarding R2Set deviations and cytoplasmic proton balance was observed. But, different reactions were involved in the process. Electrons for biomass production are taken by the NADHI reaction, and electrons for balancing the cytoplasmic protons are used in CYTBD/CYTBO3 or CYTAA32 reactions. It is worthwhile drawing attention to the fact that for TSM (Fig. 2F), better solutions for proton translocation and GAM stoichiometry values, compared to TTM (Fig. 2E) were found, specifically for ATPS5rpp, NADHI, CYTAA32, CYTRED, and GAM. This is shown by the displacement of the maximum R2Set value from the line equal to 1 (no changes from the local optimum). Furthermore, TSM showed more degrees of freedom in comparison to TTM, since R2Set solutions might be achieved at higher perturbations. It is worthwhile to notice that all of these predictions were performed assuming an external pH equal to 2. Since thermodynamic bound on proton translocation stoichiometry changes depending on the pH difference between the cytoplasm and the periplasm, different results might be obtained when changing the pH conditions.
Fig. 2.
Parameter estimation and validation of the carbon fixation and electron donor pathways results: based on the genetic algorithm model parameter estimation results, three graphs representing the experimental phenotypic data (represented in squares) and the model-predicted growth conditions (represented in lines) on ferrous ion (A), tetrathionate (B) and thiosulfate (C) as electron donors and using oxygen as an electron acceptor are shown. The corresponding R2Set (1:fe2, 2:ttton and 3:tsul) for each predicted/experimental dataset pair are shown. A R2Set sensitivity analysis for ferrous ion (D), tetrathionate (E) and thiosulfate (F) metabolism was performed. Proton translocation predicted stoichiometry values were varied along the y axis and the corresponding R2Set variation from the optimum was calculated and plotted each one of the proton translocated stoichiometry predicted reactions as a violin graph. In this case R2Set represents the prediction error for each studied metabolism independently. The maximum calculated R2Set due to parameter variations was reported and represented as a red line. For sake of simplicity GAM denotes the stoichiometry on the atp reaction consumption in the BOF due to growth associated maintenance. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.3. iMC507 model-driven analysis of electron transfer and central carbon metabolism
The electron transfer metabolism in A. ferrooxidans is mostly distributed in the periplasmic membrane. Depending on the electron donor, different reactions might be involved in the electron transfer process. In this work FIM, TTM and TSM were studied. As shown in Fig. 2A, B and C, the experimentally measured h2co3 uptake rate range varies depending on the final electron donor. When inspecting dependencies between the carbon uptake rates with the rest of the measured variables, it is clearly shown that all of them correlated linearly. In order to compare the flux states under different electron acceptors, and normalize the h2co3 uptake for each studied condition, FBA was performed by setting the h2co3 uptake rate at 2 mmol/g DW/h, and optimizing the BOF. The corresponding electron donor was allowed to freely enter the system. It is reasonable to use this specific value of carbon uptake rate for FBA simulations since it lies in the feasible uptake rate range for all electron donor conditions.
In the case of FIM, results indicated (Fig. 3) that the flow of electrons is split at the rusticyanin branch point, which has actually been suggested as the balancing point for nadh and ATP requirements in the cell by adjusting the flow of electrons (Elbehti et al., 2000). Most of the electron flow from ferrous ion goes to oxygen via cytochrome oxidase (CYTAA31) which generates a proton motive force by consuming hydrogen for the oxygen reduction. Specifically 3% of the electrons goes to the formation of nadh by the CYTBC1 and NADHI reactions. nadh is further utilized by GAPD1 (i.e. in the Calvin cycle) for carbon fixation. This result is consistent with a previous report (Ferguson and Ingledew, 2008) that shows that less than 5% of electrons go to NADHI. Furthermore, for simulations under oxygen limitation (Supplementary Fig. 11), this behavior remains valid. It has been demonstrated that before ferric compounds start precipitating in the form of jarosite, growth on ferrous ion tends to increase the culture pH (Qiu et al., 2005, Yarzabal et al., 2004). This ferrous ion oxidation might be due to A. ferrooxidans metabolism. In fact, a net inward proton flux through the extracellular membrane was calculated. Predictions show that the majority of the inward proton flux into the cytoplasm goes through the ATPS5rpp, specifically 67% of all protons entering the periplasm. The remaining proton flux is mainly distributed in equal amounts between NADHI and CYTBC1. In the cytoplasm, approximately 50% of all incoming protons are used for oxygen reduction through CYTAA31 and the rest mainly for anabolic processes. Furthermore, as shown in Supplementary Fig. 7, growth is only achievable under alkalization conditions. A sensitivity analysis using FVA also supports this fact (Supplementary Fig. 10, reaction Htex), where no proton outward flux was calculated.
For TTM and TSM a completely different set of periplasmic membranes for transferring electrons into the Calvin cycle and for proton motive force generation were found to be active in the metabolic network. TTM and TSM flux distributions have been outlined in Fig. 3. In these cases, ttton is hydrolyzed by tetrathionate hydrolase (4THASE2) generating sulfate (so4), which is excreted by the system, and tsul which is used by the thiosulfate-quinone oxireductase complex (TSQOC) to transfer electrons into the quinone pool. The 4THASE2/TSQOC cycle plays an important role at the beginning of the electron transfer system by first incorporating the electrons coming from tsul into the metabolism, and second transferring electrons from water to ttton, recycling the water generated by either CYTBO3/CYTBD or CYTRED in conjunction with CYTAA32. Interestingly, discrepancies showed that TSM produces less protons and sulfate through reactions Htex and SO4tex, specifically 40% and 13% less than TTM, respectively. Furthermore, based on a sulfur molar basis, approximately 12% less sulfur in the form of tsul is needed to sustain growth for TSM. This demonstrates that in terms of electron transfer efficiency TSM is more efficient than TTM. This is due to the amount of electrons that each electron donor is able to give on a sulfur molar basis, where tsul showed twice as many of electrons per sulfur. Furthermore, for TTM, water is needed to overcome the electron deficiency in order to sustain growth. Specifically for TTM, 70% of all incoming electrons from water goes to TSQCO via thiosulfate, the rest are excreted in the form of sulfate. Basically the tetrathionate/thiosulfate/sulfate complex worked as a shuttle electron transfer system to deliver electrons from water to the quinone pool. At this point, the rest of the network behaves similarly for TSM and TTM, where electrons are diverged analogous to the FIM, to a “RISC uphill pathway” through NADHI, and to a “RISC downhill pathway” through CYTBO3/CYTBD or CYTRED in conjunction with CYTAA32. Due to the existence of a FBA alternate optimum, for the “RISC downhill pathway” different cytochrome oxidase combinations might work as well as CYTBO3/CYTBD. By knocking-out the CYBO3/CYTBD reactions, the parallel solution regarding the CYTRED/CYTAA32 system was calculated and outlined in Fig. 3. According to the flux distribution results both systems worked similarly. Furthermore, the total amount of proton translocation stoichiometry of both systems is the same (i.e. 1.8 protons translocating through the periplasmic membrane). Approximately 10% of all incoming electrons from TSQCO go to NADHI. The rest, just like the FIM are used to balance the cytoplasmic protons and generate proton motive force through the cytochrome oxidases. It is well known that during growth A. ferrooxidans acidifies the media when using RISCs compounds as electron donors (Rohwerder and Sand, 2007). This fact is represented by predicting a flux of protons outside the external membrane through the transport reaction Htex under TSM and TTM. In a different manner, FVA analysis showed that Htex feasible bounds able to sustain growth are within the positive range of the scale (Supplementary Fig. 10), demonstrating that in order to sustain growth, the system only has to produce protons, which are generated by 4THASE. The same kind of behavior was described in Supplementary Fig. 7, where growth was only possible under acidification media conditions.
The central carbon metabolism was modeled by integrating the carbon fixation, Calvin cycle, glycolysis and incomplete TCA pathways. Since FIM, TTM and TSM shared similar reactions and pathways, only FIM was studied in detail. For this, different FBA simulations were performed to quantitatively describe and study the fluxes involved in central carbon metabolism (see Supplementary Note 4). In general, simulations showed a high flux activity through the Calvin cycle, which incorporate co2 by using the RUBISCO reaction. The predicted fluxes directionality and reactions were in concordance with previously reported expression analysis, which described the genes and pathways involved in A. ferrooxidans co2 fixation (Esparza et al., 2010). Furthermore, due to network rigidity in terms of carbon metabolism, and in order to represent the effect of carbon leaking through the cell due to metabolic co2, transport reactions allowing the metabolic co2 transport through the periplasmic (CO2tpp) and extracellular (CO2tex) membrane were added to iMC507.
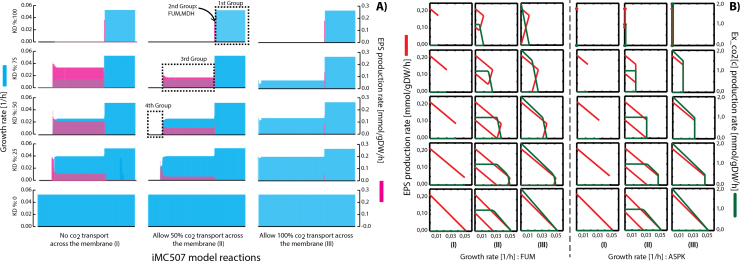
3.4. EPS production potential analysis
Extracellular polymeric substance production in A. ferrooxidans is determinant for increasing the pyrite bioleaching capacity (Gehrke et al., 1998). It basically allows the oxidative attack on the sulfur to take place (Sand and Gehrke, 2006), by mediating the bacterial adhesion to the mineral surface. In order to understand EPS production and further synthetically design strains able to couple EPS production to growth, a retrospective model driven analysis was performed. First, using FBA, the effect of the individual reaction knock-down on the predicted growth rate and EPS production was analyzed. Based on Fig. 4A, specifically in the first column, EPS production increases when reaction knock-down increases, This means that the more the metabolic network is constrained, a higher EPS production and a lower growth rate is calculated for a specific set of reactions in the network. This effect is directly related to the network rigidity in terms of how the cell metabolizes the carbon atoms. Since, no evidence of side carbon formation in A. ferrooxidans has been found, the metabolic network was modeled in a way that can only export carbon through the BOF or EPS reactions. Thus, whenever the network is perturbed, the only possible option to overcome the network imbalances is by re-routing the fluxes towards EPS production. For this reason, and in order to allow certain flexibility for the network, a metabolic co2 export reaction was added and its effect on the overall system simulated. The results of these simulations are outlined in Fig. 4A and B, specifically the second and third column, where the EPS production clearly decreases when allowing the cell to export metabolic co2. This demonstrates the preference for the cell for producing co2 instead of EPS when the network is internally perturbed. Based on this result, reactions can be clustered in 4 different groups. From right to left the first group contains only non-essential reactions, where no matter the perturbation on reactions, the cell still grows and no EPS is generated. The second group is defined only by two reactions, FUM and MDH, which are responsible for perturbing the network in a way that EPS is produced coupled to growth, independently of the presence of the co2 external transporter or the degree of reaction knock-down. This reaction knock-out allows the insilico design of A. ferrooxidans strains for growth-coupled EPS production, which is crucial for increasing the bioleaching capacity (see Supplementary Note 5 for detailed explanation). The third group, is represented by reactions where perturbations generate a network imbalance that produces EPS when no co2 external transporter was added (Fig. 4 first column), and does not generate any EPS when the co2 transporter is allowed to work at its full capacity (Fig. 4A, third column). The fourth group contains reactions which are essential for growth when the external co2 is constrained (Fig. 4A, first and second column). Based on this categorization, two reactions, FUM and ASPK, were chosen for computing their production envelopes (Fig. 4B). According to these results it is clearly shown (see Fig. 4B for FUM and ASPK, first column) how rigid the network behaves when no external co2 transport is added, where no production envelope is generated. Instead only production lines were obtained. This demonstrates how tightly coupled the EPS production and growth are under these conditions. Production envelopes shown in Fig. 4B, specifically in the second and third columns, show how the production envelopes are generated, allowing the network to export co2. EPS growth coupled production appearance, can be appreciated for the FUM reaction, which is independent from the external co2 transport. On the other hand, for reactions defined in the second group, and represented by ASPK in Fig. 4B, no growth coupled design through reaction knock was seen, still for certain knock-down values, EPS production is still coupled to growth. Furthermore, in both cases, a tradeoff between EPS and metabolic co2 production exists whenever the network is perturbed and co2 allowed to leave the network.
Fig. 4.
Individual reaction Knock-down and external metabolic co2 transport flux constraint effect on growth rate and EPS production: A) A graph describing the effect of constraining the hypothesized external metabolic co2 transport flux together with the individual reaction knock-down effect for all reactions on the predicted growth rate and EPS production is outlined. For each simulation, growth rate (specified in light blue bars in sub-graph on the left hand side y-axis) and EPS flux (specified in red bars in sub-graph on the right hand side y-axis) were plotted. In each sub-graph the corresponding biomass and EPS production rate was plotted for each one of the reactions (x-axis) in the model. Simulations were executed by adding an external co2 transport and constraining it in 0, 50% and 100% from the maximum capacity (x-axis), all reactions in the model were constraint between 0, 25, 50, 75% and 100% from their optimum capacities (y-axis). For each simulation the carbon uptake rate in the form of h2co3 was set to 2 mmol/g DW/h, oxygen and ferrous ion uptake rate were left unconstrained, and FBA was ran to maximize the flux through BOF. B) A set of graphs describing the production envelopes for reactions FUM and ASPK under the same conditions described previously. EPS (left y-axis in red lines) and co2 (right y-axis in green lines) were plotted as a function of growth rate (x-axis). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
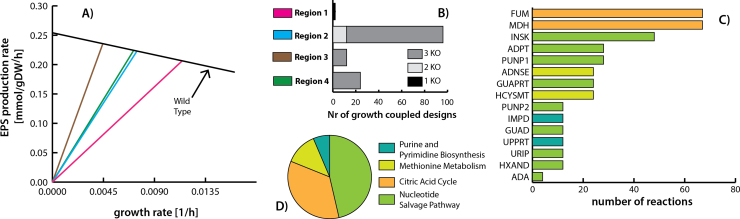
Furthermore, analysis on the EPS growth-coupled potential for additional double and triple knock-outs was evaluated. Results from this analysis are shown in Fig. 5. A total of 134 growth-coupled designs were calculated (Supplementary Table 7). Two of them correspond to single knock-outs, 12 to double knock-outs and the rest to triple knock outs. Out of the 134 growth-coupled designs, 4 different production envelopes were identified, which described the Pareto frontier in the EPS flux vs. the biomass flux plane. The four different regions were plotted in Fig. 5A. Approximately 72% of all the calculated designs correspond to region 2 (blue). Most of the knock-out designs in region 2 correspond to triple knock-outs (Fig. 5B). Region 3 shows the higher theoretical EPS achievable flux, a total of 12 designs were calculated, and all of them correspond to triple knock-outs. In all of the calculated designs two different reactions were always found, either fumarase (FUM) or malate dehydrogenase (MDH), which described the production enveloped in region 1. Actually, each of them was found independently 67 times among all designs (see Fig. 5C), representing the reactions with more instances among all growth-coupled design reactions. Despite the latter, the subsystem with more reaction instances in growth-coupled designs is the Nucleotide Salvage Pathway (see Fig. 5D), which does not contain MDH nor FUM that are part of the citric acid cycle.
Fig. 5.
Growth-coupled EPS production strain design results: A) A graph that shows the EPS production envelopes for A. ferrooxidans wild type (black) and different KO combinations that were calculated during the analysis. The EPS production rate is shown in the y-axis and the growth rate is given on the x-axis. Four different production envelopes were characterized and described in different colors according to the regions described in B. B) A bar plot describing the number of growth coupled designs associated with the each characterized region. For each region the information about the number of knock-outs reactions associated with the corresponding growth coupled designs was incorporated. C) A bar plot showing the number of instances that each reaction in the y-axis was found in all growth coupled designs. Bar colors represents the subsystem associated with the corresponding reaction. D) A pie chart descrying the major subsystems associated with the predicted knock-out reactions. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
4. Discussion
A fundamental goal in biology is to understand and predict the genotype-phenotype relationships in the cell. COBRA methods organize biochemical, genetic and genomic knowledge into a mathematical framework which enables the quantitatively description of metabolic physiology. In this work, the first genome-scale metabolic reconstruction of Acidithiobacillus ferrooxidans ATCC 23270, iMC507 is presented. The bottom-up metabolic network reconstruction of iMC507 represents a comprehensive knowledge base that summarizes and categorizes the information currently available for A. ferrooxidans, and serves as a framework for computational analysis. Based on experimental evidence together with the model-driven analysis performed in this investigation, i) key electron transfer reaction proton translocation stoichiometry were predicted, ii) quantitatively phenotypic description of specific organism key properties (i.e. uphill vs downhill pathway and effect of growth rate due to proton exchanged) in the electron transfer metabolism under aerobic growth and using ferrous ion, tetrathionate and thiosulfate as electron donors was determined, iii) quantitative phenotypic description of central carbon metabolism for aerobic growth and using ferrous ion was determined, and iv) based on the knock-down and knock-out analysis, several growth-coupled designs able to produce EPS were calculated and evaluated for further experimental implementation.
iMC507 represents the first genome-scale reconstruction of A. ferrooxidans specie and accounts for 507 ORFs (16%) of the current A. ferrooxidans genome annotation. iMC507 represent a major advance compared to the small scale metabolic network models previously developed, which only account for 62 reactions in the first case (Hold et al., 2009) and 190 in the second case (Sepúlveda et al., 2011). iMC507 average confidence score was equal to 2.02. This means that most of the network content was based on genome annotation. In fact only 58% of the reactions were based on the genome annotation and 19% of all reactions in iMC507 have been very well or well-studied. The next step in the expansion of the A. ferrooxidans metabolic network will required research efforts directed to poorly described pathways. The current knowledge reflects the traditional use of A. ferrooxidans as a chemolithoautotrophic ferrous ion and RISCs oxidation-model bacterium, where subsystems such as co2 fixation, sulfur metabolism, oxidative phosphorylation, and EPS biosynthesis have been well studied. In contrast, metabolisms involved in amino acid fixation, central carbon, transport reactions, among others are poorly described and in need of further characterization.
Since its discovery A. ferrooxidans has been extensively studied under chemolithoautotrophic aerobic condition, by using mainly ferrous ion and RISCs compounds as electron donors. In this study the proton translocation stoichiometry for key enzymes involved in the ferrous ion and RISCs metabolism together with the GAM and NGAM values were predicted. Specifically for the CYTAA31 reaction (cytochrome c oxidase aa3-type), involved in ferrous ion metabolism, much debate has been generated during the years whether it actually translocates protons across the membrane. Based on the genetic-algorithm base approach, no proton translocation stoichiometry for this reaction was predicted. Furthermore, ATP synthase proton translocation stoichiometry was determined, showing how COBRA methods in conjunction with experimental phenotypic data can be useful for elucidation of chemiosmotic parameters of the cell.
A quantitatively phenotypic description of specific organism key properties in the electron transfer metabolism (i.e. uphill vs downhill pathway and effect of growth rate due to proton exchanged) under aerobic growth and using ferrous ion, tetrathionate and thiosulfate as electron donors was determined. For FIM, a complete and accurate description of the electron transfer metabolism was performed, showing that most of the electrons (i.e. more than 90%) go to the proton neutralization by o2 reduction into water through CYTAA31. The rest of the electrons go directly to the Calvin cycle via the “uphill pathway”. Furthermore, media alkalization, a characteristic behavior during ferrous iron growth was also predicted. With these results model prediction capacity was validated for FIM. More interesting was the study of TTM and TSM, where the TSM efficiency over the TTM was quantitatively demonstrated. Furthermore, a “RISC uphill pathway” and a “RISC downhill pathway” were described and quantitatively characterized. In this work the central carbon metabolism was outlined by incorporating three major pathways: the Calvin cycle, glycolysis and the incomplete TCA cycle. The junction point was determined by 3-phosphoglyceric acid, which connects glycolysis with the Calvin cycle. The Calvin cycle showed a higher flux activity compared to the rest of the central carbon metabolism, mainly to sustain the co2 fixation through the RUBISCO reaction. Based on the simulations, results showed how rigid the metabolic network is in terms of the carbon metabolism backbone. Thus, in order to allow more degrees of freedom, regarding the internal carbon fluxes a metabolic co2 transport was hypothesized and incorporated into the metabolic network. Result showed that instead of producing EPS, the network prefers to generate co2. Although these results are reasonable, no experimental evidence regarding metabolic co2 evolution, or small compound secretions (i.e. ethanol, lactate, acetate, etc.) has been reported.
EPS production sensitivity was evaluated by inspecting two variables. The first was EPS production due to individual reaction knock-down, and the second was the presence or absence of a metabolic co2 transport. Overall, when no metabolic co2 was allowed to leave the cell, by increasing the knock-down strength, a subset of perturbed reactions was identified to promote EPS production. Due to the network rigidity in terms of carbon metabolism, this result was expected. On the other hand, when metabolic co2 was allowed to leave the cell, EPS production was observed only when FUM and MDH were perturbed. Furthermore, when these reactions were knocked-out, growth coupled EPS production was observed. Evidence regarding the incomplete TCA cycle disruption leading to EPS overproduction (Sepúlveda et al., 2011), validates this results since FUM and MDH are reactions present in the incomplete TCA cycle. The evaluation of all possible single, double and triple knock-outs for growth coupled design was performed. In total 134 growth-coupled designs were found. In all of the predicted designs either FUM or MDH were found to be involved. Due to the lack of by-product formation reactions, these network disruptions, re-routes the formate utilization towards the fatty acid biosynthesis, and further EPS production. Growth coupled designs, such as those produced here, provide an extra tool for metabolic engineers by allowing the use of selection pressure to achieve a desired production state. Nevertheless, due to the lack of genetic tools that allows the knock-out implementation in A. ferrooxidans, it is not possible to implement the design experimentally yet. Still, competitive inhibition might be used in order to disrupt the metabolism as utilized elsewhere (Sepúlveda et al., 2011).
As the field of constraint based modeling and analysis continues to expand by incorporating different cellular processes and interactions into the genome-scale space (e.g. transcription and translation in Escherichia coli (O’Brien et al., 2013) and Thermotoga maritime (Lerman et al., 2012)), iMC507 will serve as a key component for the quantitative study of A. ferrooxidans and related organisms by providing an extensive map of the cellular metabolism, enabling the incorporation of different datasets (e.g. transcriptomics, proteomics, fluxomics, metabolomics) for improving the computational outcome of the cellular metabolism. Furthermore, iMC507 represents a useful tool for the A. ferrooxidans community, by unifying and describing our knowledge of this unique species and providing a computational platform for further analysis and hypothesis formulation for environmental and biotechnological applications.
Acknowledgments
We would like to thank the Project UCH0717 National Doctoral Scholarship, CONICYT.
Footnotes
Supplementary data associated with this article can be found in the online version at 10.1016/j.meteno.2016.03.003.
Appendix A. Supplementary material
Supplementary material
.
Supplementary material
.
Supplementary material
.
References
- Boon M. 1996. Theoretical and Experimental Methods in the Modelling of Bio-oxidation Kinetics of Sulphide Minerals. [Google Scholar]
- Bordbar A., Monk J.M., King Z.A., Palsson B.O. Constraint-based models predict metabolic and associated cellular functions. Nat. Rev. Genet. 2014;15:107–120. doi: 10.1038/nrg3643. [DOI] [PubMed] [Google Scholar]
- Brandl H. second edition. 2008. Theoretical and Experimental Methods in the Modelling of Bio-oxidation Kinetics of Sulphide Minerals; pp. 191–224. [Google Scholar]
- Burgard A.P., Pharkya P., Maranas C.D. Optknock: a bilevel programming framework for identifying gene knockout strategies for microbial strain optimization. Biotechnol. Bioeng. 2003;84:647–657. doi: 10.1002/bit.10803. [DOI] [PubMed] [Google Scholar]
- Carbajosa S., Malki M., Caillard R., Lopez M.F., Palomares F.J., Martin-Gago J.A., Rodriguez N., Amils R., Fernandez V.M., De Lacey A.L. Electrochemical growth of Acidithiobacillus ferrooxidans on a graphite electrode for obtaining a biocathode for direct electrocatalytic reduction of oxygen. Biosens. Bioelectron. 2010;26:877–880. doi: 10.1016/j.bios.2010.07.037. [DOI] [PubMed] [Google Scholar]
- Caspi R., Altman T., Dreher K., Fulcher C.A., Subhraveti P., Keseler I.M., Kothari A., Krummenacker M., Latendresse M., Mueller L.A., Ong Q., Paley S., Pujar A., Shearer A.G., Travers M., Weerasinghe D., Zhang P., Karp P.D. The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic Acids Res. 2012;40:D742–D753. doi: 10.1093/nar/gkr1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- C.I.A., 2011. The World Factbook. (Retrieved from 23.11.11).
- Cox J.C., Nicholls D.G., Ingledew W.J. Transmembrane electrical potential and transmembrane pH gradient in the acidophile Thiobacillus ferro-oxidans. Biochem. J. 1979;178:195–200. doi: 10.1042/bj1780195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dale J.M., Popescu L., Karp P.D. Machine learning methods for metabolic pathway prediction. BMC Bioinform. 2010;11:15. doi: 10.1186/1471-2105-11-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drobner E., Huber H., Stetter K.O. Thiobacillus ferrooxidans, a facultative hydrogen oxidizer. Appl. Environ. Microbiol. 1990;56:2922–2923. doi: 10.1128/aem.56.9.2922-2923.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eccleston M., Kelly D.P. Oxidation kinetics and chemostat growth kinetics of Thiobacillus ferrooxidans on tetrathionate and thiosulfate. J. Bacteriol. 1978;134:718–727. doi: 10.1128/jb.134.3.718-727.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards C. Microbiology of Extreme Environments. Open University Press; 1990. [Google Scholar]
- Elbehti A., Brasseur G., Lemesle-Meunier D. First evidence for existence of an uphill electron transfer through the bc(1) and NADH-Q oxidoreductase complexes of the acidophilic obligate chemolithotrophic ferrous ion-oxidizing bacterium Thiobacillus ferrooxidans. J. Bacteriol. 2000;182:3602–3606. doi: 10.1128/jb.182.12.3602-3606.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esparza M., Cardenas J.P., Bowien B., Jedlicki E., Holmes D.S. Genes and pathways for CO2 fixation in the obligately, chemolithoautotrophic acidophile, Acidithiobacillus ferrooxidans carbon fixation in A. ferrooxidans. BMC Microbiol. 2010;10:229. doi: 10.1186/1471-2180-10-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falco L., Pogliani C., Curutchet G., Donati E. A comparison of bioleaching of covellite using pure cultures of Acidithiobacillus ferrooxidans and Acidithiobacillus thiooxidans or a mixed culture of Leptospirillum ferrooxidans and Acidithiobacillus thiooxidans. Hydrometallurgy. 2003;71:31–36. [Google Scholar]
- Feist A.M., Henry C.S., Reed J.L., Krummenacker M., Joyce A.R., Karp P.D., Broadbelt L.J., Hatzimanikatis V., Palsson B.O. A genome-scale metabolic reconstruction for Escherichia coli K-12 MG1655 that accounts for 1260 ORFs and thermodynamic information. Mol. Syst. Biol. 2007;3:121. doi: 10.1038/msb4100155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feist A.M., Nagarajan H., Rotaru A.E., Tremblay P.L., Zhang T., Nevin K.P., Lovley D.R., Zengler K. Constraint-based modeling of carbon fixation and the energetics of electron transfer in Geobacter metallireducens. PLoS Comput. Biol. 2014;10:e1003575. doi: 10.1371/journal.pcbi.1003575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feist A.M., Scholten J.C., Palsson B.O., Brockman F.J., Ideker T. Modeling methanogenesis with a genome-scale metabolic reconstruction of Methanosarcina barkeri. Mol. Syst. Biol. 2006:2. doi: 10.1038/msb4100046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson S.J., Ingledew W.J. Energetic problems faced by micro-organisms growing or surviving on parsimonious energy sources and at acidic pH: I. Acidithiobacillus ferrooxidans as a paradigm. Biochim Biophys. Acta. 2008;1777:1471–1479. doi: 10.1016/j.bbabio.2008.08.012. [DOI] [PubMed] [Google Scholar]
- Fischer-Kowalski, M., Swilling, M., 2011. Decoupling: Natural Resource Use and Environmental Impacts From Economic Growth. United Nations Environment Programme.
- Franks D.M., Boger D.V., Côte C.M., Mulligan D.R. Sustainable development principles for the disposal of mining and mineral processing wastes. Resour. Policy. 2011;36:114–122. [Google Scholar]
- Gehrke T., Telegdi J., Thierry D., Sand W. Importance of extracellular polymeric substances from thiobacillus ferrooxidans for bioleaching. Appl. Environ. Microbiol. 1998;64:2743–2747. doi: 10.1128/aem.64.7.2743-2747.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harneit K., Göksel A., Kock D., Klock J.-H., Gehrke T., Sand W. Adhesion to metal sulfide surfaces by cells of Acidithiobacillus ferrooxidans, Acidithiobacillus thiooxidans and Leptospirillum ferrooxidans. Hydrometallurgy. 2006;83:245–254. [Google Scholar]
- Hold C., Andrews B.A., Asenjo J.A. A stoichiometric model of Acidithiobacillus ferrooxidans ATCC 23270 for metabolic flux analysis. Biotechnol. Bioeng. 2009;102:1448–1459. doi: 10.1002/bit.22183. [DOI] [PubMed] [Google Scholar]
- Ingledew W. Thiobacillus ferrooxidans. The bioenergetics of an acidophilic chemolithotroph. Biochim. Biophys. Acta. 1982;683:89–117. doi: 10.1016/0304-4173(82)90007-6. [DOI] [PubMed] [Google Scholar]
- Ishigaki T., Nakanishi A., Tateda M., Ike M., Fujita M. Bioleaching of metal from municipal waste incineration fly ash using a mixed culture of sulfur-oxidizing and iron-oxidizing bacteria. Chemosphere. 2005;60:1087–1094. doi: 10.1016/j.chemosphere.2004.12.060. [DOI] [PubMed] [Google Scholar]
- Kanehisa M., Goto S., Hattori M., Aoki-Kinoshita K.F., Itoh M., Kawashima S., Katayama T., Araki M., Hirakawa M. From genomics to chemical genomics: new developments in KEGG. Nucleic Acids Res. 2006;34:D354–D357. doi: 10.1093/nar/gkj102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karp P.D., Paley S.M., Krummenacker M., Latendresse M., Dale J.M., Lee T.J., Kaipa P., Gilham F., Spaulding A., Popescu L., Altman T., Paulsen I., Keseler I.M., Caspi R. Pathway Tools version 13.0: integrated software for pathway/genome informatics and systems biology. Brief. Bioinform. 2010;11:40–79. doi: 10.1093/bib/bbp043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuenen J. Growth yields and “maintenance energy requirement in Thiobacillus species under energy limitation”. Arch. Microbiol. 1979;122:183–188. [Google Scholar]
- Lerman J.A., Hyduke D.R., Latif H., Portnoy V.A., Lewis N.E., Orth J.D., Schrimpe-Rutledge A.C., Smith R.D., Adkins J.N., Zengler K., Palsson B.O. In silico method for modelling metabolism and gene product expression at genome scale. Nat. Commun. 2012;3:929. doi: 10.1038/ncomms1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Liu Y., Zeng G., Niu Y., Xiao X., Xu W., Xia W., Zhu Y., Liu J. Direct current stimulation of Thiobacillus ferrooxidans bacterial metabolism in a bioelectrical reactor without cation-specific membrane. Bioresour. Technol. 2010;101:6035–6038. doi: 10.1016/j.biortech.2010.02.094. [DOI] [PubMed] [Google Scholar]
- Lun D.S., Rockwell G., Guido N.J., Baym M., Kelner J.A., Berger B., Galagan J.E., Church G.M. Large-scale identification of genetic design strategies using local search. Mol. Syst. Biol. 2009;5:296. doi: 10.1038/msb.2009.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahadevan R., Schilling C.H. The effects of alternate optimal solutions in constraint-based genome-scale metabolic models. Metab. Eng. 2003;5:264–276. doi: 10.1016/j.ymben.2003.09.002. [DOI] [PubMed] [Google Scholar]
- Mayer H., Krauss J.H., Urbanik-Sypniewska T., Puvanesarajah V., Stacey G., Auling G. Lipid A with 2,3-diamino-2,3-dideoxy-glucose in lipopolysaccharides from slow-growing members of Rhizobiaceae and from “Pseudomonas carboxydovorans”. Arch. Microbiol. 1989;151:111–116. doi: 10.1007/BF00414423. [DOI] [PubMed] [Google Scholar]
- McCloskey D., Palsson B.O., Feist A.M. Basic and applied uses of genome-scale metabolic network reconstructions of Escherichia coli. Mol. Syst. Biol. 2013;9:661. doi: 10.1038/msb.2013.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mykytczuk N.C., Trevors J.T., Ferroni G.D., Leduc L.G. Cytoplasmic membrane fluidity and fatty acid composition of Acidithiobacillus ferrooxidans in response to pH stress. Extremophiles. 2010;14:427–441. doi: 10.1007/s00792-010-0319-2. [DOI] [PubMed] [Google Scholar]
- O’Brien E.J., Lerman J.A., Chang R.L., Hyduke D.R., Palsson B.O. Genome-scale models of metabolism and gene expression extend and refine growth phenotype prediction. Mol. Syst. Biol. 2013;9:693. doi: 10.1038/msb.2013.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orth J.D., Thiele I., Palsson B.Ø. What is flux balance analysis? Nat. Biotechnol. 2010;28:245–248. doi: 10.1038/nbt.1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osorio H., Mangold S., Denis Y., Nancucheo I., Esparza M., Johnson D.B., Bonnefoy V., Dopson M., Holmes D.S. Anaerobic sulfur metabolism coupled to dissimilatory iron reduction in the extremophile Acidithiobacillus ferrooxidans. Appl. Environ. Microbiol. 2013;79:2172–2181. doi: 10.1128/AEM.03057-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patil K.R., Rocha I., Forster J., Nielsen J. Evolutionary programming as a platform for in silico metabolic engineering. BMC Bioinform. 2005;6:308. doi: 10.1186/1471-2105-6-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pronk J.T., Meijer W.M., Hazeu W., van Dijken J.P., Bos P., Kuenen J.G. Growth of Thiobacillus ferrooxidans on formic acid. Appl. Environ. Microbiol. 1991;57:2057–2062. doi: 10.1128/aem.57.7.2057-2062.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu M.-q, Xiong S.-y, Zhang W.-m, Wang G.-x. A comparison of bioleaching of chalcopyrite using pure culture or a mixed culture. Miner. Eng. 2005;18:987–990. [Google Scholar]
- Rawlings D.E. Heavy metal mining using microbes 1. Annu. Rev. Microbiol. 2002;56:65–91. doi: 10.1146/annurev.micro.56.012302.161052. [DOI] [PubMed] [Google Scholar]
- Rawlings D.E., Dew D., du Plessis C. Biomineralization of metal-containing ores and concentrates. Trends Biotechnol. 2003;21:38–44. doi: 10.1016/s0167-7799(02)00004-5. [DOI] [PubMed] [Google Scholar]
- Rawlings D.E., Johnson D.B. Biomining. Springer; 2007. [Google Scholar]
- Rohwerder T., Gehrke T., Kinzler K., Sand W. Bioleaching review part A: progress in bioleaching: fundamentals and mechanisms of bacterial metal sulfide oxidation. Appl. Microbiol. Biotechnol. 2003;63:239. doi: 10.1007/s00253-003-1448-7. [DOI] [PubMed] [Google Scholar]
- Rohwerder T., Sand W. Oxidation of inorganic sulfur compounds in acidophilic prokaryotes. Eng. Life Sci. 2007;7:301–309. [Google Scholar]
- Sand W., Gehrke T. Extracellular polymeric substances mediate bioleaching/biocorrosion via interfacial processes involving iron(III) ions and acidophilic bacteria. Res. Microbiol. 2006;157:49–56. doi: 10.1016/j.resmic.2005.07.012. [DOI] [PubMed] [Google Scholar]
- Sand W., Rohde K., Sobotke B., Zenneck C. Evaluation of Leptospirillum ferrooxidans for leaching. Appl. Environ. Microbiol. 1992;58:85–92. doi: 10.1128/aem.58.1.85-92.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schellenberger J., Que R., Fleming R.M., Thiele I., Orth J.D., Feist A.M., Zielinski D.C., Bordbar A., Lewis N.E., Rahmanian S., Kang J., Hyduke D.R., Palsson B.O. Quantitative prediction of cellular metabolism with constraint-based models: the COBRA Toolbox v2.0. Nat. Protoc. 2011;6:1290–1307. doi: 10.1038/nprot.2011.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schomburg I., Chang A., Schomburg D. BRENDA, enzyme data and metabolic information. Nucleic Acids Res. 2002;30:47–49. doi: 10.1093/nar/30.1.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sepúlveda, A.E.M., Cortéz, P.A.M., Abarca, M.A.B., Valdecantos, P.A.P., Iglesias, L.M.P., Roa, M.N.B., Method to increase the production of extracellular polymeric substances (eps) in a acidithiobacillus ferrooxidans culture by the inhibition of enzymes of tricarboxilic acid cycle. Google Patents, 2011.
- Shively J.M., Benson A.A. Phospholipids of thiobacillus thiooxidans. J. Bacteriol. 1967;94:1679–1683. doi: 10.1128/jb.94.5.1679-1683.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sublette K.L. Production of microbial biomass protein from autotrophic fermentation of hydrogen sulfide. Biotechnol. Bioeng. 1988;32:408–409. doi: 10.1002/bit.260320324. [DOI] [PubMed] [Google Scholar]
- Tepper N., Shlomi T. Predicting metabolic engineering knockout strategies for chemical production: accounting for competing pathways. Bioinformatics. 2010;26:536–543. doi: 10.1093/bioinformatics/btp704. [DOI] [PubMed] [Google Scholar]
- Thiele I., Palsson B.O. A protocol for generating a high-quality genome-scale metabolic reconstruction. Nat. Protoc. 2010;5:93–121. doi: 10.1038/nprot.2009.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdes J., Pedroso I., Quatrini R., Dodson R.J., Tettelin H., Blake R., 2nd, Eisen J.A., Holmes D.S. Acidithiobacillus ferrooxidans metabolism: from genome sequence to industrial applications. BMC Genomics. 2008;9:597. doi: 10.1186/1471-2164-9-597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarzabal A., Appia-Ayme C., Ratouchniak J., Bonnefoy V. Regulation of the expression of the Acidithiobacillus ferrooxidans rus operon encoding two cytochromes c, a cytochrome oxidase and rusticyanin. Microbiology. 2004;150:2113–2123. doi: 10.1099/mic.0.26966-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Supplementary material
Supplementary material