Abstract
Chemical and fuel production by photosynthetic cyanobacteria is a promising technology but to date has not reached competitive rates and titers. Genome-scale metabolic modeling can reveal limitations in cyanobacteria metabolism and guide genetic engineering strategies to increase chemical production. Here, we used constraint-based modeling and optimization algorithms on a genome-scale model of Synechocystis PCC6803 to find ways to improve productivity of fermentative, fatty-acid, and terpene-derived fuels. OptGene and MOMA were used to find heuristics for knockout strategies that could increase biofuel productivity. OptKnock was used to find a set of knockouts that led to coupling between biofuel and growth. Our results show that high productivity of fermentation or reversed beta-oxidation derived alcohols such as 1-butanol requires elimination of NADH sinks, while terpenes and fatty-acid based fuels require creating imbalances in intracellular ATP and NADPH production and consumption. The FBA-predicted productivities of these fuels are at least 10-fold higher than those reported so far in the literature. We also discuss the physiological and practical feasibility of implementing these knockouts. This work gives insight into how cyanobacteria could be engineered to reach competitive biofuel productivities.
Keywords: Cyanobacteria, Modeling, Flux balance analysis, Biofuel, MOMA, OptFlux, OptKnock
Highlights
-
•
Model-derived knockout strategies to couple growth and biofuel in Synechocystis.
-
•
Alcohols by reverse beta-oxidation require knockout of many NADH-reactions.
-
•
Fatty acid and terpene derived fuels require alteration of ATP and NADPH consumption.
1. Introduction
The engineering of microbes for the production of chemicals and fuels is a pillar in a future bio-based economy (Choi et al., 2015, Peralta-Yahya et al., 2012). Autotrophic hosts such as cyanobacteria are particularly attractive cell factories for large-volume, low-value products like biofuel, as handling of plant-based feedstock can negatively affect the cost and energy balances of heterotroph-based processes (Jiang et al., 2014). Many cyanobacteria strains have been developed which produce small amounts of chemicals and biofuels directly from CO2 (Oliver and Atsumi, 2014). However, systems-level metabolic engineering is needed to achieve industrially-relevant chemical productivities in cyanobacteria (Gudmundsson and Nogales, 2014).
Cell factory design is based on a genome-scale metabolic model (GEM), where cellular metabolic reactions are tabulated and connected into a network topology. The GEM can be subjected to flux balance analysis (FBA), which uses external nutrient uptake rates and optimization principles to estimate steady-state intracellular and extracellular reaction fluxes, including cell growth rate (O’Brien et al., 2015). A suite of algorithms have been developed which can use the GEM to calculate how cellular metabolism should be changed to achieve high productivities or yields of a given product (Machado and Herrgård, 2015). These algorithms report in silico modifications that could be manifest experimentally as genetic knockouts, knockdowns, or knockups.
One powerful algorithm is OptKnock, which seeks to maximize flux to product while simultaneously maximizing growth rate. The result is a list of knockouts, that when executed in silico, result in a strain where product synthesis occurs at maximum growth (Burgard et al., 2003). This is beneficial as there is evidence that bacterial metabolism will evolve to maximize growth (Fong and Palsson, 2004). Therefore, product-growth “coupled” strains would ensure high productivity over time. A first application of OptKnock was to predict and execute the reaction knockouts necessary to link lactate production to cell growth in E. coli (Fong et al., 2005).
In the OptGene algorithm, reaction knockouts are implemented randomly, creating a mutant population (Patil et al., 2005). The reaction fluxes of each mutant are predicted using minimization of metabolic adjustment (MOMA), which assumes a minimal deviation from wild-type fluxes (Segrè et al., 2002). Mutants having higher fluxes toward product (or some other fitness metric) are selected for subsequent mutation or crossover with other mutants. OptGene is computationally efficient because it searches for local optima; i.e. mutants are subjected to an evolutionary trajectory, and each mutant is compared only to others in the population. While global optima are not found, the mutants reveal heuristics about the most effective mutations to improve productivity. OptGene was recently used to identify knockouts in yeast that improved succinate titers 30-fold when the strategy was executed in vivo (Otero et al., 2013).
Synechocystis GEMs have been previously used to suggest engineering strategies for increasing production of ethanol, isobutanol, fumarate, and hydrogen (Sengupta et al., 2013;Erdrich et al., 2014, Nogales et al., 2013). However, strategies for product-growth coupling have been elusive in cyanobacteria, as photosynthesis metabolism is robust, with several electron “valves” such as cyclic and alternative electron flows. Here we use OptGene and OptKnock on stoichiometric Synechocystis GEMs to identify in silico gene knockouts that improve production of fermentation, fatty-acid, and terpene-derived biofuels. OptGene revealed heuristics for improving productivity, while OptKnock identified reaction knockouts that couple photoautotrophic growth and biofuel production. The underlying logic of these strategies is revealed.
2. Material and methods
2.1. Genome scale modeling
The Synechocystis sp. PCC 6803 model iJN678 (Nogales et al., 2012) was used to perform flux balance analysis using the COBRA toolbox 2.0 (Schellenberger et al., 2011) on MATLAB (Mathworks Inc., Natick, MA). The model iJN678 was downloaded from the BiGG Models database (http://bigg.ucsd.edu). The reconstruction iJN678 incorporates 678 genes, 863 reactions and 795 metabolites. Several additions were made in accordance with recent literature: the TCA cycle shunt reactions (Zhang and Bryant, 2011), the light-independent serine biosynthesis pathway (Klemke et al., 2015), and the phosphoketolase reaction (Xiong et al., 2015). Electron-transport chain reactions in the cytoplasm and thylakoid were modified to be consistent with recent literature (Lea-Smith et al., 2015). All modifications are added in the iJN678_2016 SBML file (Sourceforge).
Model iJN678_2016 can simulate heterotrophic, autotrophic and mixotrophic conditions by constraining photon, glucose, or carbonate/CO2 uptakes. All simulations were done for autotrophic conditions in light limited state (LLS); photon flux was fixed at −45 mmol/gDW h (both bounds). This photon uptake gives a maximum specific growth rate of 0.08 h−1, which is similar to that recorded under common laboratory conditions of 1% v/v CO2 and 50 uE/m2/s illumination. Carbon uptake (HCO3) was not fixed but had a limit of −3.7 mmol/gDW h, which is the estimated maximal uptake rate (Young et al., 2011). A second Synechocystis sp. PCC 6803 GEM (Knoop et al., 2013) was used for comparison. The ferredoxin plastoquinone reductase (FQR) and the Mehler-related reactions were added to this GEM. The biofuel biosynthesis reactions were added as described in Section 3.
Execution of OptKnock on genome scale models has been described (Chowdhury et al., 2015). Here, OptKnock was performed on iJN678 with scripts provided in the COBRA toolbox 2.0 (Hyduke et al., 2011). The TOMLAB/CPLEX solver was used (Tomlab Optimization Inc., San Diego, CA). The reactions available to OptKnock were reduced to a pool of approximately 325 following a published methodology (see Supplemental Matlab file (Sourceforge) and (Feist et al., 2010)). The testable subset excluded transport reactions, peripheral reactions, essential reactions, and reactions acting on high-carbon containing molecules. Simulations were allowed to run for up to 12 h. OptForce was performed on the same iJN678 reaction subset using the General Algebraic Modeling System GAMS 24.4.1 (GAMS Development Corporation, Washington, DC) as described in Ranganathan et al. (2010).
OptGene was performed on the iJN678 reaction subset using the “Evolutionary optimization” algorithm in the OptFlux software (Rocha et al., 2010).
Minimization of metabolic adjustment (MOMA) was used to predict the flux distributions of mutant strains (Segrè et al., 2002). The fitness metric was Biomass-Product Coupled Yield (BPCY), the product of biomass and biofuel fluxes divided by HCO3− uptake rate. Up to 5 reaction knockouts were allowed. The reference flux distribution for MOMA was obtained by solving iJN678 with FBA constrained with 13C MFA data from the 95% confidence intervals from the reactions in Table 1 of Young et al. (Young et al., 2011). These constraints reduced the maximum biomass formation rate to 0.0735 h−1. All simulations were performed on a MacBook Pro with 8 GB RAM and a 2.7 GHz Intel i5 processor.
Table 1.
Reactions that must be knocked out to create mutant M1 and enable 1-butanol-coupled growth in iJN678_ButFER.
| Reaction name in iJN678 | Enzyme (s) | Reaction* | Locus to target** |
|---|---|---|---|
| NDH1_2u | NAD(P)H dehydrogenase NDH-1 (thylakoid) | 4h[c]+nadh[c]+pq[u]→nad[c]+3h[u]+pqh2[u] | slr0331 (ndhD1) and slr1291 (ndhD2) |
| NDH2_syn | NdbA, NdbB, NdbC (thylakoid) | h[c]+nadh[c]+pq[u]→nad[c]+pqh2[u] | slr0851, slr1743, and sll1484 |
| NDH2_2p | NdbA, NdbB, NdbC (periplasm) | h[c]+nadh[c]+pq[p]→nad[c]+pqh2[p] | slr0851, slr1743, and sll1484 |
| GLYCTO1 | Glycolate oxidase | o2[c]+glyclt[c]→h2o2[c]+glx[c] | sll0404 (glcD2) |
| GLUSx | Glutamate synthase GOGAT (NADH-dependent) | h[c]+nadh[c]+akg[c]+ gln-L[c]→nad[c]+2 glu-L[c] | sll1502 |
| MDH | Malate dehydrogenase | nad[c]+mal-L[c]⇔h[c]+nadh[c]+oaa[c] | sll0891 |
| POR_syn | Pyruvate: ferredoxin oxidoreductase | coa[c]+pyr[c]+2 fdxo-2:2[c]→h[c]+co2[c]+accoa[c]+2 fdxr-2:2[c] | sll0741 |
| FPK | Phosphoketolase | f6p[c]+pi[c]→actp[c]+e4p[c]+h2o[c] | slr0453 |
| NADTRHD*** | NAD transhydrogenase | nad[c]+nadph[c]⇔nadp[c]+nadh[c] | slr1239 (pntA) |
[c] cytoplasmic, [u] thylakoid, [p] periplasmic compartments.
Locus to target is suggestion for gene deletion to eliminate enzyme activity. For multi-domain proteins a core subunit is given. NDH-1 (Battchikova et al., 2011), GlcD2 (Eisenhut et al., 2008).
Reaction assumed to be reversible.
3. Results
3.1. OptGene for metabolic engineering heuristics in Synechocystis
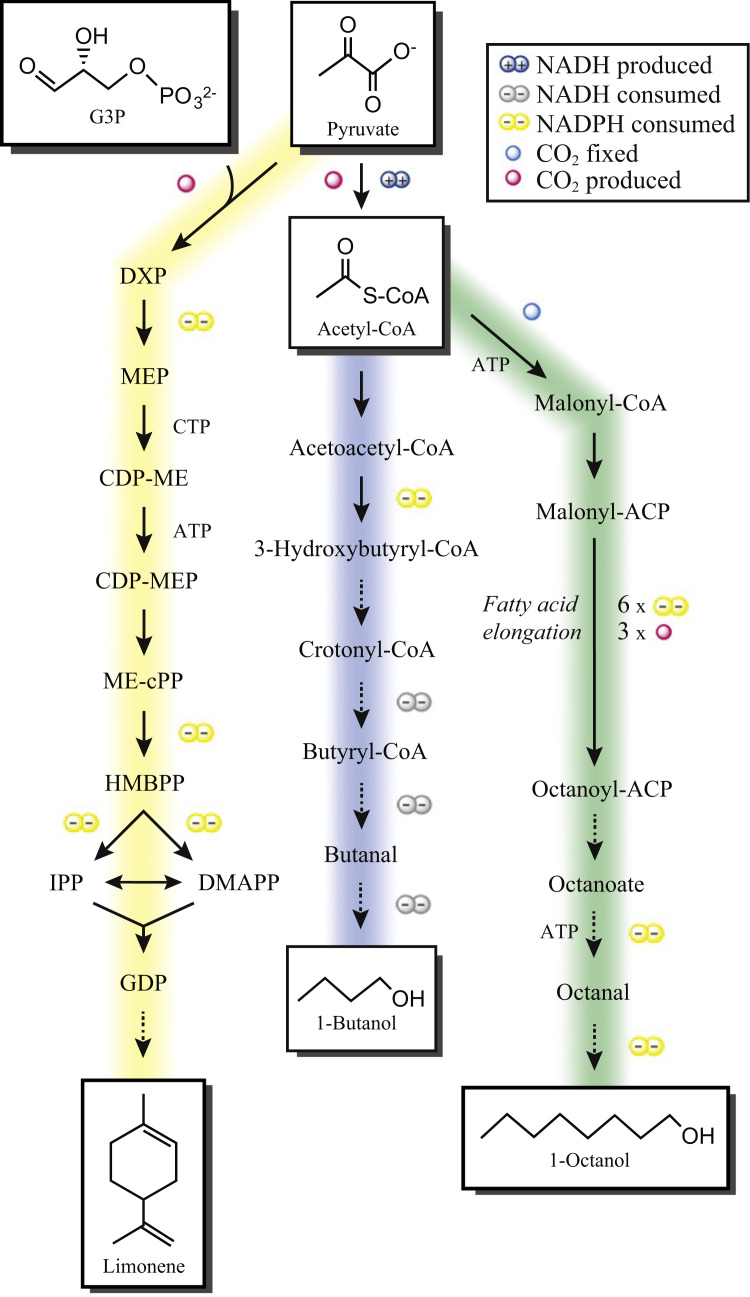
Several stoichiometric GEMs are available for Synechocystis PCC 6803 and have been recently reviewed (Baroukh et al., 2015). We chose the iJN678 GEM due to its detail of linear electron flow (LEF) and alternative electron flow (AEF) reactions (Nogales et al., 2012). We added and removed several reactions from the GEM to make it consistent with current literature (see Section 2). Three non-native biofuel synthesis pathways were considered for photoautotrophic production (Fig. 1). The first is a chimeric version of the Clostridia 1-butanol fermentation pathway (Bond-Watts et al., 2011, Shen et al., 2011) that was recently implemented in Synechococcus (Lan and Liao, 2012) and Synechocystis (Anfelt et al., 2015). The fermentation pathway is characterized by demand of 3 NADH. 1-octanol was chosen as a representative of a fatty-acid derived alcohol. Here, fatty-ACP is hydrolyzed with a chain-length specific thioesterase; the subsequent fatty acid is reduced by carboxylic acid reductase and a native alcohol dehydrogenase. This pathway has a high NADPH and ATP demand. It has recently been implemented in E. coli (Akhtar et al., 2015) but not in cyanobacteria. Limonene was included as a representative of terpene-based biofuels and is one step from geranyl-diphosphate in the native MEP pathway (Davies et al., 2014). All pathways were added separately to iJN678 to form iJN678_ButFER, iJN678_OctFA and iJN678_Limonene (Table S2). Flux balance analysis of the GEMs using a biomass objective function produced no biofuel, indicating that optimal growth in photoautotrophic conditions does not require biofuel synthesis in order to satisfy mass balances. Maximum 1-butanol, 1-octanol and limonene productivities were 0.925, 0.463 and 0.37 mmol/gDW h (17, 15, and 12 mg/L/OD730/h, respectively), as calculated by solving FBA with biofuel secretion as the objective function.
Fig. 1.
Heterologous pathways to biofuels in Synechocystis. Blue, fermentative pathway ButFER is adapted from Anfelt et al. (2015): Green, fatty-acid derived pathway Oct_FA adapted from Akhtar et al. (2015). Yellow, Lim_TER limomene pathway from Davies et al. (2014). Dashed arrows indicate reactions not present in iJN678. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
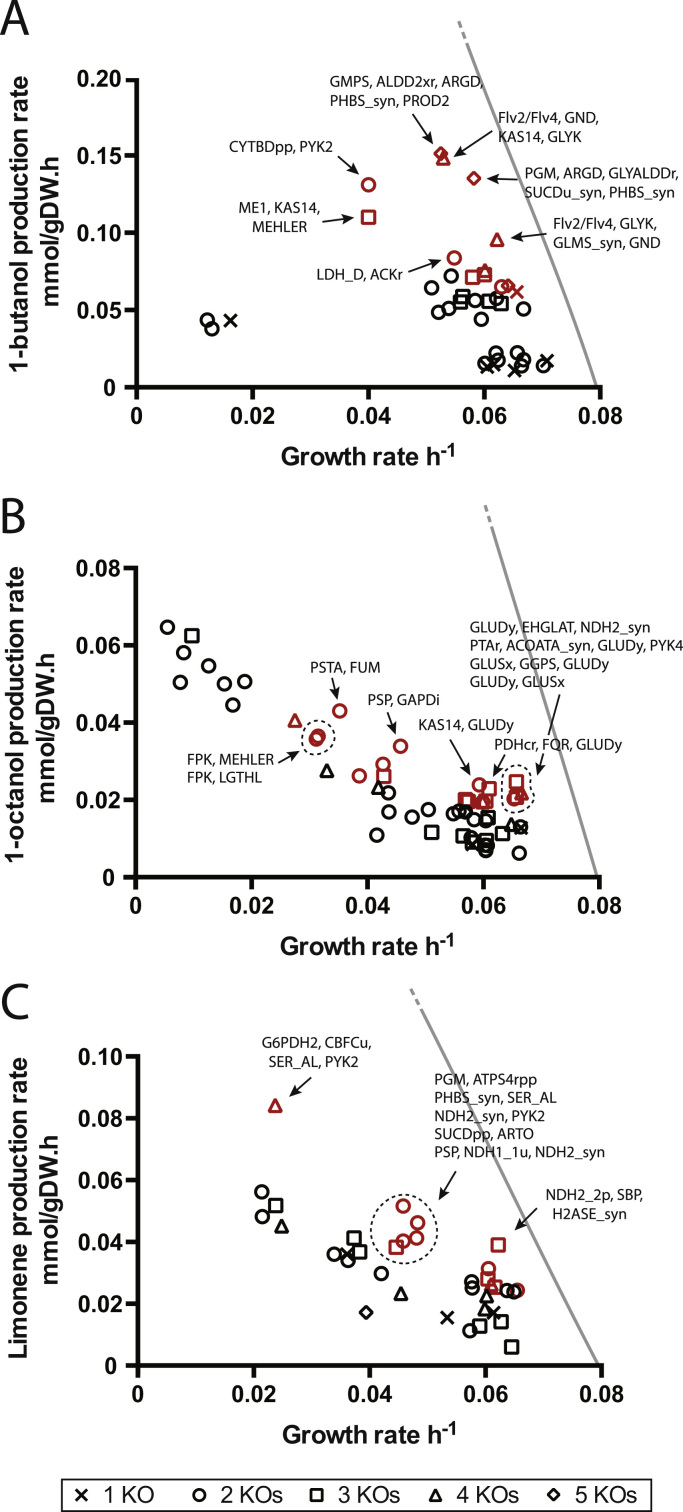
OptGene finds local optima in a given fitness metric by comparing randomly mutated strains to one another (Patil et al., 2005). We chose OptGene as a computationally fast way to uncover trends for improving productivity from the reference flux state (zero biofuel productivity). First, critical reactions were removed from consideration, resulting in Subset A (ca. 170 reactions, see Supplemental). Up to five reaction knockouts were allowed. Mutant intracellular fluxes were calculated using MOMA (minimization of metabolic adjustment) and the fitness metric was biomass-product coupled yield (BPCY). OptGene readily found mutations that increased BPCY for each biofuel. Because OptGene finds local optima from a random starting point, 3–5 sessions were run, each providing ca. 20 simulations (knockout strains). These strains were assessed for BPCY. To further avoid redundancy in the solutions, several reactions that appeared often were removed from the initial available pool, to form Subset B (ca. 150 reactions). Fig. 2 shows the production envelope for each biofuel as calculated with FBA, and selected OptGene-derived mutant strains are plotted with their biofuel and biomass fluxes as calculated by MOMA. A list of gene knockouts and corresponding BPCY calculations are in Supplemental.
Fig. 2.
Reaction knockout strategies that improve BPCY of selected biofuels in Synechocystis. OptGene simulation details are found in Section 2. In silico strains in the top 20% BPCY are in red. Selected reaction knockouts for some strains are listed. (A) iJN678_ButFER, (B) iJN678_OctFA and (C) iJN678_Limonene. The gray line is the phenotypic phase plane for butanol-biomass determined without 13C MFA constraints. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
In general, more knockouts increased BPCY, though there was a diminishing-returns effect for more than 4 knockouts. The productivities of all mutants were well below the maximal productivity for each fuel. Among the most common knockouts for 1-butanol were phosphoenolase (ENO) and phosphoglycerate mutase (PGM). These knockouts block pyruvate formation in lower glycolysis; flux is instead diverted through methylglyoxal to lactate, which is oxidized to pyruvate to generate NADH. This extra NADH is beneficial for 1-butanol production. Alternatively, the phosphoketolase (FPK) is used to produce acetyl-CoA from Calvin-cycle intermediates (Anfelt et al., 2015). In fact, ENO and PGM were such prevalent knockouts from OptGene that we were forced to exclude them from consideration to find more diverse solutions (Subset B). In the fatty-acid 1-octanol pathway, a recurring theme for increasing BPCY was knockout of alternative-electron flow reactions, such as the MEHLER reaction and Flv2/Flv4, which could have the effect of reducing NADPH consumption. Knockout of glutamate dehydrogenase (GLUDy, NADPH-utilizing) forces NH4 uptake to occur through the ATP-consuming glutamine synthase. This knockout thus reduces NADPH consumption and increases ATP consumption. Glutamate dehydrogenase was also identified by MOMA as a knockout target for increasing ethanol and butanol production in Synechococcus PCC 7002 (Hendry et al., 2016). One set of knockouts for increasing limonene BCPY involved the pyruvate synthesis reaction SER_AL (serine-ammonia lyase). This is counter-intuitive, as pyruvate is a necessary metabolite in the MEP pathway leading to limonene. However, knockout of these routes to pyruvate result in flux through other pyruvate-generating pathways. Furthermore, the BPCY objective function maximizes the product of growth and limonene synthesis, so that a tradeoff between these two is sought.
3.2. Model-derived mutants with growth-coupled biofuel production
OptGene was useful in identifying gene knockouts that could be expected to increase BPCY. However, the predicted flux distributions are calculated using MOMA, and may not be indicative of long-term flux distributions, which tend to optimize biomass formation (Fong and Palsson, 2004). We next used OptKnock to find gene knockouts that would result in biofuel production at FBA-predicted flux distributions (Burgard et al., 2003). Since OptKnock uses biomass optimization to predict fluxes, more interventions are required to get non-zero product synthesis than for MOMA-based prediction.
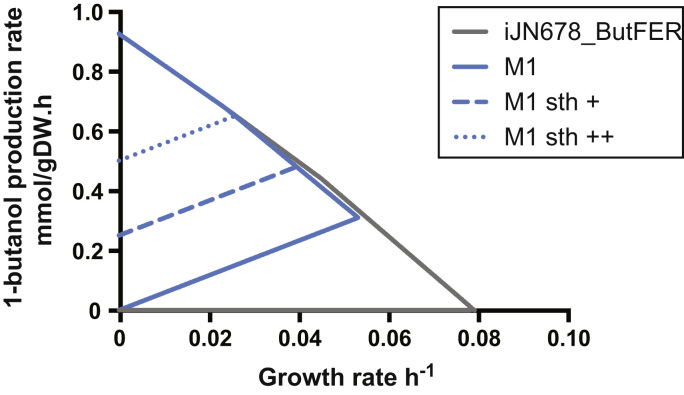
OptKnock was run first on iJN678_ButFER and up to 10 reaction knockouts were allowed, from a pool of 325 (Methods). Based on reaction knockouts proposed by OptKnock, we devised two mutants (M1 and M2) that have a strong coupling of 1-butanol and growth, i.e. 1-butanol production is predicted at all growth rates. The M1 knockouts are listed in Table 1. The production envelope of M1 is significantly constrained compared to iJN678_ButFER (Fig. 3). The biofuel-growth coupling could likely be strengthened with more knockouts. At maximum growth rate under LLS (Methods), the M1 strain is predicted to produce butanol at 0.3 mmol/gDW h, which is 33% of the theoretical maximum rate and 6-fold higher than the highest experimental report, which was estimated to be 0.05 mmol/gDW h from a butanol-producing Synechococcus PCC7942 at similar conditions (Lan et al., 2013). The M2 strategy required high flux through pyruvate: ferredoxin oxidoreductase, known to be oxygen sensitive (McNeely et al., 2011), and through methylglyoxal, a toxic electrophile (see flux map Fig. S1). For these reasons, M2 is considered to not be physiologically feasible and is not discussed further. We note that the M1 strategy also provides growth-biofuel coupling for longer-chain alcohols (Fig. S2) produced via reverse-beta oxidation (Dellomonaco et al., 2011).
Fig. 3.
In silico mutants that couple growth and 1-butanol. The iJN678_ButFER is not butanol-growth coupled. The M1 mutant has a restricted flux space and shows a strong coupling between growth and butanol. Forcing flux through a transhydrogenase reaction (Sth, NADPH+NAD+→NADH+NADP+) strengthens the coupling in M1. “Sth+” 0.25 mmol/gDW h, “Sth++” 0.50 mmol/gDW h. Simulation conditions are described in Section 2.
We next ran OptKnock for the fatty-acid derived 1-octanol pathway (iJN678_Oct_FA) but did not find growth-coupled strategies after several attempts and extended simulation time (up to 24 h). This suggests that coupling from this pathway cannot be achieved with knockouts alone, or with fewer than 10 knockouts. As an alternative to OptKnock, we used OptForce, which tests not only knockouts but also reaction “knock-ups” (increased flux) and “knock-downs” (reduced flux) (Ranganathan et al., 2010). OptForce was able to find a set of interventions that coupled 1-octanol production and growth. This mutant (M3) is described in Table 2. As discussed below, M3 contains several deletions in alternative electron flow reactions and requires upregulation of the ferredoxin: NADPH oxidoreductase reaction.
Table 2.
Reaction interventions needed to create mutant M3 and enable 1-octanol-coupled growth in iJN678_OctFA.
| Reaction name in iJN678 | Enzyme (s) | Reaction* | Locus to target** |
|---|---|---|---|
| NDH1_1u | NAD(P)H dehydrogenase NDH-1 (thylakoid) | 4h[c]+nadph[c]+pq[p]→nadp[c]+3h[p]+pqh2[p] | slr0331 (ndhD1) and slr1291 (ndhD2) |
| NDH1_2u | NAD(P)H dehydrogenase NDH-1 (thylakoid) | 4h[c]+nadh[c]+pq[u]→nad[c]+3h[u]+pqh2[u] | slr0331 (ndhD1) and slr1291 (ndhD2) |
| NDH2_syn | NdbA, NdbB, NdbC (thylakoid) | h[c]+nadh[c]+pq[u]→nad[c]+pqh2[u] | slr0851, slr1743, and sll1484 |
| NDH2_2p | NdbA, NdbB, NdbC (periplasm) | h[c]+nadh[c]+pq[p]→nad[c]+pqh2[p] | slr0851, slr1743, and sll1484 |
| NDH1_3u | Active CO2 transporter facilitator (thylakoid) | 3h[c]+h2o[c]+nadph[c]+pq[u]+co2[p]→nadp[c]+hco3[c]+3h[u]+pqh2[u] | sll1733 (ndhD3) and sll0027 (ndhD4) |
| Mehler | Flavodiiron proteins Flv1 and Flv3 | h[c]+0.5 o2[c]+nadph[c]→h2o[c]+nadp[c] | sll1521 (flv1) |
| sll0550 (flv3) | |||
| Cyo1b_syn | Cytochrome c oxidase | 4h[c]+2 focytc6[u]+0.5 o2[u]→2h[u]+2 ficytc6[u]+ h2o[u] | slr1137 |
| GLYCTO1 | Glycolate oxidase | o2[c]+glyclt[c]→h2o2[c]+glx[c] | sll0404 (glcD2) |
| GLUSx | Glutamate synthase GOGAT (NADH-dependent) | h[c]+nadh[c]+akg[c]+gln-L[c]→nad[c]+2 glu-L[c] | sll1502 |
| ACKr | Acetate kinase | atp[c]+ac[c]⇔adp[c]+actp[c] | sll1299 |
| H2ase_syn | [NiFe] Hydrogenase | h[c]+ nadph[c]⇔nadp[c]+h2[c] | sll1224 (hoxY) |
| ATPS4rpp | ATP synthase (periplasmic) | 3 adp[c]+3 pi[c]+14h[p]→3 atp[c]+11h[c]+3 h2o[c] | slr1330 (atpE) |
| FNOR*** | Ferredoxin: NADP+reductase | h[c]+nadp[c]+2 fdxr-2:2[c]⇔nadph[c]+2 fdxo-2:2[c] | slr1643 |
[c] cytoplasmic, [u] thylakoid, [p] periplasmic compartments.
Locus to target is suggestion for gene deletion to eliminate enzyme activity. For multi-domain proteins a core subunit is given. NDH-1 (Battchikova et al., 2011), GlcD2 (Eisenhut et al., 2008), Hox (Eckert et al., 2012), AtpE (Imashimizu et al., 2011).
Overexpression required.
3.3. Flux differences in OptKnock and OptForce -derived in silico mutant strains
3.3.1. NADH recycling as driving force for butanol-growth coupling in ButFER
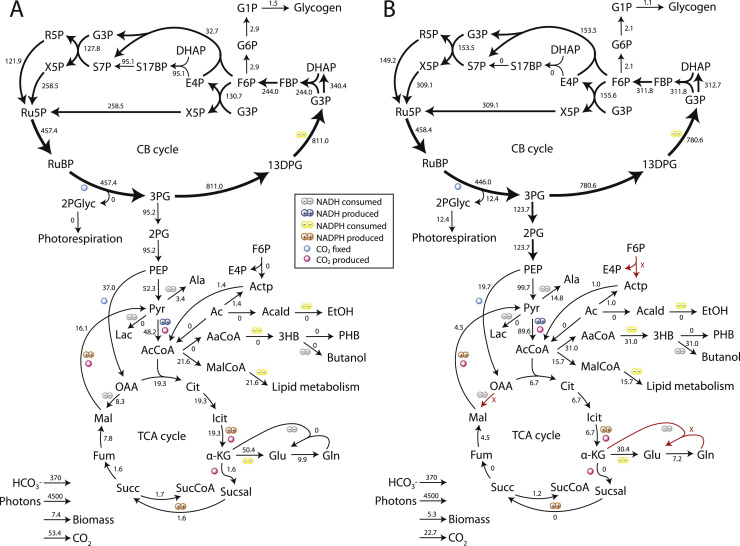
From inspection of Table 1 it is clear that most knockouts in M1 are involved in NADH metabolism. Phosphoketolase (FPK), which was recently shown to benefit butanol production during light-limited cultivation (Anfelt et al., 2015), is part of a bypass around the NADH-generating pyruvate dehydrogenase (Pdh) and must therefore be knocked out. Additionally, knockout of NADH-dependent GOGAT enzyme in M1 forces nitrogen uptake through glutamate dehydrogenase (Gdh, NH3+, NADPH-dependent). There is experimental evidence that Gdh is active in Synechocystis and is involved in nitrate uptake in light-limited conditions (Chávez et al., 1999). Comparison of the flux distributions in M1 and iJN678 at maximal growth show that M1 has increased flux through Pdh, decreased flux through the TCA cycle and respiratory electron transport chain, and an increased flux in carbon fixation (Fig. 4). The primary sources of NADH in both strains are acetyl-CoA biosynthesis by Pdh and serine biosynthesis from 3PGA. The primary NADH usage in iJN678 is alanine biosynthesis and in M1 it is butanol.
Fig. 4.
Flux distributions of iJN678_ButFER and mutant M1. Fluxes were calculated using FBA with a biomass formation objective function in light-limited condition (see Section 2). A) iJN678_ButFER, constrained with 13C MFA data (see Section 2) B) mutant M1, which was not constrained with 13C MFA data. Flux values are in mmol/gDW h (*10−2) except for the flux to biomass (h−1).
The M1 strain suggests that NADH recycling is an important lever for improving butanol productivity, which we tested in several ways. When we added a generic NADH “burning” reaction to M1, coupling was lost. When we added a generic NADH-generation reaction to iJN678 and forced flux through it at 20 mmol/gDW h, NDH-1 and NDH-2 dissipated the increased NADH. If these reactions were held at low fluxes, NADH was instead dissipated via cytochrome oxidases or glutamate synthase. These NADH dissipating reactions overlap with those suggested by OptKnock and support the theory that butanol is a necessary NADH valve in M1. Importantly, the M1 strategy provides biofuel-biomass coupling for longer chain alcohols produced via the reverse beta-oxidation pathway (Dellomonaco et al., 2011)(Fig. S2), as well as the NphT7, ATP-dependent butanol pathway (Lan and Liao, 2012).
Recent strategies to increase productivities in cyanobacteria have focused on utilizing NADPH-dependent enzymes or pathways, (Angermayr et al., 2014, Oliver and Atsumi, 2014), in order to exploit the NADP pool in Synechocystis, which is 5-fold larger than the NAD pool (Cooley and Vermaas, 2001). However, only a few knockouts are needed to force butanol productivity when NADH-dependent enzymes are used. Addition of a soluble transhydrogenase (Sth) to convert the NADPH pool to NADH could increase butanol productivity further, as reported for lactate production in Synechocystis (Angermayr et al., 2012, Varman et al., 2013). We tested the effect of forcing flux through the Sth reaction (NADPH+NAD+→NADH+NADP+) and found that this extra NADH generation strengthened the coupling and increased butanol productivity at maximal growth rate (Fig. 3). It should be noted, however, that stronger coupling reduced the maximal growth rate. Simulation of batch-phase growth and butanol production showed that weaker butanol-growth couplings would give higher final butanol titers if starting culture density was low (Suppl Note 1). A growth-biofuel tradeoff is partially captured by the BPCY metric used in OptGene.
3.3.2. ATP/NADPH ratio as a driving force for fatty-acid and terpene biofuels
Fatty-acid derived biofuels have a high NADPH requirement but since NADPH is used in anabolic reactions, it is not possible to eliminate NADPH sinks while retaining growth. An alternative strategy to force product-growth coupling is to alter fluxes of ATP and NADPH so as to create an imbalance in the ATP/NADPH ratio (Kramer and Evans, 2011) that can only be alleviated by production of fuel. The Synechocystis linear electron flow (LEF) generates ATP/NADPH at a ratio of 1.28–1.5, depending on the exact stoichiometry of the ATP synthase. CO2 fixation, biomass formation, and other maintenance reactions create an overall demand of ATP/NADPH>2 (Knoop and Steuer, 2015). Therefore, additional ATP generating reactions such as cyclic electron flow (CEF) or alternative electron flow (AEF) are employed to ensure that ATP and NADPH are supplied in the proper proportion. If these ATP-generating reactions are knocked out, fluxes of NADPH-consuming reactions, such as biofuel synthesis, must increase in order to allow growth and satisfy mass balances. For example, Erdich et al. used elementary flux mode analysis of a cyanobacteria GEM containing ethanol synthesis reactions (ATP/NADPH=1) and found that elementary metabolic modes (EMs) where ethanol and growth occurred simultaneously carried no flux through several ATP-generating AEF reactions (Erdrich et al., 2014).
The M3 reaction knockouts for fatty-acid derived 1-octanol follow this strategy. The in silico M3 strain is weakly coupled and produces 1-octanol at high growth rates (Fig. 5A). Components of CEF and AEF that could pump protons into the thylakoid lumen and thus generate ATP are required knockouts (see schematic, Fig. 6). The ATP-forming acetate kinase is a required knockout as is the periplasmic ATP synthase. The latter is not experimentally possible, since a knockout of ATP synthase genes would eliminate both periplasmic and thylakoid ATP production. We interpret this knockout as a need to further lower ATP production and ATP synthesis was reduced in M3 compared to wild type by 7.5% (5.3–4.9 mmol/DW h). This reduced ATP production lowers the LLS maximum theoretical productivity of M3 relative to iJN678_OctFA. M3 also requires knockout of several NADPH-consuming reactions, including both the CEF and CO2 uptake reactions of NDH-1 dehydrogenase. A knockout of NDH-1 has been reported, but was not viable at high light (>200 μE) due to the absence of CEF (Battchikova et al., 2011). This was confirmed in the model: no growth was observed above a simulated light intensity of 100 μE when CEF was removed (data not shown, also (Nogales et al., 2012)). OptForce also required that NADPH generation be increased so that flux through ferredoxin-NADP+ oxidoreductase (FNOR) was required to be 34% higher than in iJN678_OctFA (6.7–9.0 mmol/gDW.h). However, increase in FNOR flux above 9.5 mmol/gDW h resulted in no FBA solution, presumably due to inability to dissipate the excess NADPH. The M3 knockouts also give growth coupling for other fatty-acid derived products, including longer chain alcohols and alkanes (Fig. S5-S6).
Fig. 5.
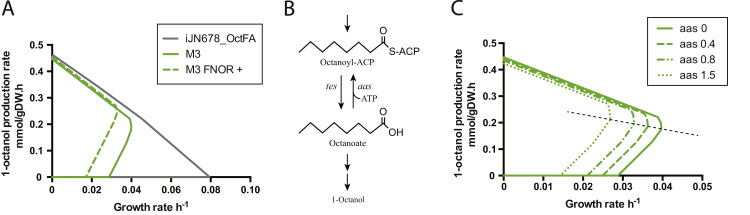
In silico mutants that couple growth and 1-octanol production. A) The M3 mutant has a restricted flux space and shows a weak coupling between growth and 1-octanol. M3 requires increased flux through the ferredoxin:NADP reductase (FNOR), relative to iJN678_OctFA (9.0 and 6.7 mmol/gDW h, respectively). Increasing flux to 9.5 mmol/gDW h (M3 FNOR +) strengthens coupling. Simulation conditions are described in Section 2. B) A proposed futile cycle using acyl-ACP synthetase (Aas) and a thioesterase (Tes) to consume ATP during production of 1-octanol. C) Forcing flux through the Aas reaction strengthens coupling in the M3 mutant.
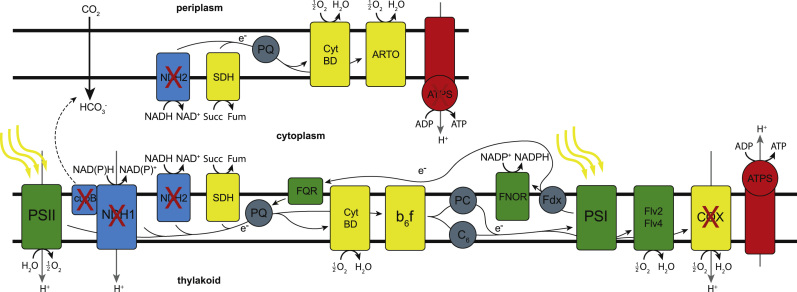
Fig. 6.
Electron transport chain in Synechocystis as modeled in iJN678. Reactions that are knocked out in M3 are indicated. Green, photosynthesis proteins; Yellow, respiratory proteins; Blue, cyclic electron flow proteins. NAD(P)H indicates that both NADH and NADPH-utilizing reactions are present in the model. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
An alternative to knockout of ATP-producing reactions is to increase ATP-consumption (Adolfsen and Brynildsen, 2015, Hädicke et al., 2015). One potential ATP-burning futile cycle for fatty-acid derived products is the ATP-dependent activation with ACP and subsequent thiolysis of the fatty acid intermediate. Thioesterases (Jing et al., 2011) and acyl-ACP synthetases (Kaczmarzyk et al., 2016) with varying chain-length specificity have been characterized. We incorporated the Tes/Aas futile cycle into the M3 model and forced flux through, which significantly strengthened the coupling between growth and 1-octanol (Fig. 5C). A similar strengthening of coupling was observed when either a phosphoenolpyruvate synthase/pyruvate kinase and adenylate kinase or a glutamine synthase/glutaminase futile cycle was implemented (Fig. S7-S8). Implementation of the former cycle increased specific productivity of lactate in E. coli, despite consuming pyruvate precursor (Hädicke et al., 2015). Together, these results indicate that the product-growth coupling can be modulated by ATP-burning futile cycles, but at the expense of growth (see also Supplemental Note 1). However, we note that futile cycles may increase susceptibility to reactive oxygen species (Adolfsen and Brynildsen, 2015).
Limonene biosynthesis requires a relatively low number of NADPH (4 NADPH/10C) compared to octanol (8 NADPH/8C). Furthermore, there are no reduction steps in the limonene pathway after the biomass/limonene branch point at geranyl diphosphate, so that flux diversion to limonene at this point does not recycle NADPH or NADH. Nevertheless, we were able to find a product-growth coupling strategy for limonene using OptForce, which requires a significant number of reaction knockouts (Table S6, Fig. 7C). Similar to the coupling strategy for fatty-acid based fuels, components of the CEF and AEF were required knockouts and the ATP synthase flux must be restricted by at least 16% relative to wild type (5.4–4.5 mmol/DW h). This significant downregulation is necessary due to the low NADPH demand of limonene; when we added an NADPH oxidation reaction to the terminal synthesis step (4 NADPH oxidized), coupling was possible without knockdown of ATP synthase.
Fig. 7.
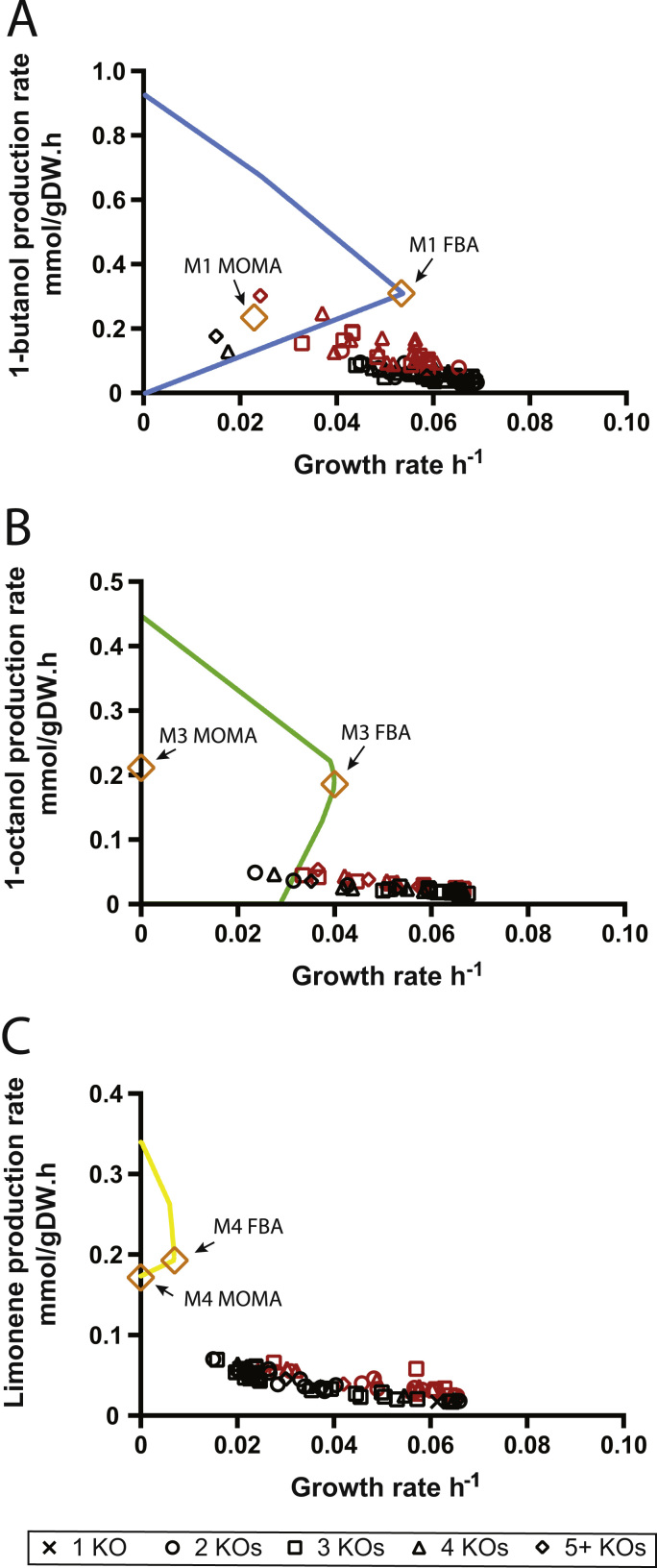
Comparison of M1, M3, and M4 phenotypes predicted by MOMA or FBA. Flux distributions for M1, M3, and M4 mutants were calculated using MOMA or FBA (yellow squares). Also pictured are OptGene-derived mutants that were found by only considering those reaction knockouts present in M1, M3, and M4 (Subset C); fluxes are predicted by MOMA. Black (bottom 80% in BPCY) and red (top 20% in BCPY). A) 1-butanol. B) 1-octanol. C) limonene. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
4. Discussion
We present genetic engineering strategies to obtain cyanobacteria strains where biofuel productivity is increased or coupled to growth. An advantage of growth-coupled production strains is that adaptive evolution can be used to improve both growth and production rate, as the strain evolves to the FBA-predicted flux distribution. To visualize an evolutionary progression for the M1 and M3 strains, we used MOMA to estimate the flux distributions after these knockouts. The MOMA-predicted flux of M1 was quite favorable, resulting in high butanol productivity. However, MOMA predicted no growth for strains M3 and M4 (Fig. 7). This is likely due to the large number of gene knockouts needed to couple growth to biofuel, which results in a massive perturbation from the reference flux. It is likely that not all knockouts are needed to improve biofuel productivity. The M1, M3, and M4 reaction knockouts could be used to limit the reactions “pool” considered by OptGene, which could then be used find the effective combinations. When we recomputed OptGene with only M1, M3, and M4 knockouts considered (Subset C), the BPCY of mutants for 1-butanol, 1-octanol, and limonene were generally higher than for when the larger subsets were considered (Fig. 7).
A combination of methods for predicting intracellular fluxes is useful since the true objective function may not be biomass formation, but rather ATP generation, a minimization of total intracellular flux, or a combination of these (Bordbar et al., 2014). Cyanobacteria in particular pose a challenge to the biomass assumption since their flexible metabolism, which provides robustness necessary for survival in a fluctuating marine environment, can be considered suboptimal for growth. Recent works have explored cyanobacteria metabolism under diurnal cycles and assumed that the cellular objective function changes depending on external conditions (Knoop et al., 2013, Rügen et al., 2015; Saha et al., 2016). With this caveat in mind, the general themes of the two coupling strategies appear to be robust and are consistent with strategies found using other methods. The M1 knockout strategy applies to other alcohols produced via reverse beta-oxidation and our work shows that modulating ATP and NADPH production can be applied to a range of products with different energy requirements. Our strategies for fatty-acid derived 1-octanol (M3) and limonene (M4) also have several overlapping knockouts with a recent work that used Elementary Mode Analysis to find knockouts which coupled ethanol production to growth in cyanobacteria (Erdrich et al., 2014). The EMA approach has the benefit of visualizing many possible flux distributions (elementary modes) within the allowed flux space, including those that are not maximal growth. A desired degree of product-biomass coupling can be defined and algorithms such as CASOP can be used to compute which reaction deletions are needed to remove all possible elementary modes outside of this cutoff (Klamt and Mahadevan, 2015).
The knockout strategies reported here were found to be somewhat model-dependent, as transfer to a second GEM did not result in coupling unless extra reactions were knocked out (Supplemental Note 2). The discrepancy among GEMs is a reflection of the incomplete knowledge of cyanobacteria metabolism, which influences the number of needed knockouts. For example, strategies that rely on co-factor imbalance require knowledge of the cofactor usage of an enzyme (King and Feist, 2013). Here the models tend to be conservative and if the co-factor preference of an enzyme is not known, then both are included. In the M1 reaction target list the NDH-1 reaction using NADH is a necessary knockout while the NDH-1 reaction using NADPH is not. The preferred co-factor for NDH-1 is presumed to be NADPH (Ma et al., 2006), though there is some experimental evidence supporting NADH activity (Ooyabu et al., 2008). The required knockout of GlcD2 glycolate oxidase (O2 requiring) in M1 is another illustrative example. The model iJN678 contains two glycolate oxidation reactions. The glycolate dehydrogenase GlcD1 (sll0404) catalyzes NAD+-dependent glycolate oxidation and does not have detectable O2-dependent oxidase activity (Eisenhut et al., 2006). A second glycolate dehydrogenase/oxidase GlcD2 (slr0806) was identified in Synechocystis, but its activity was not characterized (Eisenhut et al., 2008), so it is not known if GlcD2 has oxygen-dependent glycolate oxidase activity. The reaction may have been included in iJN678 based on the prevalence of oxygen-dependent glycolate oxidases in plants. Therefore, the knockouts of NADH-dependent NDH-1 and GlcD2 may not be necessary in practice to achieve coupling in M1. Of course, it is possible that as new reactions are experimentally verified, they will add to the reaction knockout list.
The flux capacity of reactions can also be constrained based on experimental evidence, such as enzyme kinetics and expression data (Reed, 2012) and could also obviate some knockouts. Integration of transcriptomics and proteomics data will improve the accuracy of the cyanobacteria GEM and could reduce the number of required knockouts. For example, three type-2 NADH dehydrogenases are required knockouts in the M1 strain, since they can oxidize NADH at high fluxes in silico. However, RNA-Seq showed that these type-2 NADH dehydrogenases are each expressed at less than 10% of the NDH-1 (Anfelt et al., 2013) and experimental evidence for their activity was not found (Howitt et al., 1999). A similar case can be made against pyruvate: ferredoxin oxidoreductase, which is another target in M1. This enzyme is known to be inhibited by O2 and is likely not active in photoautotrophic conditions (McNeely et al., 2011). Overall, these cofactor and flux constraints could reduce the required gene knockouts to achieve the M1 mutant butanol to 3 (FPK slr0453, GLUSx sll1502, MDH sll0891).
Even considering cofactor and flux constraints, the number of gene knockouts for the M3 (fatty alcohols) and M4 (limonene) strains is at least 6. A central question is whether these strains can be realized in practice. While there are several selection and counter-selection methods available for cyanobacteria (Begemann et al., 2013, Cheah et al., 2012, Viola et al., 2014), sequential gene knockout and antibiotic cassette curing is time-consuming and to date no strain with more than 4 knockouts has been reported. The type-II CRISPR/Cas tool is a powerful way to realize gene knockouts and has been applied recently in cyanobacteria for single knockouts (Wendt et al., 2016). A CRISPRi knockdown tool could repress four genes simultaneously at 50–90% in Synechocystis (Yao et al., 2015). One limitation of CRISPRi is that it is not possible to knockdown just one gene in an operon. Therefore, more advanced genetic engineering techniques must be developed in cyanobacteria in order to perform systems-level engineering.
5. Conclusions
We used available genome-scale models and algorithms and found metabolic engineering strategies for creating growth-coupled cyanobacteria biofuel strains. The relative dearth of NADH-utilizing reactions in Synechocystis allowed for coupling of fermentative butanol with relatively few gene knockouts. Lowering the ATP/NADPH ratio in the cell is a general approach for coupling fatty-acid derived products such as alcohols and alkanes and terpenes such as limonene. Advances in genome engineering techniques will allow testing of these genetic interventions and integration of systems-biology data will refine the models to be more accurate.
Acknowledgements
We are grateful to Markus Herrgård (DTU, NNF Center for Biosustainability) for comments and advice. This work is funded by the Swedish Foundation for Strategic Research (SSF) grant number RBP14–0013 and with a Science for Life Laboratory Fellowship (B-2013-0201to EPH).
Footnotes
Supplementary data associated with this article can be found in the online version at 10.1016/j.meteno.2016.07.003.
Contributor Information
Kiyan Shabestary, Email: kiyan@kth.se.
Elton P. Hudson, Email: paul.hudson@biotech.kth.se.
Appendix A. Supplementary material
Supplementary material
.
References
- Adolfsen K.J., Brynildsen M.P. Futile cycling increases sensitivity toward oxidative stress in Escherichia coli. Metab. Eng. 2015;29:26–35. doi: 10.1016/j.ymben.2015.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akhtar M.K., Dandapani H., Thiel K., Jones P.R. Microbial production of 1-octanol: a naturally excreted biofuel with diesel-like properties. Metab. Eng. Commun. 2015;2:1–5. doi: 10.1016/j.meteno.2014.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anfelt J., Hallström B., Nielsen J., Uhlén M., Hudson E.P. Using transcriptomics to improve butanol tolerance of Synechocystis sp. strain PCC 6803. Appl. Environ. Microbiol. 2013;79:7419–7427. doi: 10.1128/AEM.02694-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anfelt J., Kaczmarzyk D., Shabestary K., Rockberg J., Renberg B., Uhlen M., Nielsen J., Hudson E.P. Genetic and nutrient modulation of acetyl-CoA levels in Synechocystis for n-butanol production. Microb. Cell Fact. 2015;14:1–12. doi: 10.1186/s12934-015-0355-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angermayr S.A., Paszota M., Hellingwerf K.J. Engineering a cyanobacterial cell factory for production of lactic acid. Appl. Environ. Microbiol. 2012;78:7098–7106. doi: 10.1128/AEM.01587-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angermayr S.A., van der Woude A.D., Correddu D., Vreugdenhil A., Verrone V., Hellingwerf K.J. Exploring metabolic engineering design principles for the photosynthetic production of lactic acid by Synechocystis sp. PCC6803. Biotechnol. Biofuels. 2014;7:99. doi: 10.1186/1754-6834-7-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baroukh C., Muñoz-Tamayo R., Steyer J.-P., Bernard O. A state of the art of metabolic networks of unicellular microalgae and cyanobacteria for biofuel production. Metab. Eng. 2015;30:49–60. doi: 10.1016/j.ymben.2015.03.019. [DOI] [PubMed] [Google Scholar]
- Battchikova N., Eisenhut M., Aro E.M. Cyanobacterial NDH-1 complexes: novel insights and remaining puzzles. Biochim. Biophys. Acta Bioenerg. 2011;1807:935–944. doi: 10.1016/j.bbabio.2010.10.017. [DOI] [PubMed] [Google Scholar]
- Begemann M.B., Zess E.K., Walters E.M., Schmitt E.F., Markley A.L., Pfleger B.F. An organic acid based counter selection system for cyanobacteria. PLoS One. 2013;8:1–12. doi: 10.1371/journal.pone.0076594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bond-Watts B.B., Bellerose R.J., Chang M.C.Y. Enzyme mechanism as a kinetic control element for designing synthetic biofuel pathways. Nat. Chem. Biol. 2011;7:1–6. doi: 10.1038/nchembio.537. [DOI] [PubMed] [Google Scholar]
- Bordbar A., Monk J.M., King Z. a, Palsson B.O. Constraint-based models predict metabolic and associated cellular functions. Nat. Rev. Genet. 2014;15:107–120. doi: 10.1038/nrg3643. [DOI] [PubMed] [Google Scholar]
- Burgard A.P., Pharkya P., Maranas C.D. OptKnock: a bilevel programming framework for identifying gene knockout strategies for microbial strain optimization. Biotechnol. Bioeng. 2003;84:647–657. doi: 10.1002/bit.10803. [DOI] [PubMed] [Google Scholar]
- Chávez S., Lucena J.M., Reyes J.C., Florencio F.J., Candau P. The presence of glutamate dehydrogenase is a selective advantage for the Cyanobacterium synechocystis sp. strain PCC 6803 under nonexponential growth conditions. J. Bacteriol. 1999;181:808–813. doi: 10.1128/jb.181.3.808-813.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheah Y.E., Albers S.C., Peebles C. a M. A novel counter-selection method for markerless genetic modification in Synechocystis sp. PCC 6803. Biotechnol. Prog. 2012;29:23–30. doi: 10.1002/btpr.1661. [DOI] [PubMed] [Google Scholar]
- Choi S., Song C.W., Shin J.H., Lee S.Y. Biorefineries for the production of top building block chemicals and their derivatives. Metab. Eng. 2015;28:223–239. doi: 10.1016/j.ymben.2014.12.007. [DOI] [PubMed] [Google Scholar]
- Chowdhury A., Zomorrodi A.R., Maranas C.D. Bilevel optimization techniques in computational strain design. Comput. Chem. Eng. 2015;72:363–372. [Google Scholar]
- Cooley J.W., Vermaas W.F.J. Succinate dehydrogenase and other respiratory pathways in thylakoid membranes of Synechocystis sp. Strain PCC 6803: capacity comparisons and physiological function succinate dehydrogenase and other respiratory pathways in thylakoid membranes of synechoc. J. Bacteriol. 2001:4251–4258. doi: 10.1128/JB.183.14.4251-4258.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies F.K., Work V.H., Beliaev A.S., Posewitz M.C. Engineering limonene and bisabolene production in wild type and a glycogen-deficient mutant of Synechococcus sp. PCC 7002. Front. Bioeng. Biotechnol. 2014;2:21. doi: 10.3389/fbioe.2014.00021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dellomonaco C., Clomburg J.M., Miller E.N., Gonzalez R. Engineered reversal of the β-oxidation cycle for the synthesis of fuels and chemicals. Nature. 2011;476:4–10. doi: 10.1038/nature10333. [DOI] [PubMed] [Google Scholar]
- Eckert C., Boehm M., Carrieri D., Yu J., Dubini A., Nixon P.J., Maness P.C. Genetic analysis of the Hox hydrogenase in the cyanobacterium Synechocystis sp. PCC 6803 reveals subunit roles in association, assembly, maturation, and function. J. Biol. Chem. 2012;287:43502–43515. doi: 10.1074/jbc.M112.392407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenhut M., Kahlon S., Hasse D., Ewald R., Lieman-Hurwitz J., Ogawa T., Ruth W., Bauwe H., Kaplan A., Hagemann M. The plant-like C2 glycolate cycle and the bacterial-like glycerate pathway cooperate in phosphoglycolate metabolism in cyanobacteria. Plant Physiol. 2006;142:333–342. doi: 10.1104/pp.106.082982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenhut M., Ruth W., Haimovich M., Bauwe H., Kaplan A., Hagemann M. The photorespiratory glycolate metabolism is essential for cyanobacteria and might have been conveyed endosymbiontically to plants. Proc. Natl. Acad. Sci. USA. 2008;105:17199–17204. doi: 10.1073/pnas.0807043105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdrich P., Knoop H., Steuer R., Klamt S. Cyanobacterial biofuels: New insights and strain design strategies revealed by computational modeling. Microb. Cell Fact. 2014;13:128. doi: 10.1186/s12934-014-0128-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feist A.M., Zielinski D.C., Orth J.D., Schellenberger J., Herrgard M.J., Palsson B.O. Model-driven evaluation of the production potential for growth-coupled products of Escherichia coli. Metab. Eng. 2010;12:173–186. doi: 10.1016/j.ymben.2009.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong S.S., Burgard A.P., Herring C.D., Knight E.M., Blattner F.R., Maranas C.D., Palsson B.O. In silico design and adaptive evolution of Escherichia coli for production of lactic acid. Biotechnol. Bioeng. 2005;91:643–648. doi: 10.1002/bit.20542. [DOI] [PubMed] [Google Scholar]
- Fong S.S., Palsson B.Ø. Metabolic gene-deletion strains of Escherichia coli evolve to computationally predicted growth phenotypes. Nat. Genet. 2004;36:1056–1058. doi: 10.1038/ng1432. [DOI] [PubMed] [Google Scholar]
- Gudmundsson S., Nogales J. Cyanobacteria as photosynthetic biocatalysts: a systems biology perspective. Mol. BioSyst. 2014 doi: 10.1039/c4mb00335g. [DOI] [PubMed] [Google Scholar]
- Hädicke O., Bettenbrock K., Klamt S. Enforced ATP futile cycling increases specific productivity and yield of anaerobic lactate production in Escherichia coli. Biotechnol. Bioeng. 2015;9999 doi: 10.1002/bit.25623. n/a–n/a. [DOI] [PubMed] [Google Scholar]
- Hendry J.I., Prasannan C.B., Joshi A., Dasgupta S., Wangikar P.P. Biores. Technol. 2016. Metabolic model of Synechococcus sp. PCC 7002: prediction of flux distribution and network modification for enhanced biofuel production. [DOI] [PubMed] [Google Scholar]
- Howitt C. a, Udall P.K., Vermaas W.F. Type 2 NADH dehydrogenases in the cyanobacterium Synechocystis sp. strain PCC 6803 are involved in regulation rather than respiration. J. Bacteriol. 1999;181:3994–4003. doi: 10.1128/jb.181.13.3994-4003.1999. doi:0021–9193/99/$04.00+0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyduke D., Schellenberger J., Que R., Fleming R., Thiele I., Orth J., Feist A., Zielinski D., Bordbar A., Lewis N., Rahmanian S., Kang J., Palsson B. 2011. COBRA Toolbox 2.0. Protoc. Exch. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imashimizu M., Bernát G., Sunamura E., Broekmans M., Konno H., Isato K., Rögner M., Hisabori T. Regulation of F0F1-ATPase from Synechocystis sp. PCC 6803 by gamma and epsilon subunits is significant for light/dark adaptation. J. Biol. Chem. 2011;286:26595–26602. doi: 10.1074/jbc.M111.234138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y., Liu J., Jiang W., Yang Y., Yang S. Current status and prospects of industrial bio-production of n-butanol in China. Biotechnol. Adv. 2014;2014:1–9. doi: 10.1016/j.biotechadv.2014.10.007. [DOI] [PubMed] [Google Scholar]
- Jing F., Cantu D.C., Tvaruzkova J., Chipman J.P., Nikolau B.J., Yandeau-Nelson M.D., Reilly P.J. Phylogenetic and experimental characterization of an acyl-ACP thioesterase family reveals significant diversity in enzymatic specificity and activity. BMC Biochem. 2011;12:44. doi: 10.1186/1471-2091-12-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaczmarzyk D., Hudson E.P., Fulda M. Arabidopsis acyl-acyl carrier protein synthetase AAE15 with medium chain fatty acid specificity is functional in cyanobacteria. AMB Express. 2016;6:7. doi: 10.1186/s13568-016-0178-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King Z. a, Feist A.M. Optimizing cofactor specificity of oxidoreductase enzymes for the generation of microbial production strains—OptSwap. Ind. Biotechnol. 2013;9:236–246. [Google Scholar]
- Klamt S., Mahadevan R. On the feasibility of growth-coupled product synthesis in microbial strains. Metab. Eng. 2015;30:166–178. doi: 10.1016/j.ymben.2015.05.006. [DOI] [PubMed] [Google Scholar]
- Klemke F., Baier A., Knoop H., Kern R., Jablonsky J., Beyer G., Volkmer T., Steuer R., Lockau W., Hagemann M. Identification of the light-independent phosphoserine pathway as additional source for serine in the cyanobacterium Synechocystis sp. PCC 6803. Microbiology. 2015;161:1050–1060. doi: 10.1099/mic.0.000055. [DOI] [PubMed] [Google Scholar]
- Knoop H., Gründel M., Zilliges Y., Lehmann R., Hoffmann S., Lockau W., Steuer R. Flux balance analysis of cyanobacterial metabolism: the metabolic network of Synechocystis sp. PCC 6803. PLoS Comput. Biol. 2013;9:e1003081. doi: 10.1371/journal.pcbi.1003081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoop H., Steuer R. A computational analysis of stoichiometric constraints and trade-offs in cyanobacterial biofuel production. Front. Bioeng. Biotechnol. 2015;3:1–15. doi: 10.3389/fbioe.2015.00047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer D.M., Evans J.R. The importance of energy balance in improving photosynthetic productivity. Plant Physiol. 2011;155:70–78. doi: 10.1104/pp.110.166652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan E.I., Liao J.C. ATP drives direct photosynthetic production of 1-butanol in cyanobacteria. Proc. Natl. Acad. 2012;109:1–6. doi: 10.1073/pnas.1200074109. doi:10.1073/pnas.1200074109/-/DCSupplemental.www.pnas.org/cgi/doi/10.1073/pnas.1200074109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan E.I., Ro S.Y., Liao J.C. Oxygen-tolerant coenzyme A-acylating aldehyde dehydrogenase facilitates efficient photosynthetic n-butanol biosynthesis in cyanobacteria. Energy Environ. Sci. 2013:2672–2681. [Google Scholar]
- Lea-Smith D.J., Bombelli P., Vasudevan R., Howe C.J. Photosynthetic, respiratory and extracellular electron transport pathways in cyanobacteria. Biochim. Biophys. Acta Bioenerg. 2015:1–9. doi: 10.1016/j.bbabio.2015.10.007. [DOI] [PubMed] [Google Scholar]
- Ma W., Deng Y., Ogawa T., Mi H. Active NDH-1 complexes from the cyanobacterium Synechocystis sp. strain PCC 6803. Plant Cell Physiol. 2006;47:1432–1436. doi: 10.1093/pcp/pcl008. [DOI] [PubMed] [Google Scholar]
- Machado D., Herrgård M. Co-evolution of strain design methods based on flux balance and elementary mode analysis. Metab. Eng. Commun. 2015;2:85–92. doi: 10.1016/j.meteno.2015.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNeely K., Xu Y., Ananyev G., Bennette N., Bryant D. a, Dismukes G.C. Synechococcus sp. strain PCC 7002 nifJ mutant lacking pyruvate: ferredoxin oxidoreductase. Appl. Environ. Microbiol. 2011;77:2435–2444. doi: 10.1128/AEM.02792-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogales J., Gudmundsson S., Knight E.M., Palsson B.O., Thiele I. Detailing the optimality of photosynthesis in cyanobacteria through systems biology analysis. Proc. Natl. Acad. Sci. USA. 2012;109:2678–2683. doi: 10.1073/pnas.1117907109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogales J., Gudmundsson S., Thiele I. Toward systems metabolic engineering in cyanobacteria: opportunities and bottlenecks. Bioengineered. 2013;4:37–41. doi: 10.4161/bioe.22792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien E.J., Monk J.M., Palsson B.O. Primer using genome-scale models to predict biological capabilities. Cell. 2015;161:971–987. doi: 10.1016/j.cell.2015.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver J.W.K., Atsumi S. Metabolic design for cyanobacterial chemical synthesis. Photosynth. Res. 2014;120:249–261. doi: 10.1007/s11120-014-9997-4. [DOI] [PubMed] [Google Scholar]
- Ooyabu J., Ohtsuka M., Kashino Y., Koike H., Satoh K. The expression pattern of NAD(P)H oxidases and the cyclic electron transport pathway around photosystem I of Synechocystis sp. PCC6803 depend on growth conditions. Biosci. Biotechnol. Biochem. 2008;72:3180–3188. doi: 10.1271/bbb.80370. [DOI] [PubMed] [Google Scholar]
- Otero J.M., Cimini D., Patil K.R., Poulsen S.G., Olsson L., Nielsen J. Industrial systems biology of Saccharomyces cerevisiae enables novel succinic acid cell factory. PLoS One. 2013;8:e54144. doi: 10.1371/journal.pone.0054144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patil K.R., Rocha I., Förster J., Nielsen J. Evolutionary programming as a platform for in silico metabolic engineering. BMC Bioinform. 2005;6:308. doi: 10.1186/1471-2105-6-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peralta-Yahya P.P., Zhang F., del Cardayre S.B., Keasling J.D. Microbial engineering for the production of advanced biofuels. Nature. 2012 doi: 10.1038/nature11478. [DOI] [PubMed] [Google Scholar]
- Ranganathan S., Suthers P.F., Maranas C.D. OptForce: an optimization procedure for identifying all genetic manipulations leading to targeted overproductions. PLoS Comput. Biol. 2010:6. doi: 10.1371/journal.pcbi.1000744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed J.L. Shrinking the metabolic solution space using experimental datasets. PLoS Comput. Biol. 2012;8:1–5. doi: 10.1371/journal.pcbi.1002662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha I., Maia P., Evangelista P., Vilaça P., Soares S., Pinto J.P., Nielsen J., Patil K.R., Ferreira E.C., Rocha M. OptFlux: an open-source software platform for in silico metabolic engineering. BMC Syst. Biol. 2010;4:45. doi: 10.1186/1752-0509-4-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rügen M., Bockmayr A., Steuer R. Elucidating temporal resource allocation and diurnal dynamics in phototrophic metabolism using conditional FBA. Sci. Rep. 2015;5:15247. doi: 10.1038/srep15247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha R. Diurnal regulation of cellular processes in the cyanobacterium Synechocystis sp. Strain PCC 6803: insights from transcriptomic, fluxomic, and physiological analyses. mBio 7. 2016 doi: 10.1128/mBio.00464-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schellenberger J., Que R., Fleming R.M., Thiele I., Orth J.D., Feist A.M., Zielinski D.C., Bordbar A., Lewis N.E., Rahmanian S., Kang J., Hyduke D.R., Palsson B.O. Quantitative prediction of cellular metabolism with constraint-based models: the COBRA Toolbox. Nat. Protoc. 2011;6:1290–1307. doi: 10.1038/nprot.2011.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segrè D., Vitkup D., Church G.M. Analysis of optimality in natural and perturbed metabolic networks. Proc. Natl. Acad. Sci. USA. 2002;99:15112–15117. doi: 10.1073/pnas.232349399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta T., Bhushan M., Wangikar P.P. Metabolic modeling for multi-objective optimization of ethanol production in a Synechocystis mutant. Photosynth. Res. 2013;118:155–165. doi: 10.1007/s11120-013-9935-x. [DOI] [PubMed] [Google Scholar]
- Shen C.R., Lan E.I., Dekishima Y., Baez A., Cho K.M., Liao J.C. Driving forces enable high-titer anaerobic 1-butanol synthesis in Escherichia coli. Appl. Environ. Microbiol. 2011;77:2905–2915. doi: 10.1128/AEM.03034-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varman A.M., Yu Y., You L., Tang Y.J. Photoautotrophic production of D-lactic acid in an engineered cyanobacterium. Microb. Cell Fact. 2013;12:117. doi: 10.1186/1475-2859-12-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viola S., Rühle T., Leister D. A single vector-based strategy for marker-less gene replacement in Synechocystis sp. PCC 6803. Microb. Cell Fact. 2014;13:4. doi: 10.1186/1475-2859-13-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendt K.E., Ungerer J., Cobb R.E., Zhao H., Pakrasi H.B. CRISPR/Cas9 mediated targeted mutagenesis of the fast growing cyanobacterium Synechococcus elongatus UTEX 2973. Microb. Cell Fact. 2016;15 doi: 10.1186/s12934-016-0514-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong W., Lee T.-C., Rommelfanger S., Gjersing E., Cano M., Maness P.-C., Ghirardi M., Yu J. Phosphoketolase pathway contributes to carbon metabolism in cyanobacteria. Nat. Plants. 2015 doi: 10.1038/nplants.2015.187. [DOI] [PubMed] [Google Scholar]
- Yao, L., Cengic, I., Anfelt, J., Hudson, E.P., 2015. Multiple gene repression in cyanobacteria with CRISPRi. ACS Synth. Biol. http://dx.doi.org/10.1021/acssynbio.5b00264. [DOI] [PubMed]
- Young J.D., Shastri A. a, Stephanopoulos G., Morgan J. a. Mapping photoautotrophic metabolism with isotopically nonstationary (13)C flux analysis. Metab. Eng. 2011;13:656–665. doi: 10.1016/j.ymben.2011.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S., Bryant D.A. Tricarboxylic acid cycle cyanobacteria. Science. 2011;334:1551–1554. doi: 10.1126/science.1210858. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material