Abstract
The ribulose-1,5-bisphosphate (RuBP) oxygenation reaction catalyzed by Ribulose-1,5-bisphosphate carboxylase/oxygenase (RuBisCO) is competing with carboxylation, being negative for both energy and carbon balances in photoautotrophic organisms. This makes RuBisCO one of the bottlenecks for oxygenic photosynthesis and carbon fixation. In this study, RuBisCO was overexpressed in the unicellular cyanobacterium Synechocystis PCC 6803. Relative RuBisCO levels in the engineered strains FL50 and FL52 increased 2.1 times and 1.4 times, respectively, and both strains showed increased growth, photosynthesis and in vitro RuBisCO activity. The oxygen evolution rate increased by 54% and 42% on per chlorophyll basis, while the in vitro RuBisCO activity increased by 52% and 8.6%, respectively. The overexpressed RuBisCO were tagged with a FLAG tag, in strain FL50 on the N terminus of the large subunit while in strain FL52 on the C terminus of the small subunit. The presence of a FLAG tag enhanced transcription of the genes encoding RuBisCO, and, with high possibility, also enhanced the initiation of translation or stability of the enzyme. However, when using a streptavidin-binding tag II (strep-tag II), we did not observe a similar effect. Tagged RuBisCO offers an opportunity for further studying RuBisCO expression and stability. Increased levels of RuBisCO can further improve photosynthesis and growth in the cyanobacterium Synechocystis PCC 6803 under certain growth conditions.
Keywords: Synechocystis PCC 6803, RuBisCO, Increased growth, FLAG tag, Photosynthesis, Strep-tag II
Highlights
-
•
Tagged RuBisCO were expressed in Synechocystis PCC 6803.
-
•
Expressing FLAG tagged RuBisCO resulted in increased protein levels.
-
•
Synechocystis with increased RuBisCO content grow faster with enhanced photosynthesis.
1. Background
Ribulose-1,5-bisphosphate carboxylase/oxygenase (RuBisCO, EC 4.1.1.39) has attracted intensive research interest as an abundant, low efficiency but critical enzyme (Spreitzer and Salvucci, 2002) since it was firstly purified from spinach in 1947 (Wildman and Bonner, 1947). Presently, food supply becomes an outstanding social problem as world population increases rapidly. Optimizing RuBisCO performance is one of the main strategies to increase crop yield to meet the high food requirement (Durall and Lindblad, 2015, Parry et al., 2013).
RuBisCO evolved about 3.8 billion years ago. Until now, three types of RuBisCO (Form I, II, and III) and one RuBisCO like protein (RLP, also called form IV) which lacks the carboxylation ability have been demonstrated (Tabita et al., 2008). Form I hexadecameric RuBisCO exists in most plants, algae, cyanobacteria and proteobacteria. It has eight large subunits forming the catalytic core (4L2) and four small subunits capping on the top and the bottom. L2 is anti-paralleled and the catalytic site is formed by N terminus of one large subunit and C terminus of the neighbor large subunit, altogether 8 catalytic sites in one holoenzyme (Tabita et al., 2008). Significant efforts have aimed to increase RuBisCO performance. Unfortunately, only limited successes have been achieved so far due to the turnoff between CO2 affinity and catalytic ratio (Carmo-Silva et al., 2015, Durall and Lindblad, 2015). RuBisCO is an abundant protein in leaves consisting of almost half of the soluble proteins. This makes it difficult to further increase RuBisCO amount in the cells. The complex regulatory system of RuBisCO does not only make homologous expression difficult, but also make heterologous expression challenging (Parry et al., 2013). Overexpressing Synechococcus elongatus PCC 6301 rbcLS in Synechococcus elongatus PCC 7942 resulted in cells with higher in vitro RuBisCO activity and twofold isobutyraldehyde production whereas oxygen evolution remained the same as the strains without additional rbcLS expression (Atsumi et al., 2009). This is a successful example of heterologous expression of RuBisCO even though the Synechococcus elongatus PCC 6301 genome is almost identical to the Synechococcus elongatus PCC 7942 genome, except for a 188.6 kb inversion (Sugita et al., 2007). In addition, positive effects on the free fatty acid production in Synechococcus sp. PCC 7002 have been reported by heterologous expression of RuBisCO from Synechococcus elongates PCC 7942 (Ruffing, 2014). In addition, an increased RuBisCO activity in engineered cells of Synechococcus elongates PCC 7942 containing heterologous rbcLS has been observed (Iwaki et al., 2006). These positive examples of heterologous overexpression of RuBisCO may benefit from avoiding post-transcriptional regulations of the introduced, foreign RuBisCO by the native machinery (Ruffing, 2014).
Cyanobacterial and algae RuBisCO are reported to have higher efficiency compared to plant RuBisCO and are regarded as alternatives to increase crop yield (Whitney et al., 2011). Unfortunately, there is no breakthrough report on expressing cyanobacterial or algae RuBisCO in any plant for now. One of the reasons is the differences and necessary of folding and assembling chaperones. RuBisCO has complex folding and assembling processes with chaperones involved (Hauser et al., 2015). In addition, RuBisCO activation and activity maintenance require an activase that belongs to the AAA+ family. Successful expression of cyanobacterial Synechococcus elongatus RuBisCO in tobacco was reported only recently (Lin et al., 2014). Synechococcus elongatus RuBisCO were assembled in tobacco chloroplasts both with and without the assembling chaperone RbcX, or the carboxysome protein CcmM35, and replaced the tobacco RuBisCO (Occhialini et al., 2016). Cyanobacterial RuBisCO functioned in tobacco, but the transformant line could grow autotrophically only under elevated CO2 concentration (3% v/v CO2/air) (Lin et al., 2014, Occhialini et al., 2016). Even though being an important step this transformant line is far away from an ideal high yield plant.
Fusion tag (protein tag) is widely used for protein purification (Kosobokova et al., 2016). Fusion tag can be a larger protein (like Glutathione S_Transferase, GST) or short peptides containing several amino acids (like FLAG tag, Streptavidin_binding tag). The short peptide tag may have positive effects on protein expression, protein solubility, protein efficiency or even folding (Kosobokova et al., 2016). There are reports that the presence of a His tag increased slr1192 and aldehyde reductase gene (from Synechocystis PCC 6803) expression in Escherichia colicoli and trans-2-enoyl-CoA reductase gene (from Treponema denticola) efficiency in Synechococcus elongates PCC 7942 (Akhtar et al., 2013, Lan and Liao, 2011). Even though the mechanism is not elucidated, it is possible that the tag may confer some stability at (post)transcriptional and/or (post)translational level. Other short protein tags like FLAG tag and Streptavidin-binding tag II are also well studied even though there are no reports about their positive effects on protein expression (Kosobokova et al., 2016). A FLAG tag consists of eight amino acids with the sequence DYKDDDDK, only 1 kDa (Einhauer and Jungbauer, 2001). A Streptavidin-binding tag II (strep-tag II) is also an 8 amino acid tag with the sequence WSHPQFEK (Schmidt and Skerra, 2007). It was developed from the original strep tag (WSHPQFEK) to conquer the constraint that a strep tag can only be used on the C terminus of the partner protein (Korndörfer and Skerra, 2002).
In this study, we engineered Synechocystis PCC 6803 (Synechocystis hereafter) strains with higher level of RuBisCO protein and characterized these strains. RuBisCO overexpression was only observed when the gene was tagged with a FLAG tag. RuBisCO overexpressed strains had higher in vitro RuBisCO activity, growth and oxygen evolution rate under the experimental conditions. This work indicates that improving RuBisCO can further enhance photosynthesis, growth and potentially improve yield.
2. Materials and methods
2.1. Strains and culturing conditions
Escherichia coli DH 5α (E. coli hereafter) strain was cultured with LB medium (liquid or agar petri dish) under 37 °C. Synechocystis PCC 6803 (Synechocystis) wild type and engineered strains were grown with BG11 medium (liquid or agar petri dish) in 30 °C room. 50 µg/ml kanamycin was supplied for E. coli and 25 µg/ml for Synechocystis when screening stress was required.
Synechocystis strains were grown in tubes (20 cm length, 2.6 cm inner-diameter, 3 cm outer-diameter) bubbled with air under 100 µmol photons m−2 s−1 light intensity with 0.01% antifoam (polypropylene glycol 1025, VWR) from day 1. For strains having kanamycin cassette on vectors, 25 µg/ml kanamycin was added. For strains having kanamycin cassette on chromosome, BG11 was used.
2.2. Plasmids and engineered strains
The RuBisCO gene operon (rbc, slr0009-slr0011-slr0012) and PpsbA2 were amplified from wild type Synechocystis genome with corresponding primers (Supplementary Table 1) using Phusion polymerase (Thermo Fisher Science). rbc was expressed either on pPMQAK1 (Huang et al., 2010) or on the chromosome.
Synechocystis strain carrying pPMQAK1 (cutting off ccdB gene, WT+Kmr-vector) is control to engineered strains introducing another rbc gene on pPMQAK1. PpsbA2 was flanked with EcoRI and XbaI digesting sites. RuBisCO gene was flanked with XbaI and PstI digesting sites. In total, five versions of RuBisCO genes were designed and constructed, encoding large subunit N terminus tagged with FLAG tag (FLAG- slr0009-slr0011-slr0012), encoding small subunit C terminus tagged with FLAG tag (slr0009-slr0011-slr0012-FLAG), encoding large subunit C terminus tagged with FLAG tag (slr0009-FLAG-slr0011-slr0012), encoding large subunit N terminus tagged with strep-tag II (strep- slr0009-slr0011-slr0012), and encoding small subunit C terminus tagged with strep-tag II (slr0009-slr0011-slr0012-strep). To insert FLAG tag on C terminus of large subunit, overlap extension PCR was used. pPMQAK1 was digested with EcoRI and PstI and larger fragment was recovered. These three fragments were ligated using Quick ligase (New England Bio-labs). The resulting plasmids (pFL50, pFL52, pFL50C, pFL50strep, pFL52strep) were conjugated into wild type Synechocystis cells, resulting into engineered strains FL50, FL52, FL50C, FL50strep and FL52strep respectively. Conjugation was performed as described previously (Liang and Lindblad, 2016).
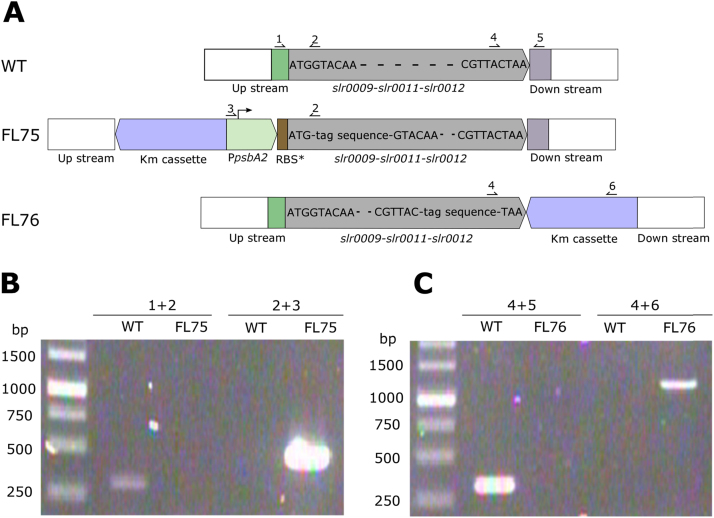
Synechocystis strain carrying a kanamycin cassette on the slr0168 site (WT+Kmr-genome) was used as a control to the engineered strains with genetic constructs on the chromosome. PpsbA2 together with the operon encoding RuBisCO (FLAG-slr0009-slr0011-slr0012 or slr0009-slr0011-slr0012-FLAG) were amplified from pFL50 and pFL52 and inserted onto pEERM3 Km (Englund et al., 2015) using EcoRI and PstI sites. The resulting plasmids were conjugated into wild type Synechocystis, resulting in the engineered strains FL50G and FL52G. Complete segregation was confirmed with PCR. Kanamycin cassette and PpsbA2 (with ribosome binding site and an ATG-FLAG tag) were flanked with upstream sequences of slr0009 and the initial part of slr0009 (without start code) before introduced into Synechocystis wild type cells. Flanking sequence was about 800 bp. The resulting strain, named FL75 (Fig. 1A), was confirmed to be completely segregated using PCR (Fig. 1B). FLAG-TAA and kanamycin cassette flanked with downstream sequences of slr0012 and the final part of slr0012 (without stop codon) were introduced into Synechocystis wild type cells, resulting in strain FL76 (Fig. 1A). Flanking sequence was about 800 bp and full segregation was confirmed using PCR (Fig. 1C). All digestion enzymes were fast digestion enzymes from Thermo Fisher Science. Synechocystis strains used in this study are summarized in Table 1.
Fig. 1.
Genomic organization in the engineered Synechocystis strains FL75 and FL76 compared to in the wild type cells (A). Complete segregation of FL75 (B) and FL76 (C) were confirmed by PCRs. In FL75 and FL76, rbc on genome was tagged with a FLAG tag on the N terminus of the large subunit and on the C terminus of the small subunit, respectively separately. Numbers indicate the specific primers used in the PCRs to examine segregation. For further details, see Table 1.
Table 1.
Synechocystis PCC 6803 strains used in this study.
| Strain | Expression construct | Expression site | Reference |
|---|---|---|---|
| WT+Kmr-vector | pPMQAK1 without ccdB | pPMQAK1 shuttle vector | (Liang and Lindblad, 2016) |
| rbc | PpsbA2-RBS*- slr0009-slr0011-slr0012 | pPMQAK1 shuttle vector | (Liang and Lindblad, 2016) |
| FL50 | PpsbA2-RBS*-FLAG- slr0009-slr0011-slr0012 | pPMQAK1 shuttle vector | This study |
| FL50C | PpsbA2-RBS*-slr0009-FLAG-slr0011-slr0012 | pPMQAK1 shuttle vector | This study |
| FL52 | PpsbA2-RBS*- slr0009-slr0011-slr0012-FLAG | pPMQAK1 shuttle vector | This study |
| FL50strep | PpsbA2-RBS*-strep II- slr0009-slr0011-slr0012 | pPMQAK1 shuttle vector | This study |
| FL52strep | PpsbA2-RBS*-slr0009-slr0011-slr0012-strep II | pPMQAK1 shuttle vector | This study |
| WT+Kmr-genome | Kanamycin cassette replaced slr0168 | Genome slr0168 | This study |
| FL50G | PpsbA2-RBS*-FLAG- slr0009-slr0011-slr0012 | Genome slr0168 | This study |
| FL52G | PpsbA2-RBS*- slr0009-slr0011-slr0012-FLAG | Genome slr0168 | This study |
| FL75 | A kanamycin cassette with PpsbA2 and FLAG tag inserted upstream of slr0009, after ATG of slr0009 | Upstream of slr0009 | This study |
| FL76 | FLAG tag and a kanamycin cassette inserted downstream of slr0012, in front of TAA of slr0012 | Downstream of slr0012 | This study |
2.3. Optical density, chlorophyll a content and oxygen evolution measurement
Synechocystis optical density was measured at 750 nm (OD750) using a spectrophotometer (Cary® 50 UV–visible Spectrophotometer, Varian). Chlorophyll a was extracted with 90% methanol. Chlorophyll a content and oxygen evolution measurement as detailed previously (Liang and Lindblad, 2016). Three independent experiments were conducted, each experiment with biological replicates and technical replicates. Obtained data were analyzed for statistical significance using one-way analysis of variance (one-way ANOVA).
2.4. Semi-quantitative reverse transcription polymerase chain reaction (RT-PCR)
Method and kit used for semi-quantitative RT-PCR were as detailed previously (Liang and Lindblad, 2016).
2.5. Crude protein extraction and relative RuBisCO content determination
Synechocystis cells were collected during log phase. Crude proteins were extracted with 50 mM Tris-HCl (pH 8.0) and concentration was determined using DC protein assay (BIO-RAD). For sodium dodecyl sulfate polyacrylamide gel (SDS-PAGE), 2.5 µg crude proteins were loaded. Western immunoblot was carried out following protocols detailed previously (Liang and Lindblad, 2016). Relative levels of RuBisCO content were determined using Quantity one.
2.6. In vitro RuBisCO activity measurement
In vitro RuBisCO activity measurement followed the protocol by Atsumi et al. (2009) with only minor modifications. Crude protein concentration was adjusted to 1 µg/µl. A 250 µl reaction mixture was used containing 125 µl 100 mM Tris-HCl (pH 8.0), 100 µl 50 mM MgCl2, 5 μl 5 mM ethylenediaminetetraacetic acid (EDTA), 2.5 μl NaH14CO3 (59 mCi/mmol), and 17.5 µl protein crude extraction. The reaction mixture was incubated for 5 min in 30 °C water bath to active the enzyme. Then 1 µl 100 mM RuBP was added and the mixture incubated for another 5 min in 30 °C water bath. 100 µl 99% propionic acid was added to stop the reaction and unfixed 14C was evaporated by heating the mixture overnight at 65 °C. Acid-stable products were resuspended in 200 µl 2 mol/L HCl and 3 ml scintillation cocktail (Optiphase “Hisafe” 3, PerkinElmer) added. Incorporation of 14C in the samples was analyzed using a scintillation counter (Tri-Carb® 2810 TR, PerkinElmer). RuBisCO activity in control strain was set as 1.0 and RuBisCO activities in the engineered strains were normalized to the activities in the control strain. Two independent experiments were conducted, each experiment with biological and technical replicates. Obtained data were analyzed for statistical significance using one-way analysis of variance (One-way ANOVA).
3. Results
3.1. Positive phenotypes of engineered Synechocystis strains with FLAG tag tagged RuBisCO
In a previous study, we demonstrated that increasing fructose-1,6/sedoheptolose-1,7-bisphosphatase, transketolase, and fructose-bisphosphate aldolase levels in Synechocystis PCC 6803 (Synechocystis) resulted in faster growth. Additionally, overexpressing the carboxysome protein CcmM resulted in higher RuBisCO level and faster growth under 100 μmol m−2 s−1 light intensity (Liang and Lindblad, 2016). In this study, we used pPMQAK1 to introduce tagged Synechocystis rbc, encoding RuBisCO, either on the large subunit N terminus or the small subunit C terminus, into Synechocystis, resulting in engineered strains FL50 and FL52, respectively. The engineered strains were grown with air under 100 μmol m−2 s−1 irradiance.
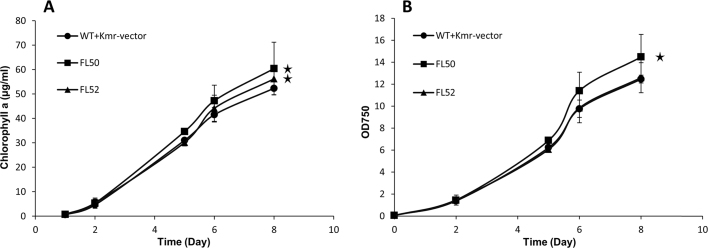
Strain FL50 grew faster than the control strain (WT+Kmr-vector), both when compared as OD750 and as chlorophyll a content (Fig. 2). However, strain FL52 showed higher chlorophyll a content than control strain but similar OD750 (Fig. 2).
Fig. 2.
Growth of engineered Synechocystis PCC 6803 strains control (WT+Kmr-vector), FL50, and FL52. A, Chlorophyll a content. B, Optical density at 750 nm (OD750). FL50 and FL52 had rbc gene, coding RuBisCO having FLAG tag fused to the large subunit N terminus or small subunit C terminus separately, overexpressed on pPMQAK1. Mean±SD is from six biological replicates, six technical replicates. Asterisks indicate that the differences observed between the respective engineered strain and the control strain are significant (One-way ANOVA, P<0.05). For strain information, see Table 1.
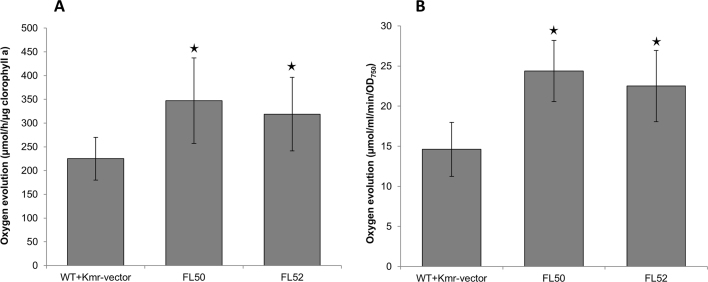
Photosynthesis, examined by measuring maximal in vivo oxygen evolution rate standardized to chlorophyll a content, which increased by 54% and 42% respectively in strains FL50 and FL52 (Fig. 3A) and oxygen evolution rates standardized to OD750, which increased by 63% and 49%, respectively, in strains FL50 and FL52 (Fig. 3B). The Calvin cycle (CBB cycle) consumes ATP and NADPH from photosynthetic light reaction. In CBB cycle, RuBisCO is the initial carboxylation enzyme. But its dual function, carboxylation and oxygenation of RuBP, as well as low turnover rate makes it one of the bottlenecks for carbon fixation in photoautotrophic organisms (Whitney et al., 2011). It is possible that enhanced RuBisCO level resulted in increased flux within the CBB cycle and as consequence, increasing the photosynthesis rate.
Fig. 3.
Photosynthesis demonstrated by maximal oxygen evolution rate of engineered Synechocystis PCC 6803 strains control (WT+Kmr-vector), FL50, and FL52. A, Oxygen evolution rate per chlorophyll a content. B, Oxygen evolution rate per OD750. FL50 and FL52 had rbc gene, coding RuBisCO having FLAG tag fused to the large subunit N terminus or small subunit C terminus, respectively, overexpressed on pPMQAK1. Mean±SD is from six biological replicates, six technical replicates. Asterisks indicate that the differences observed between the respective engineered strain and the control strain are significant (One-way ANOVA, P<0.05). For strain information, see Table 1.
3.2. FLAG tag increased both transcription and translation of RuBisCO in Synechocystis
Our earlier study showed that introducing an extra copy of rbc into Synechocystis wild type through pPMQAK1 (rbc strain) did not increase RuBisCO content even though the mRNA level of rbc increased. In addition, the engineered strain did not show any positive phenotypes (Liang and Lindblad, 2016). In the present study, we compared the mRNA and RuBisCO level of three engineered strains, rbc, FL50 and FL52, to the control strain (WT+Kmr-vector). The relative levels of the mRNA of rbc from the three engineered strains increased compared to the control strain while FL50 and FL52 showed even higher mRNA levels than rbc strain (Fig. 4). Even though the tags did not change the sequences of the 5′ untranslated region (5′-UTR) or 3′ untranslated region (3′-UTR), they changed the neighbor sequence of the two regions. Sequence changes on 5′ end of a coding gene may have further effects on DNA polymerase binding and transcription efficiency (Mutalik et al., 2013). mRNA stability can also influence the relative level of mRNA. So it is possible that RuBisCO mRNA with FLAG tag sequence was more stable than that without FLAG tag sequence.
Fig. 4.
Relative RuBisCO transcript levels of engineered Synechocystis PCC 6803 strains control (WT+Kmr-vector), rbc, FL50, FL52, FL50strep, and FL52strep measured with reverse transcription poly chain reaction (RT-PCR). FL50 and FL52 had rbc gene, coding RuBisCO having FLAG tag fused to large subunit N terminus or small subunit C terminus separately, overexpressed on pPMQAK1. In FL50strep and FL52strep, strep-tag II was used instead of FLAG. 500 ng RNA was used for cDNA synthesizing and 2 µl cDNA was used to synthesize double strand DNA. For 16 s, 18 cycles were done and for rbc, 27 cycles were used. Wild type genomic DNA (gDNA) was used in PCR as a positive control. 1 µl and 4 µl indicated the volume loaded for DNA gel. Each strain has biological duplicates. For strain information, see to Table 1.
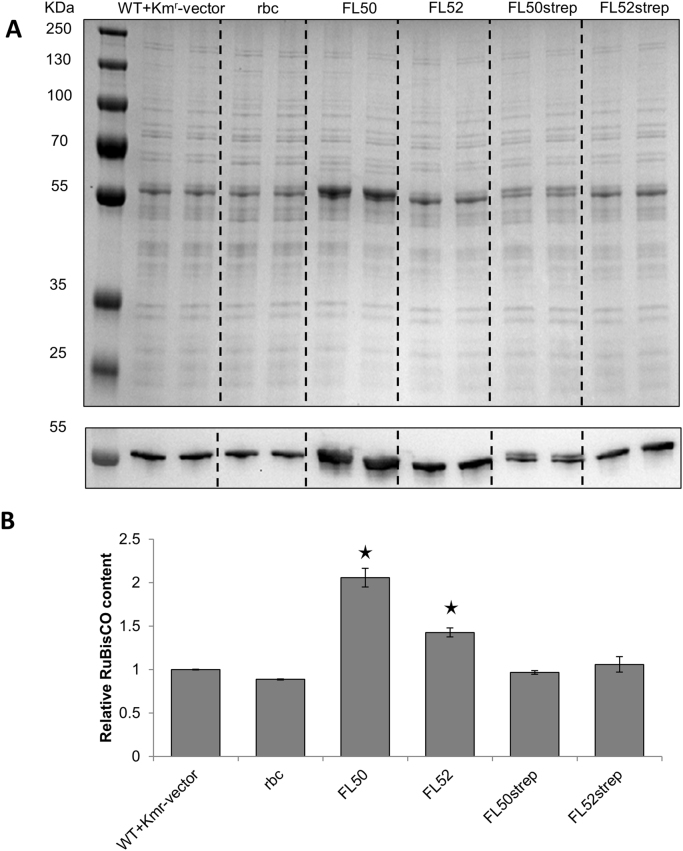
On SDS-PAGE and Western immunoblot images, it was clear that the RuBisCO protein level of rbc strain remained unchanged while FL50 and FL52 accumulated more RuBisCO protein (Fig. 5A). When the RuBisCO level of the control strain was normalized as 1.0, RuBisCO in FL50 and FL52 were 2.1 and 1.4 times of that in control strain respectively (Fig. 5B). Anti-FLAG IgG Western immunoblot showed that the tagged protein was successfully expressed (Supplementary Fig. 1A). rbc, FL50 and FL52 all had more RuBisCO mRNA. But only FL50 and FL52 accumulated more RuBisCO. It implied that the FLAG tag sequence may change the mRNA secondary structure and further affected ribosome binding and translation initiation (Mutalik et al., 2013). Since some RuBisCO large subunits from FL50 and small subunits from FL52 had extra 8 amino acids from FLAG tag, it is also possible that FLAG tag had some positive effects on protein stability and/or folding.
Fig. 5.
Relative RuBisCO content of engineered Synechocystis PCC 6803 strains control (WT+Kmr-vector), rbc, FL50, FL52, FL50strep, and FL52strep. A, SDS-PAGE and Western immunoblot, primary antibody anti-rbcL IgG was used to detect RuBisCO large subunit. B, RuBisCO content normalized to control strain determined by Quantity One. Each strain had biological duplicate. FL50 and FL52 had rbc gene, coding RuBisCO having FLAG tag fused to the large subunit N terminus or small subunit C terminus separately, overexpressed on pPMQAK1. In FL50strep and FL52strep, strep-tag II was used instead of FLAG tag. Standard derivation is from two biological replicates and two technical replicates. Asterisks indicate the differences observed between the respective engineered strain and the control strain are significant (One-way ANOVA, P<0.05). For strain information, see Table 1.
3.3. In vitro RuBisCO activity of engineered strain with FLAG tagged RuBisCO in Synechocystis
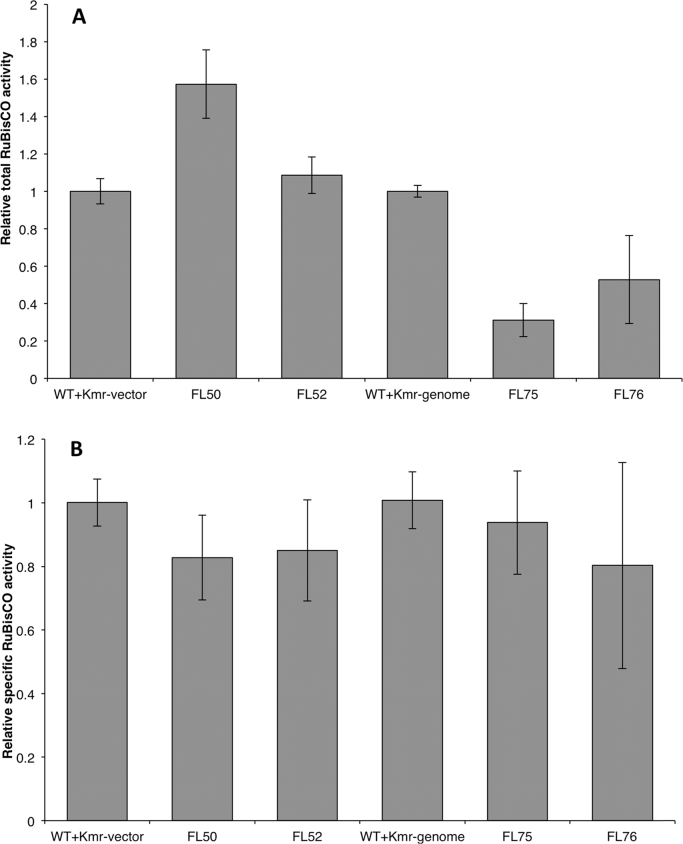
Theoretically, the small FLAG tag would not change the protein's structure and catalytic ability under general conditions (Einhauer and Jungbauer, 2001). In order to examine the level of the RuBisCO activity per crude protein (total RuBisCO activity) and per RuBisCO protein (specific RuBisCO activity), we determined the in vitro total RuBisCO activity by measuring 14C carbon fixation efficiency. When the total RuBisCO activity of the control strain (WT+Kmr-vector) was normalized to 1.0, the total RuBisCO activity of FL50 and FL52 improved by 52% and 8.6% compared to the control strain, respectively (Fig. 6A). However, the specific RuBisCO activities were similar in the three strains: WT+Kmr-vector, FL50, and FL52 (Fig. 6B).
Fig. 6.
Relative total RuBisCO activity (A) and relative specific RuBisCO activity (B) of engineered Synechocystis PCC 6803 strains control (WT+Kmr-vector), FL50, FL52, control (WT+Kmr-genome), FL75, and FL76. Total RuBisCO activity was calculated as 14C incorporation per crude protein per time. Specific RuBisCO activity was calculated as 14C incorporation per RuBisCO protein per time. FL50 and FL52 had rbc gene, coding RuBisCO having FLAG tag fused to the large subunit N terminus or small subunit C terminus separately, overexpressed on pPMQAK1. In FL75 and FL76, rbc on genome was tagged with FLAG on large subunit N terminus or small subunit C terminus separately. Mean±SD is from four biological replicates, four technical replicates. Asterisks indicate the differences between the respective engineered strain and the control strain is significant (One-way ANOVA, P<0.05). FL50 and FL52 were normalized to control strain (WT+Kmr-vector) while FL75 and FL76 were normalized to control strain (WT+Kmr-genome). Even though the respective control strains, WT+Kmr-vector and WT+Kmr-genome, both were set to 1.0, it does not mean that the RuBisCO activity was the same in the two strains. For strain information, see Table 1.
RuBisCO has complex activation and catalysis processes (Stec, 2012, Mueller-Cajar et al., 2014). It is easily inhibited by sugar phosphate as well as its own substrate ribulose-1,5-bisphosphate (RuBP). To reactive inhibited RuBisCO, an activase which belongs to the AAA+(ATPases associated with various cellular activities) family is required (Mueller-Cajar et al., 2014). In this study, we did not engineer the activase. Total RuBisCO activity measurement using crude protein extracts without adding extra activase increased in FL50 and FL52 indicating that already available activase was not limiting the RuBisCO activity under our experimental conditions.
3.4. Strep-tag II did not have the same effect as FLAG tag on RuBisCO expression in Synechocystis
Flag tag and modified streptavidin binding tag (strep-tag II) (Ayala et al., 2013) both contain eight amino acids. Therefore, we explored if strep-tag II will have similar effects as the FLAG tag on RuBisCO engineering to see if the effects were sequence specific. In Synechocystis strains FL50strep and FL52strep, the introduced rbc was tagged with strep-tag II on the large subunit N terminus and the small subunit C terminus, respectively (Table 1). RuBisCO mRNA levels in strains FL50strep and FL52strep were higher than in the control strain (WT+Kmr-vector), but similar to the rbc strain whose extra rbc was not tagged (Fig. 4). This meant that the presence of the strep-tag II did not have positive effects on transcription or mRNA stability in comparison to the effect of the FLAG tag. Moreover, the RuBisCO level in the FL50strep and FL52strep strains remained unchanged, like the rbc strain (Fig. 5). This implies that the strep-tag II does not have any positive effects on translation, post-translation modification, or RuBisCO stability.
3.5. Expressing tagged rbc on the chromosome did not result in a changed phenotype in Synechocystis
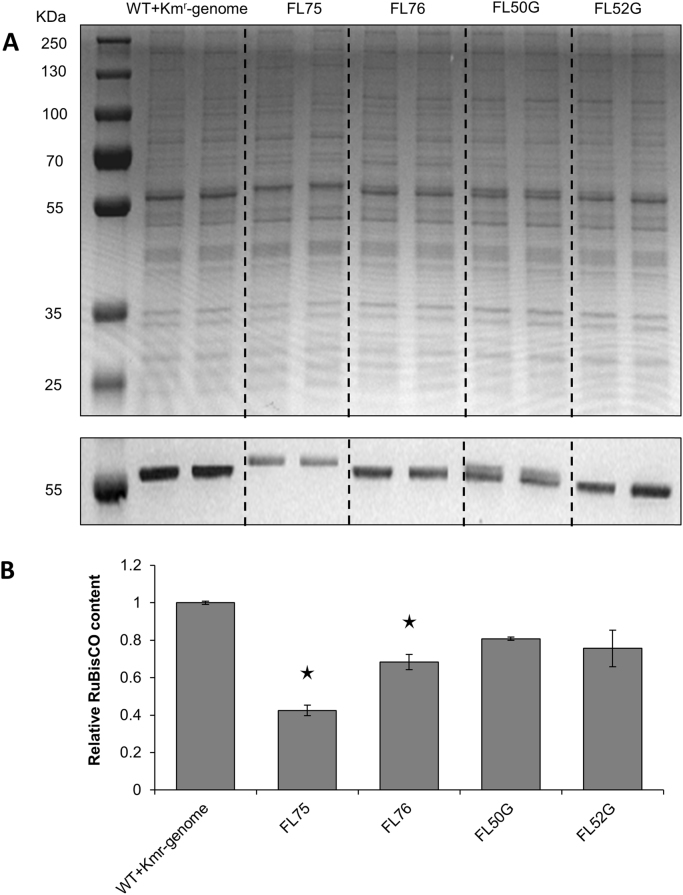
Continuous antibiotic selection pressure is a widely used method to maintain genetically engineered microbial cells, specifically when introducing the non-native genetic construct using a self-replicating vector. However, it is neither economically and environmentally friendly nor practically feasible to use antibiotics in large-scale cultivation systems. Instead of using antibiotic free technologies (Liu et al., 2011, Tan et al., 2013), inserting the target gene together with antibiotic cassette into the chromosome and selecting for complete segregation is another option to avoid using antibiotics during cultivation. In the present study, the engineered Synechocystis strains FL50G, FL52G, FL75 and FL76 (Table 1) with insertion into the chromosome, were confirm by PCR to be still completely segregated after two weeks of growth in media without any antibiotica (data not shown).
Synechocystis strain FL50G expressed the 5′ end FLAG tag tagged rbc gene on chromosome while FL52G expressed the 3′ end FLAG tag tagged rbc gene. The genetic constructs were identical to that in strains FL50 and FL52 (Table 1). However, strains FL50G and FL52G did not show any increased level of RuBisCO (Fig. 7). It has been reported that heterologous gene expression on plasmids may be more efficient than the expression on the chromosome (Ng et al., 2015). Another explanation may be some unexpected effect caused by neighbor sequences in the chromosome.
Fig. 7.
Relative RuBisCO content of engineered Synechocystis PCC 6803 strains, control (WT+Kmr-genome), FL75, FL76, FL50G, and FL52G. A, SDS-PAGE and Western immunoblot, primary antibody anti-rbcL IgG was used to detect RuBisCO large subunit. B, RuBisCO content normalized to control strain determined by Quantity One. In FL75 and FL76, rbc on genome was tagged with FLAG on large subunit N terminus and small subunit C terminus separately. FL50G and FL52G had rbc gene, coding RuBisCO having FLAG tag fused to the large subunit N terminus or small subunit C terminus separately, overexpressed on chromosome slr1608 site. Each strain had biological duplicate. Standard derivation is from two biological replicates and two technical replicates. Asterisks indicate the differences observed between the respective engineered strain and the control C strain are significant (One-way ANOVA, P<0.05). For strain information, see Table 1.
Even though the total RuBisCO activity increased in strains FL50 and FL52, the specific RuBisCO activity was the same as that in the control strain (WT+Kmr-vector) (Fig. 6). In fact, there are several versions of the RuBisCO holoenzyme in strains FL50 and FL52 based on numbers and positions of the tagged subunits. It is difficult to determine specific RuBisCO activity of each single version. Here, we measured the native version and the introduced tagged versions (all eight large subunits were tagged or all eight small subunits were tagged). The strains Synechocystis FL75 and FL76 (Table 1) were designed and constructed as only expressing tagged RuBisCO. Complete segregation was confirmed with PCR (Fig. 1B and 1C). All large subunits in strain FL75 were tagged with FLAG tag while all small subunits in strain FL76 were tagged. Interestingly, the specific RuBisCO activity was similar in control (WT+Kmr-genome), FL75 and FL76 strains (Fig. 6B). This indicates that the FLAG tag did not result in enhanced specific RuBisCO activity, and as a consequence, the increased total RuBisCO activity observed in strains FL50 and FL52 was due to the increased level of RuBisCO.
Except for the native rbc gene, strains rbc and FL52 both possessed an extra copy of rbc gene on shuttle vector. On mRNA level of rbc gene, FL52 had more mRNA compared to rbc strain. This meant rbc gene with FLAG on 3′ end resulted into more mRNA than the version without tag on shuttle vector. When comparing control strain (WT+Kmr-genome) and strain FL76, the rbc gene transcription and translation machineries were identical, meaning the promoter, ribosome binding sites and the neighbor sequences were identical. The only difference was that rbc gene in FL76 had FLAG on 3′ end. However, the mRNA level in FL76 was not higher than that in control strain (WT+Kmr-genome). This phenomenon was not the same as that on shuttle vector and indicated different expression pattern on shuttle vector and chromosome.
4. Discussion
Even though RuBisCO is one of the most abundant proteins in photoautotrophic organisms, it may be considered as a bottleneck in photosynthesis and carbon fixation due to its low efficiency. Here we experimentally demonstrated and increased the level of RuBisCO as well as RuBisCO activity in Synechocystis with subsequently increased photosynthesis and growth by introducing a FLAG tag.
Here we demonstrated that the introduction of a FLAG tag increased transcription and maybe even translation or the stability after translation, when adding a FLAG tag on the N terminus of large subunit or the C terminus of small subunit of RuBisCO. Interestingly, even though being of similar length, eight amino acids, strep-tag II in the same position did not have the same effect. Affinity tags may enhance recombinant protein yield as stated before (Walls and Loughran, 2011), either because of N terminus tags are changing the mRNA secondary structure and thereby interferes in the interaction with the ribosome binding site and/or that the tag inhibits degradation (Waugh, 2005). More work is needed to elucidate the mechanism(s) behind the increased expression of RuBisCO by introducing a FLAG tag in Synechocystis.
The FLAG tag may also have effects on the RuBisCO protein level. A recent report observed that the C terminus extension of red-type form I RuBisCO small subunit allowed red-type form I RuBisCO chaperone-independent assembly. This extension of 4 small subunits formed an 8-strand β-barrel which inserted into the rbcL8 complex core and stabilized it (Joshi et al., 2015). There is a possibility that the FLAG tag may transmit a similar function. However, the C terminus extension of the red-type RuBisCO is 25 amino acids, which is much longer than the FLAG tag. In addition, a FLAG tag is a rigid tag with a low possibility to form a β-hairpin. It would be interesting to examine if there is any similarity on the amino acid interaction between the RuBisCO protein and the FLAG tag or the β-barrel. In wheat leaves, the RuBisCO large subunit was degraded after incubation in darkness. The degradation occurred on the N terminus of the large subunit (Kokubun et al., 2002). In the present study, an increased RuBisCO level was found in Synechocystis strain FL50 where the FLAG tag was on N terminus of large subunit but not in strain FL50C where the FLAG tag was on C terminus of large subunit (Supplementary Fig. 2). These results indicated that the degradation of RuBisCO large subunit in Synechocystis might also occur from the N terminus. Specific RuBisCO activity measurement indicated that the FLAG tag did not increase fusion protein activity (Fig. 6B). Similar results have been reported for glutamate dehydrogenase (GDH). A FLAG tag on the N terminus of mouse GDH did not change the kinetic parameters while positioned on the C terminus, its sensitivity to ADP activation decreased (Pajęcka et al., 2014). Cyanobacterial RuBisCO generally has a higher Sc/o (RuBisCO specific factor) compared to plant RuBisCO. However, correct folding, assembly and stability are issues to be solved when introducing cyanobacterial RuBisCO into plants (Occhialini et al., 2016). The observations in the present study offer another approach to examine RuBisCO assembly and stability.
RuBisCO may be one of the key targets to increase efficiency of carbon fixation in photoautotrophic organisms. The present work offers an additional strategy to overexpress native RuBisCO and other versions of RuBisCO for further e.g. stability studies. In this work we developed engineered strains of Synechocystis with increased level of RuBisCO, which grew faster (Fig. 2) and had higher oxygen evolution rate (Fig. 3). This implies that increasing the performance of RuBisCO is one strategy to increase photosynthesis and carbon fixation in cyanobacterial cells.
Acknowledgement
This work supported by the Knut and Alice Wallenberg Foundation (Project MoSE, #2011.0067), the Swedish Energy Agency (#11674-5), and a Chinese scholarship to Feiyan Liang (201304910361) from The Chinese Service Center for Scholarly Exchange.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.meteno.2017.02.002.
Appendix A. Supplementary material
Supplementary material
.
References
- Akhtar M.K., Turner N.J., Jones P.R. Carboxylic acid reductase is a versatile enzyme for the conversion of fatty acids into fuels and chemical commodities. Proc. Natl. Acad. Sci. USA. 2013;110:87–92. doi: 10.1073/pnas.1216516110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atsumi S., Higashide W., Liao J.C. Direct photosynthetic recycling of carbon dioxide to isobutyraldehyde. Nat. Biotechnol. 2009;27:1177–1180. doi: 10.1038/nbt.1586. [DOI] [PubMed] [Google Scholar]
- Ayala J.C., Pimienta E., Rodríguez C., Anne J., Vallin C., Milanes M.T., King-Batsios E., Huygen K., Van Mellaert L. Use of Strep-tag II for rapid detection and purification of Mycobacterium tuberculosis recombinant antigens secreted by Streptomyces lividans. J. Microbiol. Methods. 2013;94:192–198. doi: 10.1016/j.mimet.2013.06.004. [DOI] [PubMed] [Google Scholar]
- Carmo-Silva E., Scales J.C., Madgwick P.J., Parry M.A.J. Optimizing rubisco and its regulation for greater resource use efficiency. Plant Cell Environ. 2015;38:1817–1832. doi: 10.1111/pce.12425. [DOI] [PubMed] [Google Scholar]
- Durall C., Lindblad P. Mechanisms of carbon fixation and engineering for increased carbon fixation in cyanobacteria. Algal Res. 2015;11:263–270. [Google Scholar]
- Einhauer A., Jungbauer A. The FLAG™ peptide, a versatile fusion tag for the purification of recombinant proteins. J. Biochem Biophys. Methods. 2001;49:455–465. doi: 10.1016/s0165-022x(01)00213-5. [DOI] [PubMed] [Google Scholar]
- Englund E., Andersen-Ranberg J., Miao R., Hamberger B., Lindberg P. Metabolic engineering of Synechocystis sp. PCC 6803 for production of the plant diterpenoid manoyl oxide. ACS Synth. Biol. 2015;4:1270–1278. doi: 10.1021/acssynbio.5b00070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauser T., Popilka L., Hartl F.U., Hayer-Hartl M. Role of auxiliary proteins in Rubisco biogenesis and function. Nat. Plants. 2015;1:150–165. doi: 10.1038/nplants.2015.65. [DOI] [PubMed] [Google Scholar]
- Huang H.-H., Camsund D., Lindblad P., Heidorn T. Design and characterization of molecular tools for a synthetic biology approach towards developing cyanobacterial biotechnology. Nucleic Acids Res. 2010;38:2577–2593. doi: 10.1093/nar/gkq164. (doi: 0.1093/nar/gkq164) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwaki T., Haranoh Km Inoue N., Kojima K., Satoh R., Nishino T., Wada S., Ihara H., Tsuyama S., Kobayashi H., Wadano A. Expression of foreign type I ribulose-1,5-bisphosphate carboxylase/oxygenase (EC 4.1.1.39) stimulates photosynthesis in cyanobacterium Synechococcus PCC 7942 cells. Photosynth. Res. 2006;88:287–297. doi: 10.1007/s11120-006-9048-x. [DOI] [PubMed] [Google Scholar]
- Joshi J., Mueller-Cajar O., Tsai Y.C., Hartl F.U., Hayer-Hartl M. Role of small subunit in mediating assembly of red-type Form I Rubisco. J. Biol. Chem. 2015;290:1066–1074. doi: 10.1074/jbc.M114.613091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kokubun N., Ishida H., Makino A., Mae T. The degradation of the large subunit of ribulose-1,5-bisphosphate carboxylase/oxygenase into the 44-kDa fragment in the lysates of chloroplasts incubated in darkness. Plant Cell Physiol. 2002;43:1390–1395. doi: 10.1093/pcp/pcf159. [DOI] [PubMed] [Google Scholar]
- Korndörfer I.P., Skerra A. Improved affinity of engineered streptavidin for the Strep-tag II peptide is due to a fixed open conformation of the lid-like loop at the binding site. Protein Sci. 2002;11:883–893. doi: 10.1110/ps.4150102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosobokova E.N., Skrypnik K.A., Kosorukov V.S. Overview of fusion tags for recombinant proteins. Biochem. 2016;81:187–200. doi: 10.1134/S0006297916030019. [DOI] [PubMed] [Google Scholar]
- Lan E.I., Liao J.C. Metabolic engineering of cyanobacteria for 1-butanol production from carbon dioxide. Metab. Eng. 2011;13:353–363. doi: 10.1016/j.ymben.2011.04.004. [DOI] [PubMed] [Google Scholar]
- Liang F., Lindblad P. Effects of overexpressing photosynthetic carbon flux control enzymes in the cyanobacterium Synechocystis PCC 6803. Metab. Eng. 2016;38:56–64. doi: 10.1016/j.ymben.2016.06.005. [DOI] [PubMed] [Google Scholar]
- Lin M.T., Occhialini A., Andralojc P.J., Parry M.A.J., Hanson M.R. A faster Rubisco with potential to increase photosynthesis in crops. Nature. 2014;513:547–550. doi: 10.1038/nature13776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Sheng J., Curtiss R. Fatty acid production in genetically modified cyanobacteria. Proc. Natl. Acad. Sci. USA. 2011;108:6899–6904. doi: 10.1073/pnas.1103014108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller-Cajar O., Stotz M., Bracher A. Maintaining photosynthetic CO2 fixation via protein remodelling: the Rubisco activases. Photosynth. Res. 2014;119:191–201. doi: 10.1007/s11120-013-9819-0. [DOI] [PubMed] [Google Scholar]
- Mutalik V.K., Guimaraes J.C., Cambray G., Mai Q.A., Christoffersen M.J., Martin L., Yu A., Lam C., Rodriguez C., Bennett G., Keasling J.D., Endy D., Arkin A.P. Quantitative estimation of activity and quality for collections of functional genetic elements. Nat. Methods. 2013;10:347–353. doi: 10.1038/nmeth.2403. [DOI] [PubMed] [Google Scholar]
- Ng A.H., Berla B.M., Pakrasi H.B. Fine-Tuning of photoautotrophic protein production by combining promoters and neutral sites in the cyanobacterium Synechocystis sp. strain PCC 6803. Appl. Environ. Microbiol. 2015;81:6857–6863. doi: 10.1128/AEM.01349-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Occhialini A., Lin M.T., Andralojc P.J., Hanson M.R., Parry M.A. Transgenic tobacco plants with improved cyanobacterial Rubisco expression but no extra assembly factors grow at near wild-type rates if provided with elevated CO2. Plant J. 2016;85:148–160. doi: 10.1111/tpj.13098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pajęcka K., Nielsen C.W., Hauge A., Zaganas I., Bak L.K., Schousboe A., Plaitakis A., Waagepetersen H.S. Glutamate dehydrogenase isoforms with N-terminal (His)6- or FLAG-tag retain their kinetic properties and cellular localization. Neurochem. Res. 2014;39:487–499. doi: 10.1007/s11064-013-1042-z. [DOI] [PubMed] [Google Scholar]
- Parry M.A.J., Andralojc P.J., Scales J.C., Salvucci M.E., Carmo-Silva A.E., Alonso H., Whitney S.M. Rubisco activity and regulation as targets for crop improvement. J. Exp. Bot. 2013;64:717–730. doi: 10.1093/jxb/ers336. [DOI] [PubMed] [Google Scholar]
- Ruffing A.M. Improved free fatty acid production in cyanobacteria with Synechococcus sp. PCC 7002 as host. Front Bioeng. Biotechnol. 2014;2:17. doi: 10.3389/fbioe.2014.00017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt T.G., Skerra A. The Strep-tag system for one-step purification and high-affinity detection or capturing of proteins. Nat. Protoc. 2007;2:1528–1535. doi: 10.1038/nprot.2007.209. [DOI] [PubMed] [Google Scholar]
- Spreitzer R.J., Salvucci M.E. Rubisco: structure, regulatory interactions, and possibilities for a better enzyme. Annu. Rev. Plant Biol. 2002;53:449–475. doi: 10.1146/annurev.arplant.53.100301.135233. [DOI] [PubMed] [Google Scholar]
- Stec B. Structural mechanism of RuBisCO activation by carbamylation of the active site lysine. Proc. Natl. Acad. Sci. USA. 2012;109:18785–18790. doi: 10.1073/pnas.1210754109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugita C., Ogata K., Shikata M., Jikuya H., Takano J., Furumichi M., Kanehisa M., Omata T., Sugiura M., Sugita M. Complete nucleotide sequence of the freshwater unicellular cyanobacterium Synechococcus elongatus PCC 6301 chromosome: gene content and organization. Photosynth. Res. 2007;93:55–67. doi: 10.1007/s11120-006-9122-4. [DOI] [PubMed] [Google Scholar]
- Tabita F.R., Satagopan S., Hanson T.E., Kreel N.E., Scott S.S. Distinct form I, II, III, and IV Rubisco proteins from the three kingdoms of life provide clues about Rubisco evolution and structure/function relationships. J. Exp. Bot. 2008;59:1515–1524. doi: 10.1093/jxb/erm361. [DOI] [PubMed] [Google Scholar]
- Tan X., Liang F., Cai K., Lu X. Application of the FLP/FRT recombination system in cyanobacteria for construction of markerless mutants. Appl. Microbiol. Biotechnol. 2013;97:6373–6382. doi: 10.1007/s00253-013-4837-6. [DOI] [PubMed] [Google Scholar]
- Walls D., Loughran S.T. Tagging recombinant proteins to enhance solubility and aid purification. Methods Mol. Biol. 2011;681:151–175. doi: 10.1007/978-1-60761-913-0_9. [DOI] [PubMed] [Google Scholar]
- Waugh D.S. Making the most of affinity tags. Trends Biotechnol. 2005;23:316–320. doi: 10.1016/j.tibtech.2005.03.012. [DOI] [PubMed] [Google Scholar]
- Whitney S.M., Houtz R.L., Alonso H. Advancing our understanding and capacity to engineer nature's CO2-sequestering enzyme, Rubisco. Plant Physiol. 2011;155:27–35. doi: 10.1104/pp.110.164814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wildman S.G., Bonner J. The proteins of green leaves. 1. Isolation, enzymatic properties and auxin content of spinach cytoplasmic proteins. Arch. Biochem. 1947;14:381–413. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material