Abstract
Background
The ubiquitin-dependent protein degradation pathway is essential for the proteolysis of intracellular proteins and peptides. Deubiquitinating enzymes constitute a complex protein family involved in a multitude of cellular processes. The ubiquitin-specific proteases (UBP) are a group of enzymes whose predicted function is to reverse the ubiquitinating reaction by removing ubiquitin from a large variety of substrates. We have lately reported the characterization of human USP25, a specific-ubiquitin protease gene at 21q11.2, with a specific pattern of expression in murine fetal brains and adult testis.
Results
Database homology searches at the DNA and protein levels and cDNA library screenings led to the identification of a new UBP member in the human genome, named USP28, at 11q23. This novel gene showed preferential expression in heart and muscle. Moreover, cDNA, expressed sequence tag and RT-PCR analyses provided evidence for alternatively spliced products and tissue-specific isoforms. Concerning function, USP25 overexpression in Down syndrome fetal brains was shown by real-time PCR.
Conclusions
On the basis of the genomic and protein sequence as well as the functional data, USP28 and USP25 establish a new subfamily of deubiquitinating enzymes. Both genes have alternatively spliced exons that could generate protein isoforms with distinct tissue-specific activity. The overexpression of USP25 in Down syndrome fetal brains supports the gene-dosage effects suggested for other UBP members related to aneuploidy syndromes.
Background
Ubiquitin modification of protein substrates has a major role in a variety of cellular processes such as cell-cycle progression, DNA repair, antigen presentation, differentiation and development, transcriptional activation and selective degradation of damaged proteins (for review see [1,2,3]). Covalent attachment of ubiquitin molecules is the first step for degradation of the tagged protein via the 26S proteasome pathway. This is a finely regulated and highly specific process as an error in substrate recognition might compromise cell survival.
Studies on the specificity and regulation of ubiquitin-dependent protein degradation have mainly focused on the ubiquitinating enzymes. Nonetheless, there is increasing evidence that other enzymes that remove ubiquitin from the ubiquitin-conjugates (deubiquitinating enzymes, DUBs) not only affect the fate and degradation of intracellular proteins but seem to be essential in the maintenance of cell-free ubiquitin pools [3,4,5]. Failures of the ubiquitin system have been implicated in many human diseases, among them some important neurodegenerative disorders and several carcinomas. The accumulation of ubiquitin adducts has been described in patients with Alzheimer's, Huntington's, and Parkinson's diseases although the direct involvement of ubiquitinated protein aggregates in the pathological condition has not been proved. On the other hand, a missense mutation in the ubiquitin carboxy-terminal hydrolase L1 (UCHL1) has been found in a German family with Parkinson's disease [6].
Two classes of deubiquitinating enzymes have been described: the UCHs (ubiquitin carboxy-terminal hydrolases) and the UBPs (ubiquitin processing proteases) [4,7,8]. The UCH family members share a small size, cleave ubiquitin from small peptides and amino acids and appear well conserved (40%) across species. In contrast, the UBP family gathers larger and distantly related enzymes that release ubiquitin from a wide range of ubiquitin-protein conjugates [5]. As the human UBP family is likely to be quite large, a systematic nomenclature has been proposed for these enzymes based on the abbreviation USP, for ubiquitin-specific protease [9].
In the UBP family, protein sequence comparisons have shown that homology is restricted to the regions encompassing the active site cysteine and histidine residues and other peptide segments putatively involved in catalysis or substrate binding [7,10]. More than 90 deubiquitinating enzymes have been characterized after data from genome sequencing projects [5]; as many as 16 are encoded in yeast, which is in agreement with the high specificity attributed to this family [2,3]. Although at present the overall homology among UBPs is low, subfamilies of closely related members (60% to 88% amino acid identities) begin to emerge [11,12,13].
Lately, we have identified a novel UBP member, named USP25, in the gene-poor region of human chromosome 21q11.2 and characterized the full cDNA sequence [14]. Isolation of the mouse homolog cDNA allowed us to perform northern and in situ hybridization analyses. Higher levels of USP25 expression in mouse were detected in the proliferative compartments of fetal brain and in maturating spermatocytes of adult testis, allowing the correlation of gene expression with high protein turnover.
Here we describe a human USP25 homolog, USP28, which maps to 11q23 and is preferentially expressed in heart and muscle. Tissue-specific alternatively spliced exons of USP25 and USP28 have been identified, in agreement with several isoforms described for vertebrate UBPs [13,15]. The functional deubiquitinating assay has been performed for the two members of the newly described UBP subfamily. Overexpression of USP25 as measured by real-time PCR in Down versus control fetal brains supports the gene-dosage effects reported for other UBP members.
Results
Cloning of USP28
A TBlastN comparison of USP25 sequence against nr database at the National Center for Biotechnology Information (NCBI) server revealed a homologous USP sequence on PAC clone pDJ356d6 (GenBank accession number AC002036) located at 11q23. Specific probes designed following this genomic sequence were used to screen 106 recombinant phages of a human fetal brain cDNA library (Clontech). Six positive clones were isolated, subcloned and sequenced. Four clone sequences were chimeric and were thus discarded. Two overlapping cDNA clones, 5A11 (2.3 kb) and 3A11 (1.5 kb), showed an exact match with chromosome 11 PAC clone pDJ356d6. Evidence that clones 5A11 and 3A11 did not encode the full-length cDNA was based on the protein alignment between the deduced amino acid sequence and USP25, the mRNA size obtained from northern analysis when probed with cDNA clone 5A11, and the absence of the strictly conserved UBP cysteine domain in either clone 5A11 or 3A11.
Three different strategies were followed to isolate the 5' end of USP28 cDNA. First, a chromosome 11-specific forward primer (5.2 cr11F) was designed from the TBlastN analysis of USP25 (Figure 1). This sequence corresponded to the 5'-terminal sequence of the predicted exon encoding the cysteine domain in the chromosome 11 PAC DNA. A reverse oligonucleotide, 11race4, was also designed on the basis of the 3A11 cDNA clone sequence (Figure 1). An RT-PCR reaction using heart mRNA as template was subsequently performed with the forward and reverse primers. The expected 958 bp amplified fragment was subcloned in pUC18. Its sequence overlapped with 3A11 and 5A11 cDNA clones, thus allowing the characterization of an additional 908 bp. The overall cDNA sequence, although still incomplete, contained the cysteine domain, together with all other UBP reported signatures.
Figure 1.

USP28 nucleotide and amino acid sequences (GenBank accession number AF266283). Nucleotides and residues are numbered from the presumptive first ATG (methionine) codon. Specific primers used for cloning are also indicated. The alternative polyadenylation signal is underlined.
Second, 106 recombinant phages from kidney and placenta cDNA libraries (Clontech) were screened using three different 5'-USP28-specific probes obtained after PCR amplification with primers deduced from the TBlastN comparison to USP25 (Figure 1). Five different positive clones were isolated: four from kidney and one from placenta. All inserts were subcloned in pBLUESCRIPT SK+ (Stratagene). Sequence analysis revealed a further 318 bp, although the cDNA was still incomplete.
Third, a 5' RACE experiment was performed on 109 phages from the kidney cDNA library with two specific reverse primers (11race6 and 11race7) (Figure 1) and two λgt10 vector-derived forward primers (GR1, GR2). The comparison with the PAC clone PDJ105h16 sequence (htgs database) allowed us to identify the first presumptive methionine. However, as no further in-frame stop codons were detected, the use of another initiation codon could not be ruled out. The search for a putative TATA box in the upstream genomic sequence rendered no positive results.
No cDNA clones with a poly(A)+ tail could be isolated, but BlastN comparisons against the dbEST database allowed the identification of an overlapping EST (AI337094) containing the polyadenylation signal.
USP28 cDNA (accession number AF266283) is at least 3,624 bp long and encodes 1,077 amino acids with a relative molecular mass of 122.4 kDa. The gene is organized in 25 exons and exon-intron boundaries were determined after comparison with the human htgs database (Table 1). Average exon length was 130 bp, the shortest being 71 bp and the longest 229 bp. Nucleotide and amino acid identitiesbetween USP28 and USP25 (Figure 2) are 55.77% and 51.36%, respectively. USP28 is more similar to USP25 than to any other known UBP. Besides, exon-intron boundaries between USP28 and USP25 are highly conserved (Figure 2). The homology between USP28 and USP25 with other UBP family members appears to be confined to the reported conserved domains.
Table 1.
Genomic organization of USP28
| Exon | Acceptor splice site | Donor splice site | Intron length (bp) | |
| 1 | 57 | GCTCG/gtgggctccg | >14,000 | |
| 2 | 58-135 | aaatatagag/AGCTA | TGAAG/gttggtttcc | 1,635 |
| 3 | 136-268 | tctcccatag/GCCAG | AGCAA/gtaggtacca | 10,737 |
| 4 | 269-374 | tatcttacag/AAGTT | AACAG/gttattgatt | 905 |
| 5 | 375-534 | cacaatatag/GATGC | TTCAG/gtatgatatt | 6,262 |
| 6 | 535-621 | tttatttcag/TCTCT | ATACA/gtaagttggt | 691 |
| 7 | 622-759 | ttttctacag/GAAAA | AGCAG/gtacaaaaga | 1,426 |
| 8 | 760-833 | tctcttatag/CAAGA | GTTAA/gtgagtgttg | 976 |
| 9 | 834-910 | ctaaaagcag/CAGTC | TGAAG/gtaaattctc | 1,515 |
| 10 | 911-1,059 | ctttgcctag/GAAAA | AAGAG/gtacgttgga | 1,836 |
| 11 | 1,060-1,187 | ttgacactag/CGTTG | GACAG/gtgagttctt | 3,532 |
| 12 | 1,188-1,283 | atttacttag/GTACA | GAAAG/gtgagttttg | 5,763 |
| 13 | 1,284-1,463 | tgtcgcttag/GTATG | GAAAG/gtaatgaaaa | 2,282 |
| 14 | 1,464-1,672 | tcagtacaag/TACAA | ACAAG/gttggctctt | 1,213 |
| 15 | 1,673-1,743 | ttccaaacag/ATTTA | GTCAG/gtagaatgaa | 1,380 |
| 16 | 1,744-1,972 | tgacttacag/GTGCC | TGCAG/gtaaaagtat | 3,021 |
| 17 | 1,973-2,164 | ttttcctcag/AGGCA | ACAAG/gtatctgaga | 625 |
| 18 | 2,165-2,304 | tctttatcag/AGCCT | GTGAG/gtgagaatga | 1,713 |
| 19 | 2,305-2,400 | cttgtggtag/GTGAT | TTAAG/gtatctctgt | 1,443 |
| 20 | 2,401-2,579 | cccttaatag/GCATT | GAAAG/gtgagaagag | 116 |
| 21 | 2,580-2658 | tcccacacag/ATCAA | ACAAG/gtaaagtgtt | 795 |
| 22 | 2659-2,738 | tctttttcag/AAGTG | GGAAA/gtatgttggc | 516 |
| 23 | 2,739-2,862 | tttttctaag/GTACC | TTCTG/gtttgtactg | 1,479 |
| 24 | 2,863-3,058 | ttatctgtag/GAGCT | TGCAG/gtatatgctc | 2,068 |
| 25 | 3,059 | ccctccacag/AAAAT |
The donor and acceptor splice site signals are indicated in bold.
Figure 2.

Protein alignment of USP25 and USP28. Amino acid identities are boxed in black and conserved amino acid changes are boxed in gray. The exon-intron boundaries in each gene are marked by arrowheads.
Expression analysis of USP28 in human tissues
Northern analysis for USP28 was performed using the cDNA clone 5A11 as probe (Figure 3). A single transcript of approximately 4.5 kb was identified in heart and skeletal muscle after short film exposure. At longer exposures a transcript of the same size was detected in all assayed tissues, albeit at a very low level. To compare the amount of RNA poly(A)+ loaded in each lane an actin probe was used as a control.
Figure 3.
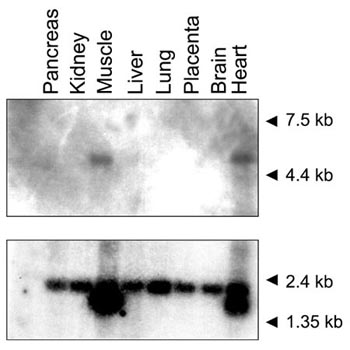
Northern analysis of USP28 in adult human tissues. Molecular weight marker sizes are indicated. Hybridization with an actin probe used as control is shown below.
Alternative splicing of USP25 and USP28
Isoforms generated by alternative splicing have been described for some vertebrate UBPs [13,15]. Sequence analysis of the isolated USP25 cDNA clones revealed four alternatively spliced exons. Three of those generated an in-frame stop codon in some human fetal brain cDNA clones and were located between exons 3 and 4, 10 and 11, and 17 and 18 (exons numbered according to [14]. The fourth alternatively spliced product corresponded to exon 19 and added 32 amino acids to the protein product (exon 19 has been renamed 19 b in this work). It had been previously identified in a single I.M.A.G.E consortium neuroepithelium cDNA clone (id: AA209364 [16]). In order to verify all these putative exons, specific sets of primers (Table 2) were designed for RT-PCR amplification on 24 different tissues (Multiple Tissue cDNA Panels, Clontech).
Table 2.
Primers used in the splicing analysis of USP25 and USP28
| Primer | Sequence 5'-3' | Position in |
| name | cDNA | |
| (nucleotides) | ||
| 261F | GCCATGACCGTGGAGCAGAACG | 367-388 |
| 261R | CAGCACTAAACCAACAAGTATTGCCA | 891-916 |
| 1.2F | GAAGCCAGCATAGCAGAGAATAAAGC | 769-794 |
| 1.2R | CACAGGTGGTAATTCAGTAAACCAATG | 1,450-1,476 |
| 321F | GGGATGCACAACTTGCCCAG | 2,478-2,496 |
| 321R | CCCTGCTTCAGGGCCACACCTG | 2,772-2,793 |
| N1 | AGGAGACCCAGAATAT | 2,574-2,589 |
| 121R | CAACCTTGCATATTCCAACT | 2,810-2,829 |
| *5A11.1 | GCTGCTTTGGCATTCAGC | 1,947-1,968 |
| *5A11.2 | TGACAAACTACCCTACTTCAATG | 2,804-2,882 |
*These primers were used in the splicing analysis of USP28.
The additional sequences between exons 3-4 and 17-18 were not further observed in any tissue tested. However, the alternativesequence between exons 10 and 11 (named exon 10b) was clearly detected and verified by sequencing in testis (Figure 4a), where both transcripts (exons 10-11 and 10-10b-11) had been amplified at similar levels. Exon 10b was also detected, albeit faintly, in small intestine, spleen and peripheral leukocytes. This exon encompasses 124 bp and generates a stop codon in the open reading frame (ORF) at nucleotide 1,453. Therefore, this transcript would produce a 361 amino acid truncated protein where the cysteine, but not the histidine UBP domain, would be preserved.
Figure 4.
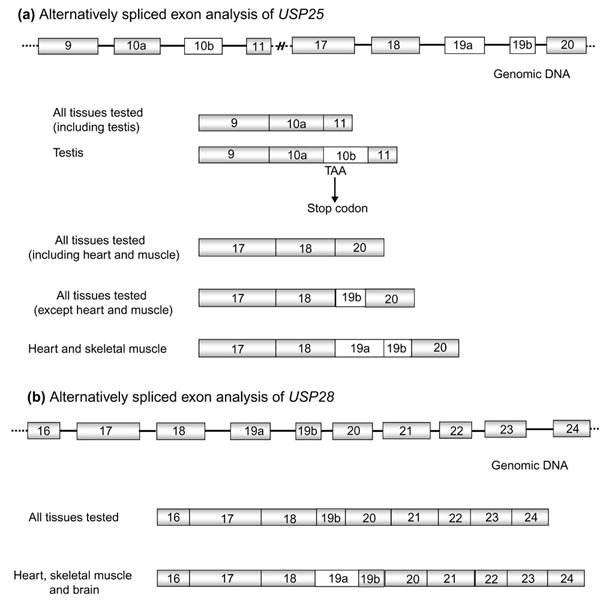
Alternatively spliced exon analysis of USP25 and USP28. (a) The specific USP25 isoforms of testis (exons 10a-10b-11) and heart and muscle (18-19a-19b-20). (b) The specific USP28 isoform of heart, muscle and brain (18-19a-19b-20).
RT-PCR experiments performed with specific primers, 321F-321R (located in exons 17 and 19b, respectively) and N1-121R (in exons 18 and 20, respectively) (Table 2), allowed us to visualize three mRNA isoforms. The shortest, already identified in many expressed sequence tags (ESTs), showed exon 18 directly fused to exon 20 and was detected in all the 24 different tissues tested (Multiple Tissue cDNA Panel, Clontech). The second isoform contained exons 18-19b-20, was of intermediate size and corresponded to the reported USP25 [14]. This isoform was detected in all the assayed tissues except fetal and adult muscle and heart (Figure 4a). Finally, the longest isoform was only detected in fetal and adult muscle and heart mRNAs and included a new exon (named 19a) which was always fused to 19b (18-19a-19b-20) in these tissues (Figure 4a). Exon 19a contained 114 bp that added 38 amino acids to the reported USP25 protein.
Primers 5A11.2 and 5A11.1 from USP28 (Table 2), located respectively in exons 16 and 24, were used to verify the homolog of the USP25 muscle and heart isoform (exons 18-19a-19b-20). RT-PCRs were performed in different tissues and the amplified products were subcloned and sequenced. Two USP28 sequences were detected in adult and fetal muscle, heart and brain. The shortest fragment corresponded to the reported USP28 cDNA containing exon 19b (Figure 4b). The longest contained a new exon (named 19a) (Figure 4b), which introduced 62 amino acids to the reported ORF. The tissue-specific exon 19a from USP25 (38 amino acids) and USP28 (62 amino acids) showed 41.6% amino acid identity. In contrast to USP25 exon 19b, the exon 19b of USP28 (sharing 53.1% amino acid identity) was not alternatively spliced, as it was always present in the transcripts from all tissues assayed.
Assay for ubiquitin-specific protease activity
To test the ubiquitin-specific protease activity of USP28, USP25 and the muscle and heart USP25 isoform (iUSP25), these enzymes were synthesized as inducible fusion proteins in the pGEX-4T-1 vector (pGEX-USP25; pGEX-iUSP25 and pGEX-USP28). Escherichia coli XL1blue cells were cotransformed with each construct and a plasmid expressing the fusion protein Ub-M-β-gal, a synthetic substrate for UBP. Immunoblot analysis using anti-β-galactosidase (β-gal) antibodies showed an efficient cleavage of ubiquitin from the Ub-M-β-gal fusion protein (Figure 5, lanes 5, 6 and 7). As expected, cells with the control pGEX-4T-1 vector failed to cleave Ub-M-β-gal (lane 4). The endogenous E. coli XL1-blue β-galactosidase (lacking the α peptide, and thus producing a shorter protein) was also detected (lanes 2 and 8).
Figure 5.
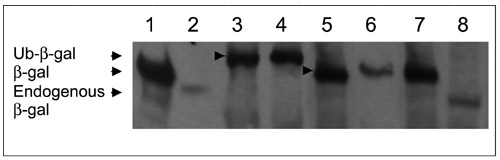
Deubiquitinating activity assay for USP25, the muscular USP25 isoform and USP28. The immunoblot was detected using anti-β-galactosidase (β-gal) antibodies. The sizes corresponding to Ub-β-gal, β-gal and endogenous deleted β-gal are shown by black arrows. Arrowheads on the lanes show the size of the Ub-β-gal fusion protein and the deubiquitinated β-gal. Lane 1, molecular weight marker containing wild-type β-gal; lane 2, untransformed XL1blue (negative control); lanes 3 and 4, XL1blue cells transformed, respectively, with pAC-M-β-gal alone or pAC-M-β-gal and empty pGEX vector; lanes 5, 6 and 7, XL1blue cells transformed, respectively, with pAC-M-β-gal and USP25, pAC-M-β-gal and muscular USP25 isoform, and pAC-M-β-gal and USP28; lane 8, XL1blue cells transformed with the empty pGEX vector.
Overexpression of USP25 in Down syndrome patients
As USP25 is located on chromosome 21, its overexpression could be presumed in Down syndrome (DS) patients. In order to assess the expression level of USP25 in DS versus control samples, we analyzed eight independent fetal brain samples (four trisomic and four disomic) by real-time quantitative PCR. An average of 1.7-fold overexpression of USP25 was shown in trisomic versus disomic samples.
Discussion
Database homology searches with the reported USP25 [14] have led to the characterization of a new UBP member in the human genome, named USP28, which maps at 11q23. Structural comparisons at the genomic and protein levels of USP25 and USP28, and the deubiquitinating enzymatic assays, allowed the definition of a new UBP subfamily.
Sequence alignments of UBPs have been hindered by the few shared conserved segments, which need specific computer programs to be identified. Nonetheless, several UBP subfamilies have been reported so far. DUB1 and DUB2, with 88.4% amino acid identities, is one of those. Similarly, alignments of UBP41, UBP46, UBP52 and UBP66 from chick skeletal muscle suggest a new subfamily. In our case, the alignment of the newly reported USP28 with USP25 showed homologies beyond the conserved UBP domains and amino acid identities amounting to 51%. In addition, the extensive alignment at the amino-terminal segment supports the proposed USP28 translation initiation site. USP25 and USP28 share the exon-intron distribution and those intron positions with an inaccurate match, located between exons 1-2, 13-14 and 17-18 (Figure 2), could be explained by slippage of the donor or acceptor splice sites. All these data strongly support a common ancestry for USP25 and USP28, and suggest that the minor variations observed may contribute to functional differences.
Northern analysis showed abundant USP28 expression in adult skeletal muscle and heart (Figure 3). The transcript size (4.5 kb approximately) was longer than expected from cDNA analysis, although this variability could be due to distinct polyadenylation and transcription initiation sites, as reported for USP25 and other UBPs [17].
The high specificity attributed to individual members of the ubiquitin proteolytic system and the structural complexity of USP25 and USP28 prompted us to analyze the tissue-specific mRNA isoforms and the cellular localization of the proteins. Alternative splicing had been also reported for other UBP members, such as USP3 [17], USP4 (previously named UnpEL-UnpES [13]), USP5 (previously named ISOT-1/2 [18,19,20]), USP9X (previously named DFFRX [15]) and USP15 [9]. Some cDNA clones of USP25 and USP28 contained additional exons that introduced in-frame stop codons, similarly to what has been reported for USP15 and USP3. These transcripts most probably originated from splicing errors and were fortuitously cloned, as they did not appear after northern or RT-PCR analyses. Besides, the absence of the conserved domains in the truncated proteins would compromise functionality. Nonetheless, the USP25 RT-PCR assays in testis produced comparable amplification levels of sequences containing either exons 10-10b-11 (10b introduces an in-frame stop codon) or exons 10-11. This argues in favor of a tissue-specific function for the truncated protein, possibly related to substrate availability and/or enzyme activity.
Exon 19b is present in all USP28 transcripts in all the assayed tissues. In constrast, exon 19b from USP25 is present in all tissues but not in all transcripts, thus constituting an alternatively spliced exon. The high degree of sequence homology of exon 19b from both genes supports its functional relevance. In addition, the tissue specificity shown for exon 19a of USP25 (muscle and heart) and USP28 (muscle, heart and brain) could confer the enzyme a tissue specificity to deubiquitinate a ubiquitous substrate. This would apply to a widely expressed gene whose function was only relevant in some tissues, as suggested for Fam [20]. Alternatively, the tissue-specific exon would bestow on the enzyme the ability to recognize a tissue-specific substrate, as suggested for Faf [20].
The specific subcellular localization reported for some deubiquitinating enzymes may imply spatial restriction of either the locus of action or the accessibility to the substrate but it might also indicate regulation of cellular processes where ubiquitylation plays a role unrelated to protein degradation (for a recent review see [21]). In our case, preliminary subcellular localization experiments with protein fusions to GFP showed that USP25 was cytosolic (data not shown) and did not support an involvement outside the ubiquitin-proteasome pathway.
Although several deubiquitinating enzymes have been shown to contribute to development and differentiation (that is, Faf (Drosophila Fat facets) [15] and UBP43 [22]), the specific function of most family members remains unknown. Homology searches of the domains conserved between different UBPs would help to elucidate the function of new members and define the substrate-specific domains. The contribution of USP25 to Down syndrome pathogenesis is still unclear. However, its overexpression (1.7-fold ± 0.13, P < 0.05 according to the Mann-Whitney test) in Down syndrome with respect to control fetal brain samples would support its involvement in the pathology. In fact, several UBPs have shown gene-dosage effects, such as USP9Y (whose gene is located in the Y-chromosome pseudoautosomal region and is involved in male azoospermia [23]) and other USPs related to aneuploidy syndromes: DFFRX in Turner syndrome [15] and USP18 in DiGeorge syndrome [24]. On the other hand, in vitro overexpression or inhibition of some ubiquitin-specific proteases has led to programmed cell death, supporting the idea that their activity is dose dependent [25].
Materials and methods
USP28 cDNA cloning
To screen cDNA libraries from human fetal brain, placenta and kidney (Clontech), specific primers of the USP-like region on chromosome 11 were designed (Figure 1) after comparison of USP25 cDNA against AC002036, a PAC that contained genomic DNA from chromosome 11. Approximately, 106 phages of each library were plated and transferred onto Nylon membranes (HybondN, Amersham-Pharmacia Biotech).
Probes were labeled by random hexamer priming with [α-32P]dCTP. A solution containing 50% (v/v) formamide, 5× SSC, 5× Denhart's, 0.1% (w/v) SDS and 100 μg/ml denatured salmon sperm was used to pre-hybridize and hybridize the filters for 2 h and 18 h, respectively. Washes were performed as follows: two washes in 2× SSC/0.1%SDS at 65°C for 10 min and two times in 1× SSC/0.1%SDS at 65°C for 10 min. Filters were exposed during two days with double screen in order to amplify the positive signals. Positive clones were subcloned into pBLUESCRIPT SK+ (Stratagene), sequenced in an ABI377 automatic sequencer (Applied Biosystems) and analyzed at the NCBI BLAST server [26]. As only truncated cDNA clones were initially isolated, additional screenings were needed to identify the full-length cDNA.
Northern blot analysis of USP28
The cDNA clone 5A11 (from nucleotides 1,473 to 3,618, Figure 1) was used to probe a human multiple tissue northern blot (Clontech). Hybridization and washes were carried out according to the manufacturer's protocol. A control actin probe was used to assess the amount of RNA poly(A)+ loaded in each lane.
Specificity of alternative splicing events
A set of primers corresponding to exon sequencesof USP25 and USP28 (Table 2) (USP25 primers numbered according to [14]) were designed for RT-PCR experiments on 24 samples from different tissues (Multiple Tissue cDNA Panels, Clontech). Amplification was performed in a total volume of 25 μl containing: 2.5 μl of template cDNA, 200 μM dNTPs, 5 pmol of each primer, 1× Taq Platinum buffer, 1.5 mM MgCl2 and 1U Taq Platinum (GIBCO-BRL). After denaturing at 94°C for 3 min, two-step PCR was carried out for 35 cycles at 94°C for 30 sec and 58°C for 30 sec. Final extension was for 5 min at 72°C. Bands differing from the expected size were subcloned in pBLUESCRIPT SK+ (Stratagene) and subsequently sequenced in an ABI377 automatic sequencer (Applied Biosystems). Sequences were analyzed using the BLAST software at the NCBI [26,27].
Assay for ubiquitin-specific protease activity
The ubiquitin-specific protease activity of USP25, USP25-specific heart and muscle isoform, which contains the additional exon 19a (USP25 isoform), and USP28 was determined as described [10,11]. The three corresponding cDNAs were cloned in-frame in pGEX-4T-1 (Amersham Pharmacia Biotech) downstream from the glutathione-S-transferase (GST) coding region. Plasmid pACY184 Cmr expressing Ub-Met-β-gal protein fusion (pAC-M-β-gal) was kindly provided by M. Hoschtrasser. E. coli XL1blue bacteria were co-transformed with pAC-M-β-gal and either pGEX-4T-1-USP25 or pGEX-4T-1-USP25 isoform or pGEX-4T-1-USP28. Ampr and Cmr colonies were grown and induced for 3 h with isopropyl-β-thiogalactopyranoside (final concentration 1 mM). Total protein extracts were analyzed by western blotting with anti-β-galactosidase rabbit polyclonal antibody (Cappel).
Overexpression of USP25 in Down syndrome patients
Total RNA from 5 mg of fetal brains of Down and control samples (three samples each) were obtained using ABI PRISM 6700 Automated Nucleic Acid workstation. RT-PCRs were produced according to Taqman Reverse transcription reagents (Roche Molecular Systems). Quantitative PCRs were performed using the Universal Master Mix (Applied Biosystems) following the manufacturer's specifications. The real-time amplification was analyzed by the ABI Prism 7700 Sequence detection system. USP25 primers and the Taqman probe (using FAM as reporter and TAMRA as quencher) were designed according to the Primer Express software. Forward primer: 5'-GATGAAAGGTGTCACAACATAATGAAA-3'; reverse primer: 5'-CCACTCCTCATATTCCTCCAAGTTT-3'; TaqMan probe: 5'-TCAAGCCAAACTGGAAATGATAAAACCTGAAGA-3'. To normalize the USP25 quantitative determinations, GAPDH (Applied Biosystems) was used as endogenous control. The standard deviation of the disomic and trisomic samples was 0.08 and 0.13, respectively. The overexpression (1.7 fold) of USP25 in trisomic fetal brains shows statistical significance according to the Mann-Whitney test (P < 0.05).
Acknowledgments
Acknowledgements
We thank the Serveis Científico-Tècnics de la Universitat de Barcelona for the use of the 377 ABI PRISM and Robin Rycroft for revising the English version. We are very grateful to M. Hochstrasser for the gift of the pAC-M-β-gal plasmid. This study was funded by Fundació Síndrome de Down/Marató de TV3-1993, European Union Grant BIO4-CT97-2123 and Ministerio de Ciencia y Tecnología PM99-0168 to R.G-D.
References
- Hershko A, Ciechanover A. The ubiquitin system for protein degradation. Annu Rev Biochem. 1992;61:761–807. doi: 10.1146/annurev.bi.61.070192.003553. [DOI] [PubMed] [Google Scholar]
- Hochstrasser M. Ubiquitin-dependent protein degradation. Annu Rev Genet. 1996;30:405–439. doi: 10.1146/annurev.genet.30.1.405. [DOI] [PubMed] [Google Scholar]
- D'Andrea A, Pellman D. Deubiquitinating enzymes: A new class of biological regulators. Crit Rev Biochem Mol Biol. 1998;33:337–352. doi: 10.1080/10409239891204251. [DOI] [PubMed] [Google Scholar]
- Hochstrasser M. Ubiquitin, proteasomes, and the regulation of intracellular protein degradation. Curr Opin Cell Biol. 1995;7:215–223. doi: 10.1016/0955-0674(95)80031-x. [DOI] [PubMed] [Google Scholar]
- Chung CH, Baek SH. Deubiquitinating enzymes: their diversity and emerging roles. Biochem Biophys Res Commun. 1999;266:633–640. doi: 10.1006/bbrc.1999.1880. [DOI] [PubMed] [Google Scholar]
- Leroy E, Boyer R, Auburger G, Leube B, Ulm G, Mezey E, Harta G, Brownstein MJ, Jonnalagada S, Chernova T, et al. The ubiquitin pathway in Parkinson's disease. Nature. 1998;395:451–452. doi: 10.1038/26652. [DOI] [PubMed] [Google Scholar]
- Baker RT, Tobias JW, Varshavsky A. Ubiquitin-specific proteases of Saccharomyces cerevisiae. Cloning of UBP2 and UBP3, and functional analysis of the UBP family. J Biol Chem. 1992;267:23364–23375. [PubMed] [Google Scholar]
- Wilkinson KD. Regulation of ubiquitin-dependent processes by deubiquitinating enzymes. FASEB J. 1997;11:1245–1256. doi: 10.1096/fasebj.11.14.9409543. [DOI] [PubMed] [Google Scholar]
- Baker RT, Wang XW, Woollatt E, White JA, Sutherland GR. Identification, functional characterization, and chromosomal localization of USP15, a novel human ubiquitin specific protease related to UNP oncoprotein, and a systematic nomenclature for human ubiquitin-specific proteases. Genomics. 1999;59:264–274. doi: 10.1006/geno.1999.5879. [DOI] [PubMed] [Google Scholar]
- Papa FR, Hochstrasser M. The yeast DOA4 gene encodes a deubiquitinating enzyme related to a product of the human tre-2 oncogene. Nature. 1993;366:313–319. doi: 10.1038/366313a0. [DOI] [PubMed] [Google Scholar]
- Zhu Y, Lambert K, Corless C, Copeland NG, Gilbert DJ, Jenkins NA, D'Andrea A. DUB-2 is a member of a novel family of cytokine-inducible deubiquitinating enzymes. J Biol Chem. 1997;272:51–57. doi: 10.1074/jbc.272.1.51. [DOI] [PubMed] [Google Scholar]
- Baek SH, Park KC, Lee JI, Kim KI, Yoo YJ, Tanaka K, Baker RT, Chung CH. A novel family of ubiquitin-specific proteases in chick skeletal muscle with distinct N- and C- terminal extensions. Biochem J. 1998;334:677–684. doi: 10.1042/bj3340677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frederick A, Rolfe M, Chiu MI. The human UNP locus at 3p21.31 encodes two tissue-selective, cytoplasmic isoforms with deubiquitinating activity that have reduced expression in small cell lung carcinoma cell lines. Oncogene. 1998;16:153–165. doi: 10.1038/sj.onc.1201537. [DOI] [PubMed] [Google Scholar]
- Valero R, Marfany G, Gonzalez-Angulo O, Gonzalez-Gonzalez G, Puelles L, Gonzalez-Duarte R. USP25, a novel gene encoding a deubiquitinating enzyme is located in the gene-poor region 21q11.2. Genomics. 1999;62:395–405. doi: 10.1006/geno.1999.6025. [DOI] [PubMed] [Google Scholar]
- Jones MH, Furlong RA, Burkin H, Chalmers IJ, Brown GM, Khwaja O, Affara N. The Drosophila developmental gene fat facets has a human homologue in Xp11.4 which escapes X-inactivation and has related sequences on Yq11.2 A. Hum Mol Genet. 1996;5:1695–1701. doi: 10.1093/hmg/5.11.1695. [DOI] [PubMed] [Google Scholar]
- Lennon G, Auffray C, Polymeropoulos M, Soares MB. The IMAGE consortium: an integrated molecular analysis of genomes and their expression. Genomics. 1996;33:151–152. doi: 10.1006/geno.1996.0177. [DOI] [PubMed] [Google Scholar]
- Sloper MK, Eyre HJ, Wang XW, Sutherland GR, Baker RT. Characterization and chromosomal localization of USP3, a novel human ubiquitin-specific protease. J Biol Chem. 1999;274:26878–26884. doi: 10.1074/jbc.274.38.26878. [DOI] [PubMed] [Google Scholar]
- Falquet L, Paquet N, Frutiger S, Hughes GJ, Hoang-Van K, Jaton JC. A human de-ubiquitinating enzyme with both isopeptidase and peptidase activities in vitro. FEBS Lett. 1995;359:73–77. doi: 10.1016/0014-5793(94)01451-6. [DOI] [PubMed] [Google Scholar]
- Wilkinson KD. Roles of ubiquitinylation in proteolysis and cellular regulation. Annu Rev Nutr. 1995;15:161–189. doi: 10.1146/annurev.nu.15.070195.001113. [DOI] [PubMed] [Google Scholar]
- Chen X, Overstreet E, Wood SA, Fischer JA. On the conservation of function of the Drosophila Fat facets deubiquitinating enzyme and Fam, its mouse homolog. Dev Genes Evol. 2000;210:603–610. doi: 10.1007/s004270000109. [DOI] [PubMed] [Google Scholar]
- Weissman AM. Themes and variations on ubiquitylation. Nature Rev Mol Cell Biol. 2001;2:169–178. doi: 10.1038/35056563. [DOI] [PubMed] [Google Scholar]
- Liu LQ, Ilaria R, Jr, Kingsley PD, Iwama A, van Etten RA, Palis J, Zhang DE. A novel ubiquitin-specific protease, UBP43, cloned from leukemia fusion protein AML1-ETO-expressing mice, functions in hematopoietic cell differentiation. Mol Cell Biol. 1999;19:3029–3038. doi: 10.1128/mcb.19.4.3029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun C, Skaletsky H, Birren B, Devon K, Tang Z, Silber S, Oates R, Page DC. An azoospermic man with a de novo point mutation in the Y-chromosomal gene USP9Y. Nat Genet. 1999;23:429–432. doi: 10.1038/70539. [DOI] [PubMed] [Google Scholar]
- Schwer H, Liu LQ, Zhou LM, Little MT, Pan Z, Hetherington CJ, Zhang DE. Cloning and characterization of a novel human ubiquitin-specific protease, a homologue of murine UBP43 (USP18). Genomics. 2000;65:44–52. doi: 10.1006/geno.2000.6148. [DOI] [PubMed] [Google Scholar]
- Monney L, Otter I, Olivier R, Ozer HL, Haas AL, Omura S, Borner C. Defects in the ubiquitin pathway induce caspase-independent apoptosis blocked by Bcl-2. J Biol Chem. 1998;273:6121–6131. doi: 10.1074/jbc.273.11.6121. [DOI] [PubMed] [Google Scholar]
- NCBI BLAST server http://www.ncbi.nlm.nih.gov/BLAST/
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman D. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]


