Abstract
Please cite this paper as: Allard et al. (2012) Invasive bacterial infections following influenza: a time‐series analysis in Montréal, Canada, 1996–2008. Influenza and Other Respiratory Viruses 6(4), 268–275.
Background Shared seasonal patterns, such as between influenza and some respiratory bacterial infections, can create associations between phenomena not causally related.
Objectives To estimate the association of influenza with subsequent bacterial infections after full adjustment for confounding by seasonal and long‐term trends.
Methods Time series of weekly counts of notified cases of invasive infections with Haemophilus influenzae, Neisseria meningitidis, Streptococcus pneumoniae and Streptococcus pyogenes, in Montréal, Canada, 1996–2008, were modelled by negative binomial regression, with terms representing seasonal and long‐term trends and terms for numbers of positive laboratory tests for influenza A and B.
Results The associations of S. pneumoniae, H. influenzae and N. meningitidis with influenza disappeared after seasonal terms were added to the model. However, the influenza B count remained associated with the S. pyogenes counts for the same week and the following week: S. pyogenes incidence rate ratios were 1.0376 (95% CI: 1.0009–1.0757) and 1.0354 (0.9958–1.0766), respectively, for each increase of 1 in the influenza count.
Conclusions Influenza B accounts for about 8percnt; of the incidence of invasive S. pyogenes infections, over and above any effect associated with modellable seasonal and long‐term trends. This association of influenza B with S. pyogenes infections can be attributed largely to the years 1997, 2001, 2007 and 2008, when late peaks in influenza B counts were followed by peaks in S. pyogenes notifications. This finding reinforces the case for universal immunization against influenza, as partial protection against the ‘flesh eating disease’.
Keywords: Bacterial infections, incidence, influenza
At the time of the 1889 influenza pandemic, there was still much uncertainty about the disease being infectious 1 but after the discovery of Haemophilus influenzae by Pfeiffer in 1892, the belief in its bacterial aetiology became widespread. 2 However, invasive bacterial infections have been recognized as complications of influenza at least since the 1918 pandemic 3 , 4 when some, like Goodpasture, believed the disease itself to be ‘caused by an unknown virus’. 5 Bacteria were well confirmed as secondary invaders by the 1930s. 6
The association between viral and subsequent bacterial infections result from a great variety of mechanisms, including the breakdown of physical barriers, increased bacterial adhesion, increased apoptosis of antibacterial immune cells and decreased phagocytosis by alveolar macrophages. 7 Specifically, there is evidence that the hemagglutinin of the influenza virus promotes the internalization Streptococcus pyogenes (group A beta‐haemolytic streptococci) 8 and that the neuraminidase promotes the adherence of Streptococcus pneumoniae to human respiratory cells. 9
At a population level, over several decades and in various populations, strong and consistent associations have been found between influenza and secondary pneumonia caused by Staphylococcus aureus, S. pneumoniae, Neisseria meningitidis, 10 S. pyogenes and Hemophilus influenzae, 10 , 11 and also with mortality. This applies to seasonal 6 , 12 and pandemic 10 influenza A, including the 2009 pH1N1 strain, 13 and to influenza B. 6 About 95% of deaths associated with influenza are considered to be caused by secondary bacterial pneumonia, the susceptibility to which is thought to peak 1–2 weeks after exposure to influenza. 11 , 14 However, the peak may be later for pneumococcal infections, up to 4 weeks in adults and 8 weeks in children. 15
The seasonality of transmissible diseases may be due to the following factors: (i) changes in host susceptibility, possibly related to light–dark cycles or photoperiodicity 12 , 16 , 17 , 18 , 19 and to melatonin production 16 , 20 ; (ii) latitude, 16 , 19 solar radiation, 21 humidity 19 , 22 and temperature 19 , 23 ; (iii) air pollution 23 ; (iv) crowding of susceptible hosts 16 , 23 ; and (v) the circulation of other, predisposing or triggering, infectious agents. 17 To what extent any of the above actually causes seasonal variations in the incidence of a given transmissible disease, or is merely a confounder, 16 , 19 , 23 , 24 lies at the heart of the seasonality problem.
Seasonality has been studied mostly by (i) aggregating data by season, 12 (ii) by modelling time series of numbers of cases using regression or autoregression 17 , 25 or Poisson models with terms representing seasonal and secular trends, 26 or (iii) by dynamic (compartmental, SEIR) modelling. 19 , 24 , 27 Conformity of the observations to the seasonal pattern expected on the basis of such models tells us little about the processes behind the pattern, there being so many synchronous processes whose effects are jointly subsumed under the seasonal model terms. However, non‐systematic deviations of the observations from the predicted pattern may be easier to associate with a particular cause. 18
Our goal in the present study was therefore twofold. First, to model the reported incidence of four invasive bacterial infections, those caused by S. pneumoniae, S. pyogenes, H. influenzae and N. meningitidis, as functions of earlier numbers of positive tests for influenza A and B. Second, in doing so, to represent as fully as possible seasonality and long‐term trends in the reported incidences of bacterial infections by means of dedicated model terms, leaving only the remaining variance to be explained by earlier numbers of positive influenza tests.
Methods
We studied all laboratory‐confirmed cases of invasive S. pneumoniae, S. pyogenes, H. influenzae and N. meningitidis infection among residents of Montréal, Canada, notified to the Public Health Department of the Montréal Health and Social Services Agency between 1 January 1996 and 7 June 2008 (with the exception of S. pneumoniae, whose notification began on January 5, 1997). The notifications of each infection were aggregated into weekly case counts. Numbers of positive influenza A and B test results for residents of Montréal during the same period were provided by the Québec Public Health Laboratory from a sentinel laboratory surveillance network database and were similarly aggregated into weekly influenza A or B counts. The number of hospital laboratories in the sentinel network varied from six in 1996 to seven in 2008, with a stable core of five laboratories throughout the study period. Testing practices changed after the introduction of commercial rapid tests in 1996 and after improvement in the access to antiviral drugs, contributing to an overall increase in the total number of positive results.
A binary indicator variable (short work week) was created to identify weeks that include fewer than five working days, when fewer notifications are to be expected, because non‐emergency medical care is less accessible or notification is delayed. About 6 weeks per year include other holidays that were not taken into account because their dates vary from year to year and could not be recovered.
Descriptive analyses used frequency distribution histograms, correlation matrices and time series of counts. Modelling of the bacterial infection counts used multivariate negative binomial regression, rather than Poisson regression, because it does not require the variance of observations to be equal to their mean. The parameters βi of the models are most easily understood after transformation into incidence rate ratios (IRR). The IRR for any predictor x i is equal to e βi; it represents the ratio of the expected value of the count when x i is in the model over its expected value when x i is not in the model. If x i is an influenza count, the IRR is the ratio of the risk associated with a count of x i + 1 compared to the risk for a count of x i; the IRR for a count of x i + n compared to x i is IRRn.
Overall trends in the time series of counts were accounted for by including in the model restricted cubic spline terms to remove long‐term (secular) trends, and sine‐cosine (periodic) terms to remove seasonal trends. 28 The restricted cubic spline function used had six knots 29 and therefore five parameters, 30 referred to as spline1–spline5. The small number of knots was chosen so that the spline terms would reflect only the long‐time trends, while the periods for the sine‐cosine terms were 1 year, plus the first five harmonics (1/2 year, 1/3, 1/4, 1/5, 1/6) to account for seasonality and any <1 year periodicities. Periodic terms had the forms sin(xt) and cos(xt), where x = 1 to 6 and t = (2π × week number × 12.438)/649; the week number ranged from 1 to 649, representing 12.438 years, the length of our time series of counts.
Comparisons of the overall closeness‐of‐fit of nested models (all the terms in the smaller model being included in the larger model) used the deviance (−2 × difference between log‐likelihoods), tested by chi‐square. The deviance served as an overall test of whether a model including a set of terms (for instance the influenza counts) is statistically superior to the same model without these terms.
As any effect of influenza on the risk of subsequent bacterial infections should occur within a few weeks, 11 , 14 , 15 and relying on preliminary results, we decided to estimate the effects on the bacterial infection counts for any given week (called week 0) of influenza counts for the same week and for the five preceding weeks (called weeks −1 to −5).
All analyses were carried out with stata 10.1 (Stata Corp., Cary, NC, USA). The data used are all publicly available aggregate surveillance results, and no ethical review was required.
Results
During the study period, there were 3027 positive tests for influenza A, with a mean weekly count of 4.664 (range 0–85) and 745 positive tests for influenza B (mean: 1.148, range 0–33). There were 107 infections caused by H. influenzae (mean: 0.165, variance: 0.212), 156 by N. meningitidis (mean: 0.240, var.: 0.248), 2920 by S. pneumoniae (over the last 596 weeks, mean: 4.899, var.: 11.190) and 667 by S. pyogenes (mean 1.028, var.: 1.292). Weekly bacterial infection counts were roughly Poisson distributed, but variances being consistently larger than means justified the choice of negative binomial regression.
The time series of influenza A and B counts (Figure 1) shows that type B varies proportionately more from year to year than type A and usually peaks some weeks later. The smoothed N. meningitidis and H. influenzae counts (Figure 2) show a seasonality that is irregular in period and amplitude; in addition, H. influenzae counts increased markedly between 2003 and 2004, when all invasive H. influenzae infections became reportable, not only those of type B. The smoothed S. pneumoniae and S. pyogenes counts (Figure 3) show yearly peaks that are particularly marked for S. pneumoniae; however, the peaks are not synchronous, those for S. pyogenes tending to occur later in the year.
Figure 1.
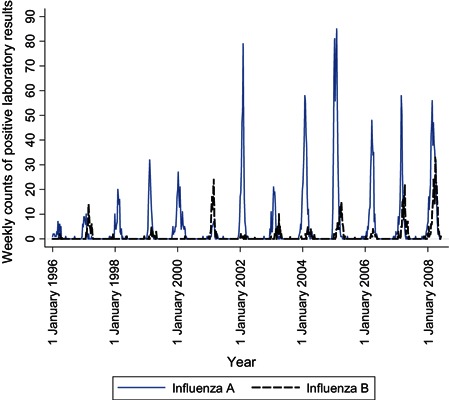
Time series of weekly numbers of positive tests for influenza A and B, Montréal, 1996–2008.
Figure 2.
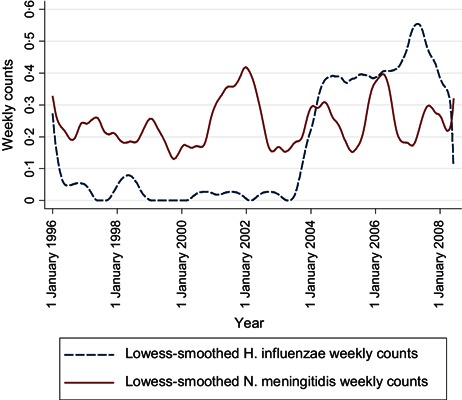
Time series of weekly numbers of notified cases of Haemophilus influenzae and Neisseria meningitidis infections, Montréal, 1996–2008.
Figure 3.
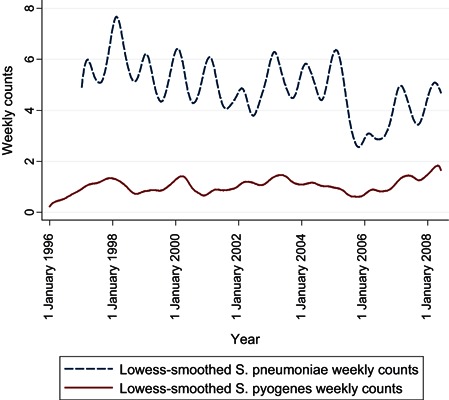
Time series of weekly numbers of notified cases of Streptococcus pneumoniae and Streptococcus pyogenes infections, Montréal, 1996–2008.
Correlations between weekly influenza counts and later bacterial counts were generally high, as expected (results not shown), but the corresponding IRR adjusted for short work weeks, long‐term and seasonal trends, in Table 1, show much weaker associations. Only two IRR, between influenza B and subsequent S. pyogenes infections, approach significance.
Table 1.
Incidence rate ratios (IRR) between each of the bacterial infection counts for a given week (week 0) and the influenza A and B counts for weeks 0 to −5. Negative binomial models including influenza A and B counts, plus the short work week, long‐term and seasonal terms (not shown)
| Independent variables | Dependent variables | |||||
|---|---|---|---|---|---|---|
| Influenza type | Week | Streptococcus pneumoniae | Streptococcus pyogenes | Haemophilus influenzae | Neisseria meningitidis | |
| A | 0 | 1·0052* | 1·0114 | 0·9850 | 0·9937 | |
| −1 | 0·9963 | 0·9882 | 1·0290 | 0·9994 | ||
| −2 | 1·0083 | 1·0068 | 0·9768 | 1·0039 | ||
| −3 | 0·9924 | 1·0118 | 0·9894 | 1·0053 | ||
| −4 | 1·0022 | 0·9836 | 1·0288 | 0·9782 | ||
| −5 | 1·0015 | 1·0075 | 0·9781 | 1·0006 | ||
| B | 0 | 1·0060 | 1·0376a | 0·9344 | 0·9660 | |
| −1 | 1·0080 | 1·0354b | 1·0228 | 1·0195 | ||
| −2 | 1·0034 | 0·9753 | 1·0123 | 0·9726 | ||
| −3 | 1·0056 | 0·9808 | 1·0001 | 0·9069c | ||
| −4 | 0·9860 | 0·9875 | 1·0233 | 1·0908d | ||
| −5 | 1·0102 | 0·9998 | 0·9930 | 1·0143 | ||
| Number of weeks | 596 | 644 | 644 | 644 | ||
*The probability associated with the IRR is >0·05 unless indicated in a footnote. n = 644. The first 5 weeks are excluded from the analysis, as influenza counts for earlier weeks up to −5 are unavailable for them.
a P = 0·045; b P = 0·080; c P = 0·093; d P = 0·051.
The full model for influenza B and S. pyogenes, in Table 2, shows several effects, all adjusted for each other. (i) Short work weeks are associated with a 27% decrease [(1−0.7337)*100] in S. pyogenes notifications. (ii) The large coefficient for sine1 reflects the yearly notification peaks, but there is also a weaker six‐monthly (cosine2) and two‐monthly (cosine6) periodicity. (iii) All the long‐term‐trend terms are highly significant. (iv) Influenza B counts for a given week (week 0) are associated with increases in S. pyogenes notifications in the same week and in the following one (weeks 0 and +1), of about 3.8% [(1.0376−1)*100] and 3.5% [(1.0354−1)*100] per additional positive influenza B test, respectively. They are also associated, although not statistically significantly, with decreases in S. pyogenes notifications in weeks +2 to +5. These two phenomena together are suggestive of ‘harvesting’ 31 , 32 : at least some of the S. pyogenes infections following influenza B would have occurred even without influenza, but a few weeks later, having merely been brought forward in time by influenza B. However, a model containing influenza B counts for all 6 weeks does not show a better fit than one containing only the counts for weeks 0 and −1 (deviance = 6.96, P = 0.1381), indicating that this apparent ‘harvesting’ of cases may well be in fact random variations in the parameters. (Note that the N. meningitidis terms marked c and d in Table 1, although large and almost significant, are in the wrong sequence for harvesting, which is why they are not reported as a finding.)
Table 2.
Full negative binomial regression model for the associations between Streptococcus pyogenes infection counts for a given week (week 0) and the influenza B counts for weeks 0 to −5*
| Independent variable | Regression coefficient | 95% CI of coefficient | IRR | 95% CI of IRR | |
|---|---|---|---|---|---|
| Short work week | – | – | 0·7337 | 0·5547–0·9705 | |
| Seasonal trends | Sine1 | 0·3113 | 0·1758–0·4469 | – | – |
| Cosine1 | −0·0646 | −0·1824–0·0532 | – | – | |
| Sine2 | −0·0414 | −0·1631–0·0804 | – | – | |
| Cosine2 | 0·1464 | 0·0192–0·2736 | – | – | |
| Sine3 | 0·0100 | −0·1141–0·1341 | – | – | |
| Cosine3 | 0·0546 | −0·0618–0·1710 | – | – | |
| Sine4 | −0·0069 | −0·1236–0·1098 | – | – | |
| Cosine4 | 0·0223 | −0·1039–0·1486 | – | – | |
| Sine5 | 0·0340 | −0·0820–0·1500 | – | – | |
| Cosine5 | 0·0230 | −0·0947–0·1407 | – | – | |
| Sine6 | −0·0166 | −0·1297–0·0965 | – | – | |
| Cosine6 | 0·1954 | 0·0758–0·3151 | – | – | |
| Long‐term trends | Spline1 | 0·0071 | 0·0024–0·0117 | – | – |
| Spline2 | −0·0654 | −0·1064 to −0·0243 | – | – | |
| Spline3 | 0·2113 | 0·0868–0·3359 | – | – | |
| Spline4 | −0·3157 | −0·4843 to −0·1471 | – | – | |
| Spline5 | 0·3303 | 0·1665–0·4941 | – | – | |
| Influenza B, week 0 | – | – | 1·0376 | 1·0009–1·0757 | |
| Influenza B, week ‐1 | – | – | 1·0354 | 0·9958–1·0766 | |
| Influenza B, week ‐2 | – | – | 0·9753 | 0·9346–1·0178 | |
| Influenza B, week ‐3 | – | – | 0·9808 | 0·9386–1·0249 | |
| Influenza B, week ‐4 | – | – | 0·9875 | 0·9440–1·0331 | |
| Influenza B, week ‐5 | – | – | 0·9998 | 0·9605–1·0408 |
*The regression coefficients and the bounds of their confidence intervals (CI) have been transformed into incidence rate ratios (IRR) when they are interpretable as such. n = 644. The first 5 weeks are excluded from the analysis, as influenza B counts for earlier weeks up to ‐5 are unavailable for them.
The comparison of observed and expected S. pyogenes counts before and after influenza counts are added to the model (4, 5) shows a visible improvement in fit. A comparison of the two dark lines shows that, with influenza B in the model, four transient increases in S. pyogenes infections are now modelled that were not before: those in 1997, 2001, 2007 and especially 2008. Looking back at the time series of influenza B counts (dark line in Figure 1), one notes that these were years with late, high peaks in the weekly numbers of influenza B‐positive tests. An additional indication that this association may be causal is that the average age of known influenza B cases in these 4 years is significantly higher, 33.9 years, than in other years, 24.6 years (P < 0.000), and closer to the average ages of S. pyogenes cases, 45.9 years.
Figure 4.
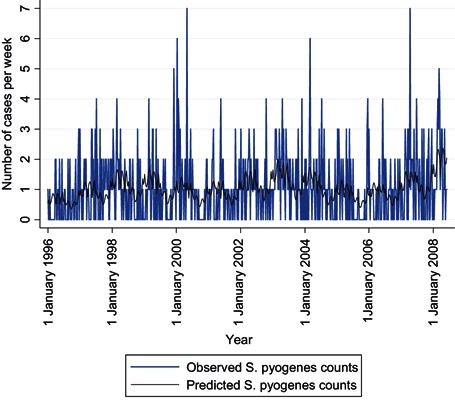
Time series of weekly Streptococcus pyogenes counts, observed and predicted by a binomial regression model including only short work week, seasonal and long‐term trend terms, Montréal, 1996–2008.
Figure 5.
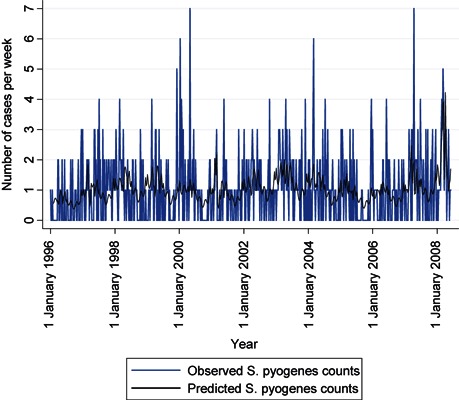
Time series of weekly Streptococcus pyogenes counts, observed and predicted from the full binomial regression model (as described in Table 2), Montréal, 1996–2008.
Comparisons of the closeness‐of‐fit of various nested models quantify rigorously the relative contributions of long‐term trends, seasonal trends and influenza (Table 3). All infections except those caused by N. meningitidis display significant long‐term trends (model 1), and both streptococcal infections also show significant seasonality (model 2). These findings confirm what has been observed in Figure 3. When long‐term trends but not seasonality are controlled for (models 3 and 4), influenza A and B counts significantly improve the model fit for both streptococcal infections, but the improvement is greater for S. pneumoniae than for S. pyogenes. When seasonality is also controlled for by dedicated terms, however, the only effect remaining is that of influenza B counts on S. pyogenes counts, as mentioned earlier (models 5 and 6). This effect remains essentially the same if one uses Poisson regression, includes earlier S. pyogenes counts in the model, splines with five or seven knots, or fewer harmonics for the periodic functions (results not shown).
Table 3.
Deviances between nested negative binomial models, with their probabilities*
| Models | Dependent variables | ||||
|---|---|---|---|---|---|
| Composition | Deviance*, probability | Neisseria meningitidis | Haemophilus influenzae | Streptococcus pneumoniae | Streptococcus pyogenes |
| 1 Long‐term trends | From constant: | 6·16 | 115·58 | 31·31 | 18·28 |
| pr = | 0·4051 | 0 | 0·0000 | 0·0055 | |
| 2 Long‐term and seasonal trends | From model 1: | 9·90 | 6·45 | 230·91 | 41·19 |
| pr = | 0·6249 | 0·8905 | 0 | 0·0000 | |
| 3 Long‐term trends and influenza A | From model 1: | 0·72 | 2·45 | 49·57 | 17·72 |
| pr = | 0·9940 | 0·8739 | 0 | 0·0069 | |
| 4 Long‐term trends and influenza B | From model 1: | 6·64 | 1·59 | 26·35 | 19·83 |
| pr = | 0·3554 | 0·9536 | 0·001 | 0·0029 | |
| 5 Long‐term and seasonal trends and influenza A | From model 2: | 3·18 | 3·60 | 6·78 | 7·01 |
| pr = | 0·7863 | 0·7312 | 0·3412 | 0·3201 | |
| 6 Long‐term and seasonal trends and influenza B | From model 2: | 10.06 | 2·39 | 5·85 | 13·56 |
| pr = | 0·1221 | 0·8807 | 0·4405 | 0·0349 | |
*Deviances were estimated using the last 596 weeks, when counts for all bacterial infections were available, including those due to Streptococcus pneumoniae. Those that are significant at the 0.05 level are in bold type.
Discussion
Associations between two phenomena that share a seasonal pattern are intrinsically difficult to interpret as causal because one can rarely exclude that they are both consequences of other phenomena displaying the same pattern (a form of confounding). Our analyses shows that such associations may well disappear once confounding by long‐term or seasonal trends has been controlled. We submit that the associations that have resisted such control provide the clearest information about true effects, in the absence of data on specific confounders. This may be especially true if these associations can be attributed to particular deviations from the usual patterns, as observed in explaining the differences between 4, 5 by means of Figure 1. Our results illustrate that the opportunities provided by modelling aberrations are different from those provided by modelling regularities: in Table 2, had the observed seasonality of S. pyogenes always been that predicted by the seasonal trend terms, no IRR for influenza B would have emerged as significant.
The S. pyogenes count for any given week is multiplied by 1.0376 for each additional influenza B‐positive test result that same week and by 1.0354 for the same increase the preceding week (Table 1). The combined effect of both weeks is the product of these IRR, 1.0743. The average weekly influenza B count being 1.1479, its effect over these 2 weeks is to multiply the S. pyogenes case count by 1.07431.1479 = 1.0858, an 8.6% increase. With an average of 59 S. pyogenes cases per year in Montréal, this represents about five extra cases every year.
All the estimates in the previous paragraph are approximations, being valid only to the extent that numbers of positive influenza B tests are a constant proportion of all incident cases of the disease. The reported incidence of invasive bacterial infections better reflects their true incidence, especially for those caused by N. meningitidis and S. pyogenes, because of their severity and their requiring an urgent public health intervention.
Other limitations include delays in the measurement of the variables. Ideally, one would use time series of dates of acquisition of all infections under study. In reality, for influenza we had the dates when the specimens were taken, and for the bacterial infections the dates when notifications reached the public health department (as this is the only date we have for all notified cases). Thus, the effects observed over weeks 0 and −1 probably occurred after only a few days, because the median delay between onset of symptoms because of S. pyogenes infection and its notification is 6 days, in the 489 cases for which it is known.
Another limitation in estimating the effects of influenza is the lack of control for (i) confounding by other triggering factors for invasive S. pyogenes infections, such as varicella and respiratory syncytial virus infections, temperature or humidity levels, and (ii) the variations in predominant influenza B subtype or in vaccine formulation and uptake. Point ii) might explain why the 2005 influenza B peak was not followed by a S. pyogenes peak. Effect modification, especially by age, also remains to be investigated: because of our relatively small bacterial infection counts, an age‐stratified analysis would have resulted in losing too much statistical power.
Finally, because we tried to model all trends, secular or periodic, as much as possible through dedicated terms, the absence of significant effects of influenza A, or of effects on other bacterial infections than those because of S. pyogenes, does not mean such effects do not exist. These effects may simply be so similar in strength and timing to the overall effects modelled by periodic and spline terms that their contributions are entirely subsumed by these terms.
Both types of influenza have been associated with secondary infections and an increased risk of mortality, 33 but the risk associated with type B has been less well documented than for type A. There are a few reports concerning the association of influenza B with unspecified or pneumococcal pneumonia, or other respiratory infections, 34 , 35 , 36 , 37 , 38 fewer with S. pyogenes infections, 39 , 40 , 41 and no attempt, to our knowledge, to quantify any of these associations.
The demonstration of a specific contribution of influenza B to the occurrence of invasive S. pyogenes infections, which include the widely feared ‘flesh eating disease’, reinforces the case for immunizing against influenza. There is already experimental evidence in mice that influenza A vaccine protects against S. pyogenes superinfection, 42 and influenza immunization has been recommended as protection against bacterial superinfections in humans, 14 including those caused by S. pyogenes. 43 , 44 Our rough estimate of the proportion of S. pyogenes infections in the general population attributable to influenza B (8%) is understandably smaller than the 50–77% preventable by influenza immunization among army trainees, 45 among whom the infection pressure for both diseases is very high. Yet, its greater generalizability provides additional support for the recent recommendation to immunize all persons aged ≥ 6 months against influenza. 46 The ability of invasive S. pyogenes infections to cripple or kill previously healthy adults makes even partial protection against them an appreciable argument in the promotion of universal influenza vaccination.
Acknowledgements
This work received no outside funding. The authors thank Ben J. Armstrong and Anthony Gasparini for their advice on time‐series modelling and Céline Plante for her technical help.
References
- 1. J.F.P. The epidemic of influenza. Nature 1889; 41:145–146. [Google Scholar]
- 2. Taubenberger JK, Hultin JV, Morens DM. Discovery and characterization of the pandemic influenza virus in historical context. Antivir Ther 2007; 12:581–591. [PMC free article] [PubMed] [Google Scholar]
- 3. Stone WJ, Swift GW. Influenza and influenzal pneumonia at Fort Riley, Kansas from Sept. 18 to Nov. 1, 1918. JAMA 1919; 72:487–493. [Google Scholar]
- 4. Goodpasture EW. Bronchopneumonia due to hemolytic streptococci following influenza. JAMA 1919; 72:724–725. [Google Scholar]
- 5. Goodpasture EW. The significance of certain pulmonary lesions in relation to the etiology of influenza. Am J Med Sci 1919; 158:863–870. [DOI] [PubMed] [Google Scholar]
- 6. McCullers JA. Insights into the interaction between influenza virus and pneumococcus. Clin Microbiol Rev 2006; 19:571–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wissinger E, Goulding J, Hussell T. Immune homeostasis in the respiratory tract and its impact on heterologous infection. Semin Immunol 2009; 21:147–155. [DOI] [PubMed] [Google Scholar]
- 8. Okamoto S, Kawabata S, Nakagawa I et al. Influenza A virus‐infected hosts boost an invasive type of Streptococcus pyogenes infection in mice. J Virol 2003; 77:4104–4112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Peltola VT, Murti KG, McCullers JA. Influenza virus neuraminidase contributes to secondary bacterial pneumonia. J Infect Dis 2005; 192:249–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hament JM, Kimpen JLL, Fleer A, Wolfs TFW. Respiratory viral infection predisposing for bacterial disease: a concise review. FEMS Immunol Med Microbiol 1999; 26:189–195. [DOI] [PubMed] [Google Scholar]
- 11. Brundage JF. Interactions between influenza and bacterial respiratory pathogens: implications for pandemic preparedness. Lancet Infect Dis 2006; 6:303–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Danai PA, Sinha S, Moss M, Haber MJ, Martin GS. Seasonal variation in the epidemiology of sepsis. Crit Care Med 2007; 35:410–415. [DOI] [PubMed] [Google Scholar]
- 13. Jean C, Louie JK, Glaser CA et al. Invasive group A streptococcal infection concurrent with 2009 H1N1 influenza. Clin Infect Dis 2010; 50:e59–e62. [DOI] [PubMed] [Google Scholar]
- 14. Beadling C, Slifka MK. How do viral infections predispose patients to bacterial infections? Curr Opin Infect Dis 2004; 17:185–191. [DOI] [PubMed] [Google Scholar]
- 15. Kim PE, Musher DM, Glezen WP, Rodriguez‐Barradas MC, Nahm WK, Wright CE. Association of invasive pneumococcal disease with season, atmospheric conditions, air pollution, and the isolation of respiratory viruses. Clin Infect Dis 1996; 22:100–106. [DOI] [PubMed] [Google Scholar]
- 16. Dowell SF. Seasonal variation in host susceptibility and cycles of certain infectious diseases. Emerg Infect Dis 2001; 7:369–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Talbot TR, Poehling KA, Hartert TV et al. Seasonality of invasive pneumococcal disease: temporal relation to documented influenza and respiratory syncytial virus. Am J Med 2005; 118:285–291. [DOI] [PubMed] [Google Scholar]
- 18. Dowell SF, Ho MS. Seasonality of infectious diseases and severe acute respiratory syndrome – what we don’t know can hurt us. Lancet Infect Dis 2004; 4:704–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Fisman DN. Seasonality of infectious diseases. Annu Rev Public Health 2007; 28:127–143. [DOI] [PubMed] [Google Scholar]
- 20. Parienti JJ, Carrat F. Viral pneumonia and respiratory sepsis: association, causation, or it depends? Crit Care Med 2007; 35:639–640. [DOI] [PubMed] [Google Scholar]
- 21. Hope‐Simpson RE. The role of season in the epidemiology of influenza. J Hyg Camb 1981; 86:35–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Shaman J, Pitzer VE, Viboud C, Grenfell BT, Lipsitch M. Absolute humidity and the seasonal onset of influenza in the continental United States. PLoS Biol 2010; 8:e1000316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Dowell SF, Whitney CG, Wright C, Rose CE, Schuchat A. Seasonal patterns of invasive pneumococcal disease. Emerg Infect Dis 2003; 9:573–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Altizer S, Dobson A, Hosseini P, Hudson P, Pascual M, Rohani P. Seasonality and the dynamics of infectious diseases. Ecol Lett 2006; 9:467–484. [DOI] [PubMed] [Google Scholar]
- 25. Moineddin R, Upshur REG, Crighton E, Mamdani M. Autoregression as a means of assessing the strength of seasonality in a time series. Popul Health Metr 2003; 1:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Thompson WW, Shay DK, Weintraub E et al. Mortality associated with influenza and respiratory syncytial virus in the United States. JAMA 2003; 289:179–186. [DOI] [PubMed] [Google Scholar]
- 27. Pascual M, Dobson A. Seasonal patterns of infectious diseases. PLOS Med 2005; 2:18–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Armstrong B. Models for the relationship between ambient temperature and daily mortality. Epidemiology 2006; 17:624–631. [DOI] [PubMed] [Google Scholar]
- 29. Muggeo VMR. Estimating regression models with unknown break points. Stats Med 2003; 22:3055–3071. [DOI] [PubMed] [Google Scholar]
- 30. Harrell FR Jr. Regression Modeling Strategies with Applications to Linear Models, Logistic Regression and Survival Analysis. New York: Springer, 2001. [Google Scholar]
- 31. Hajat S, Armstrong BG, Gouveia N, Wilkinson P. Mortality displacement in heat‐related deaths. Epidemiology 2005; 16:613–620. [DOI] [PubMed] [Google Scholar]
- 32. Schwartz J. Is there harvesting in the association of airborne particles with daily deaths and hospital admissions? Epidemiology 2001; 12:55–61. [DOI] [PubMed] [Google Scholar]
- 33. Thompson MG, Shay DK, Zhou H et al. Estimates of deaths associated with seasonal influenza—United States, 1976‐2007. MMWR 2010; 59:1057–1062. [PubMed] [Google Scholar]
- 34. Jackson WPU. Influenza B among West Indians. Lancet 1946; 248:631–635. [DOI] [PubMed] [Google Scholar]
- 35. Maxwell ES, Ward TG, Van Metre TE. The relation of influenza virus and bacteria in the etiology of pneumonia. J Clin Invest 1949; 28:307–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Stuart‐Harris CH, Laird J, Tyrrell DA, Kelsall MH, Franks ZC, Pownall M. The relationship between influenza and pneumonia. J Hyg 1949; 47:434–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Tyrrell DAJ. The pulmonary complications of influenza as seen in Sheffield in 1949. Q J Med 1952; 21:291–306. [PubMed] [Google Scholar]
- 38. Mancini DAP, Alves RCB, Mendonça RMZ et al. Influenza virus and proteolytic bacteria co‐infection in respiratory tract from individuals presenting respiratory manifestations. Rev Inst Med trop S Paulo 2008; 50:41–46. [DOI] [PubMed] [Google Scholar]
- 39. Glezen WP, Couch RB, Taber LH et al. Epidemiologic observations of influenza B virus infections in Houston Texas, 1976–1977. Am J Epidemiol 1980; 111:13–22. [DOI] [PubMed] [Google Scholar]
- 40. Parola P, Colson P, Dubourg G et al. Letter to the editor. Group A streptococcal infections during the seasonal influenza outbreak 2010/11 in South East England. Euro Surveill 2011; 16:pii:19816. [DOI] [PubMed] [Google Scholar]
- 41. Scaber J, Saeed S, Ihekweazu C, Efstratiou A, McCarthy N, O’Moore É. Group A streptococcal infection during the seasonal influenza outbreak 2010/11 in South East England. Euro Surveill 2011; 16:2–5. [PubMed] [Google Scholar]
- 42. Okamoto S, Kawabata S, Fujitaka H, Uehira T, Okuno Y, Hamada S. Vaccination with formalin‐inactivated influenza vaccine protects mice against lethal influenza Streptococcus pyogenes superinfection. Vaccine 2004; 22:2887–2893. [DOI] [PubMed] [Google Scholar]
- 43. Crum NF, Hale BR, Bradshaw DA et al. Outbreak of group A streptococcal pneumonia among marine corps recruits—California, November 1–December 20, 2002. MMWR 2003; 52:106–109. [PubMed] [Google Scholar]
- 44. Thigpen MC, Richards CL, Lynfield R et al. Invasive group A streptococcal infection in older adults in long‐term care facilities and the community, United States, 1998‐2003. Emerg Infect Dis 2007; 13:1852–1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lee S, Eick A, Bloom MS, Brundage JF. Influenza immunization and subsequent diagnoses of group A streptococcus‐illnesses among U.S. army trainees, 2002–2006. Vaccine 2008; 26:3383–3386. [DOI] [PubMed] [Google Scholar]
- 46. Fiore AE, Uyeki TM, Broder K et al. Prevention and control of influenza with vaccines: recommendations of the Advisory Committee on Immunization Practices (ACIP), 2010. MMWR 2010; 8:1–62. [PubMed] [Google Scholar]


