Abstract
Please cite this paper as: Wu et al. (2012) Severity of pandemic H1N1 2009 influenza virus infection may not be directly correlated with initial viral load in upper respiratory tract. Influenza and Other Respiratory Viruses 6(5), 367–373.
Background Recent studies have demonstrated that rapid influenza diagnostic tests (RIDTs) have a relatively low sensitivity in detecting severe cases of pandemic H1N1 2009 influenza virus (pH1N1) infection. We hypothesized that viral load in upper respiratory specimens obtained on presentation may not be correlated with disease severity.
Methods We conducted a prospective study to compare patterns of viral shedding using nasopharyngeal swab specimens, according to the number of days of post‐symptom onset and post‐antiviral therapy, between patients with and without complications.
Results From July 15, 2009 through July 23, 2010, we collected and processed a total of 141 nasopharyngeal swab specimens from 64 inpatients and outpatients with laboratory‐confirmed pH1N1 infection. These included 46 patients without any complications (uncomplicated group) and 18 patients who required hospital admission (complicated group). The mean initial viral load was higher in the uncomplicated group than in the complicated group (3·4 ± 1·6 log10 copies/μl versus 1·9 ± 1·7, P = 0·02). However, prolonged viral shedding was only detected in the complicated group (44% by day 7 of antiviral therapy). By multivariate analysis, we found that age (OR, 1·1; 95% CI, 1·0–1·1) and initial nasopharyngeal viral load (OR, 0·5; 95% CI, 0·3–0·8) were significant factors associated with complications.
Conclusion Given that patients with severe pH1N1 infection may have relatively lower initial viral load in the upper respiratory tract, cautious interpretation of negative RIDT results is particularly warranted in this patient population.
Keywords: Influenza, pandemic H1N1, rapid tests, real‐time PCR, viral load
Introduction
In April 2009, the pandemic H1N1 2009 influenza virus (pH1N1) emerged in Mexico and rapidly spread to countries worldwide. 1 While previous investigations regarding seasonal (H1N1, H3N2) and H5N1 influenza have shown that the level of viral replication influences outcome, 2 , 3 prospective studies illustrating the correlation between disease severity and both initial viral load and serial change of viral shedding among patients with pH1N1 influenza remain limited.
Recent studies, as well as our own observation, have demonstrated relatively low sensitivities of rapid influenza diagnostic tests (RIDT) in severe cases of pH1N1 infection. 4 , 5 , 6 , 7 Based on the fact that viral load is a major determinant of the sensitivity of RIDT, 8 , 9 we hypothesized that at the point when patients with pH1N1 infection arrive at hospital, the severity of the disease may not be well correlated with viral load in the upper respiratory tract.
We therefore conducted a prospective study to assess the profile of viral shedding among patients with complicated and uncomplicated pH1N1 infection and to determine whether there was a correlation between viral loads and disease severity.
Methods
Patients and data collection
This was a prospective observational study on patients with pH1N1 infections who presented to National Taiwan University Hospital (NTUH), a 2200‐bed teaching hospital, which provided both primary and tertiary medical care including acute medical service. From July 15, 2009 to July 23, 2010, both inpatients and outpatients aged 16 years or older with influenza‐like illness (ILI) were enrolled for evaluation of eligibility after the informed consent was obtained. The flu‐like symptoms were defined according to the Centers for Disease Control of Taiwan with (1) any of the following: myalgia, headache, or malaise; plus (2) all of the following: (i) sudden onset of fever with a body temperature equal to or higher than 38°C by a tympanic thermometer, plus respiratory symptoms and (ii) exclusion of simply rhinorrhea, tonsillitis, and bronchitis. A standardized case report form was used to collect patient data including demographics, comorbidities, influenza vaccination status, date of fever onset, clinical presentation and outcomes, use of antiviral agents, and laboratory and radiological findings. A RIDT (QuickVue; Quidel, San Diego, CA, USA) and nasopharyngeal swab for viral quantitative real‐time reverse transcriptase PCR (RT‐PCR) (Applied Biosystems, Foster City, CA, USA) were collected at first hospital visit. In the case of a positive RT‐PCR result, nasopharyngeal swabs for quantitative RT‐PCR were sequential collected daily for the first 5 days and then every 2 days until viral genomes became undetectable for two successive specimens. The nasopharyngeal swabs were transported in 1·2 ml of DMEM medium to our laboratory immediately after specimens were obtained and then stored at −80°C before processing. Based on recommendations by local health authorities during the study period, antiviral treatment would be considered for cases with ILI who tested positive of RIDTs or those with risk factors for complicated influenza. This study was approved by the ethical board of NTUH.
Virological analysis
Extraction of viral RNA from clinical specimens
Viral RNA from 400 μl of specimen suspension was extracted with the MagMAXTM Viral RNA isolation Kit (Applied Biosystem) according to the manufacturer’s instructions, and finally eluted in 50 μl ddH2O.
Real‐time quantitative RT‐PCR
Real‐time RT‐PCR (rRT‐PCR) amplification and detection was performed in a Step One real‐time PCR machine (Applied Biosystems) using the AgPath‐ID one‐step RT‐PCR kit (Amion, Austin, TX, USA). The 25‐μl reaction volume for each sample contained 5 μl of extracted RNA, 12·5 μl of AgPath Kit 2× buffer, 1 μl of AgPath 25× enzyme mix, and 5 pmol of Taqman probe, and 10 pmol of each of the forward and reverse primers, and 6 μl of ultrapure DNase–RNase‐free distilled water. All of the PCR were performed in duplicates. The cycling conditions included a reverse transcription step at 50°C for 30 minutes, and the amplification was performed for 45 cycles as follows: 94°C for 15 seconds, 55°C for 30 seconds, 55°C for 30 seconds. For all PCR testing, a cycle threshold ≤40 was interpreted as positive. The detection limit was 10 copies/μl.
Primer and probe
We used the Inf A (H1N1) MGB Assay (Applied Biosystems). The rRT‐PCR protocol including designs of primers and probes were applied for pandemic 2009 H1N1 influenza virus detection according to CDC recommendations. 10 TaqMan dual‐labeled probes were designed based on CDC probe sequences with 6‐carboxyfluorescein (FAM) reporter and non‐fluorescent quencher coupled with minor grove binder (MGB). The ribonuclease P (RNaseP) RT‐PCR was used both as an internal control and to check for the presence of human nucleic acids and indicates the presence of a sufficient amount of cells in the sample.
Standard construction and sensitivity test
The amplified influenza M1 gene was cloned into pGEM‐Teasy and amplified in Escherichia coli. Plasmid‐insert DNA sequences were verified by sequencing in both directions using dye‐labeled dideoxy‐terminator cycle sequencing. Sequences were analyzed using an ABI Model 3730. The concentration of the plasmid DNA was calculated by measuring the Qubit Fluorometer (Invitrogen, Carlsbad, CA, USA). Plasmid DNA was then serially diluted tenfold in ddH2O, ranging from 107 to 100 copies/μl, and used for the sensitivity test. Standard curves were produced with each assay.
Statistical analysis
All statistical analyses were performed using the Statistical Package for Social Sciences (version 17·0; SPSS, Chicago, IL, USA). Patients with confirmed pH1N1 infections were classified into complicated group if hospitalization was required because of the development of pneumonia, neurological complications, invasive bacterial infection, myocarditis, or pericarditis. Otherwise, patients treated at outpatient clinics or emergency department for mild illness were categorized into uncomplicated group. Categorical variables were compared using chi‐square or Fisher’s exact test when appropriate; continuous variables were analyzed with Student’s t‐test. Potential predictors of development of complications among patients with pH1N1 were identified by univariate and then multivariate analysis. Variables with P value ≤0·05 in the univariate analysis were further tested by means of logistic regression using the backward conditional method. Two‐tailed P values of <0·05 will be considered to be statistically significant.
Results
Patient characteristics
Between July 15, 2009 and July 23, 2010, a total of 149 patients with ILI were enrolled in our study. Of these, 64 patients (21 from the emergency department and 43 from outpatient clinics) tested positive for both influenza A virus M gene and pandemic H1N1 2009 influenza virus H1 gene by RT‐PCR. The remaining patients’ infections were caused by H3N2 (26) and influenza B (6), respectively. Among those with confirmed pH1N1 influenza, 18 (28%) were hospitalized owing to complications (Table 1) and nine of them required ICU admission. One patient had concomitant Klebsiella pneumoniae bacteremia at presentation. Two patients required extracorporeal membrane oxygenation support. One of them, a 33‐year‐old male patient with no underlying medical conditions, died of multiorgan failure after having been hospitalized for 3 months.
Table 1.
Complications and clinical outcomes of hospitalized patients with pandemic H1N1 2009 influenza virus infection
| Complication/outcome | Complicated patients (n = 18) | |
|---|---|---|
| General ward (n = 9) | ICU (n = 9) | |
| Complication | ||
| Pneumonia | 9 (100%) | 8 (100%) |
| Encephalopathy | 0 | 1 (13%) |
| Myocarditis | 0 | 1 (13%) |
| Shock | 0 | 3 (38%) |
| Mechanical ventilation | 0 | 7 (88%) |
| Bloodstream infection | 0 | 1 (13%) |
| ECMO support | 0 | 2 (25%) |
| Outcome | ||
| LOS (mean days ± SD) | 6·5 ± 5·6 | 37·5 ± 27·4 |
| Death | 0 | 1 (12·5%) |
ICU, intensive care unit; ECMO, extracorporeal membrane oxygenation, LOS, length of hospital stay; SD, standard deviation.
Demographic data and clinical characteristics of uncomplicated and complicated patients are illustrated in Table 2. A substantial proportion of patients (80%) in the uncomplicated group had no underlying medical conditions, whereas half of the patients in the complicated group had one or more comorbidities. All patients received RIDTs on presentation. The positive rates in patients with and without complications were 91% and 67%, respectively (P = 0·02).
Table 2.
Demographics and clinical features of 64 patients with pandemic H1N1 2009 influenza virus infection
| Characteristics | Uncomplicated (n = 46) | Complicated (n = 18) | P value |
|---|---|---|---|
| Demographic | |||
| Age (mean ± SD), years | 26·8 ± 10·2 | 42·0 ± 17·7 | <0·001 |
| Male gender | 18 (39%) | 12 (67%) | 0·05 |
| Pregnancy | 0 | 0 | |
| BMI ≥35 | 0 | 0 | |
| Influenza vaccination | 7 (15%) | 0 | 0·08 |
| Underlying disease | |||
| DM | 0 | 1 (6%) | 0·28 |
| Hypertension | 1 (2%) | 2 (11%) | 0·19 |
| Cardiac diseases | 0 | 2 (11%) | 0·08 |
| Chronic lung disease | 1 (2%) | 2 (11%) | 0·19 |
| Chronic renal disease | 0 | 2 (11%) | 0·08 |
| Liver cirrhosis | 1 (2%) | 2 (11%) | 0·19 |
| Malignancy | 1 (2%) | 2 (11%) | 0·19 |
| Smoking | 5 (11%) | 3 (17%) | 0·68 |
| Systemic corticosteroid use | 0 | 2 (11%) | 0·08 |
| Any of the above | 9 (20%) | 9 (50%) | 0·02 |
| Presenting symptom | |||
| Myalgia | 35 (76%) | 7 (39%) | 0·005 |
| Headache | 36 (78%) | 8 (44%) | 0·009 |
| Altered consciousness | 0 | 4 (22%) | 0·005 |
| Rhinorrhea | 33 (72%) | 11 (61%) | 0·41 |
| Cough | 44 (96%) | 14 (78%) | 0·05 |
| Sputum production | 25 (54%) | 15 (83%) | 0·04 |
| Sore throat | 31 (67%) | 14 (78%) | 0·55 |
| Chest pain | 3 (7%) | 8 (44%) | 0·001 |
| Dyspnea | 11 (24%) | 14 (78%) | <0·001 |
| Nausea and vomiting | 6 (13%) | 8 (44%) | 0·006 |
| Abdominal pain | 4 (9%) | 4 (22%) | 0·21 |
| Diarrhea | 4 (9%) | 5 (28%) | 0·10 |
| Laboratory finding (mean ± SD) | |||
| WBC (cells/μl) | 5904 ± 1573 | 6234 ± 2662 | 0·55 |
| Lymphocyte (cells/μl) | 1555 ± 764 | 741 ± 418 | <0·001 |
| Platelet (K/μl) | 209 ± 69 | 172 ± 56 | 0·06 |
| Creatinine kinase (U/l) | 99 ± 74 | 468 ± 474 | <0·001 |
| C‐reactive protein (mg/dl) | 1·2 ± 0·9 | 10·7 ± 11·3 | <0·001 |
| RIDT positivity | 42 (91%) | 12 (67%) | 0·02 |
| Mean initial viral load (log10 copies/μl) | 3·4 ± 1·6 | 1·9 ± 1·7 | 0·02 |
| Days from fever to seeking medical help | 2·1 ± 0·7 | 3·6 ± 2·6 | 0·001 |
| Days from fever to oseltamivir use | 2·2 ± 0·8 | 4·5 ± 3·9 | <0·001 |
| Days to defervescence (mean ± SD) | 2·6 ± 1·3 | 5·3 ± 3·9 | <0·001 |
BMI, body mass index; DM, diabetes mellitus; WBC, white blood cell; RIDT, rapid influenza diagnostic test.
Despite antiviral therapy, we frequently observed prolonged duration of fever in the complicated group (mean, 5·3 days). Classical influenza symptoms including cough, myalgia, and headache were presented more commonly in the uncomplicated group, while altered consciousness, chest pain, dyspnea, nausea, and vomiting were more frequently reported in patients experiencing complications on presentation. Patients with complications were also more likely to have a low lymphocyte count, rhabdomyolysis, and an elevated serum C‐reactive protein (CRP) level.
The length of time between onset of fever and seeking medical help was significantly greater for patients in the complicated group. Although all patients received oseltamivir immediately after diagnosis, the mean time from onset of fever to initiation of oseltamivir treatment was significantly longer in the complicated group (4·5 ± 3·9 versus 2·2 ± 0·8 days, P < 0·001).
Comparisons of viral loads and duration of viral shedding
Overall, we collected 161 nasopharyngeal swab specimens from 64 patients with laboratory‐confirmed pH1N1 infection and tested them for viral load. On presentation, the mean initial viral load of the uncomplicated group was higher than the complicated group (3·4 ± 1·6 log10 copies/μl versus 1·9 ± 1·7, P = 0·02). We further compared the nasopharyngeal viral loads between the two groups stratified by the day after symptom onset (Figure 1). Although statistical insignificance, nasopharyngeal viral loads were higher in the uncomplicated group for the first 3 days. Peak viral shedding occurred within 2 days of symptom onset in both groups. A slower decline and prolongation of viral shedding were observed in the complicated group, whereas all uncomplicated patients had undetectable viral RNA by RT‐PCR by day seven after onset of symptoms.
Figure 1.
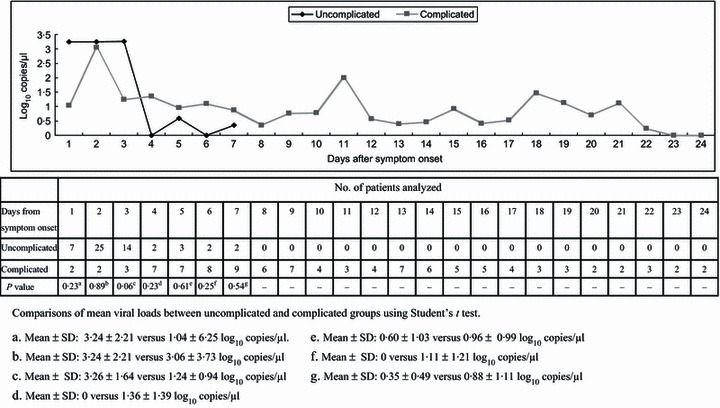
Mean viral load in patients with different disease severity according to days after symptom onset.
Irrespective of disease severity, viral shedding started to decrease after oseltamivir was initiated (Figure 2). RT‐PCR results of nasopharyngeal swabs became negative within 5 days of initiation of oseltamivir in all uncomplicated patients. In the complicated group, however, viral genome could still be detected on day 5 and day 7 after initiation of antiviral treatment in 61% and 44% of the patients, respectively.
Figure 2.
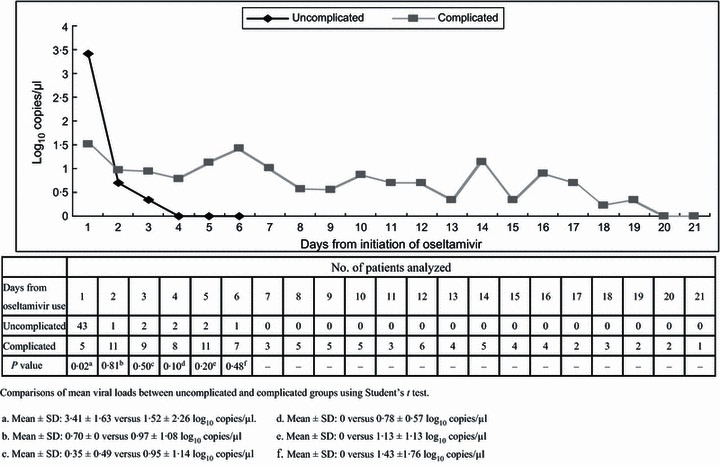
Mean viral load in patients with different disease severity according to days after initiation of oseltamivir.
The results of univariate and multivariate analysis are shown in Table 3. After adjustments, the only factors associated with the development of complications among patients with pH1N1 infection were age (OR, 1·1; 95% CI, 1·0–1·1) and initial nasopharyngeal viral load (OR, 0·5; 95% CI, 0·3–0·8). Although all patients received oseltamivir at presentation, delayed antiviral therapy were frequently observed among patients in complicated group.
Table 3.
Univariate and multivariate analyses of the predictors of complications
| Variable | Univariate | Multivariate | ||
|---|---|---|---|---|
| OR (95% CI) | P value | OR (95% CI) | P value | |
| Age | 1·1 (1·0–1·1) | 0·002 | 1·1 (1·0–1·1) | 0·007 |
| Male gender | 3·1 (1·0–9·8) | 0·05 | – | – |
| Underlying diseases* | 4·1 (1·3–13·3) | 0·02 | – | – |
| Initial viral load (log10 copies/μl) | 0·50 (0·3–0·8) | 0·004 | 0·5 (0·3–0·8) | 0·01 |
| Days from fever to oseltamivir use | 1·8 (1·2–2·8) | 0·009 | – | – |
| Days from fever to hospital visit | 1·7 (1·2–2·6) | 0·008 | – | – |
OR, odds ratio; CI, confidence interval.
*Included any one of the underlying diseases specified in Table 2.
Discussion
This prospective study demonstrates that initial viral load in the upper respiratory tract of patients with complicated pH1N1 influenza may not be higher than that of uncomplicated patients upon presentation. Given that RIDT is a diagnostic tool more widely used than PCR in clinical setting, and the sensitivity is proportional to viral load in the respiratory specimens, 8 , 9 , 11 our study highlights the difficulty and pitfall in the diagnosis of pH1N1 influenza in patients with complications using this tool. Consistent with other investigations, the time from symptom onset to hospital visit or antiviral therapy are longer in patients with complicated diseases than those without. 12 , 13 , 14 However, our findings cannot be simply explained by the delay in seeking medical help among patients with complicated diseases because the inverse correlation between viral load and level of disease severity could be observed among those presented within the first 3 days after onset of the disease. One plausible explanation is that pH1N1 virus preferentially infects the lower respiratory tract, especially among those with severe respiratory illness, as illustrated in both human and animal models. 15 While replication of seasonal H1N1 virus was confined to the nasal cavity in ferrets, pH1N1 virus also replicated in the trachea, bronchi, and bronchioles. 16 In the lungs of infected mice, ferrets and non‐human primates, pH1N1 caused more severe pathological lesions than seasonal H1N1 virus. 17 Yeh et al. 18 reported that pH1N1 virus was detected predominantly in specimens from the lower respiratory tract of a child, but was absent or at very low levels in nasopharyngeal swab samples. These observations can be attributed to a greater binding ability of pH1N1 to α2‐3‐linked sialyl glycans, which are predominant in the lower respiratory tract, than to α2‐6‐linked receptors, which are more common in the upper respiratory tract instead. 19 Hence, the upper respiratory tract may not be the optimal site for detecting or monitoring replication of the virus in severe cases of pH1N1 influenza.
While adults with seasonal influenza are generally considered infectious from the day before onset of symptoms until approximately 5 days after onset, 20 data regarding the duration and profile of viral shedding in adult patients with pH1N1 influenza, especially those with complications, are limited. Our observation that patients with complicated diseases had slower decline of viral load and prolonged viral shedding are consistent with a recent retrospective study. 13 These may be caused by failure to mount an effective innate and adaptive response, and delay of antiviral treatment. 21 As we did not test for oseltamivir resistance, we were unable to determine whether prolonged viral shedding in patients with complications was attributed to oseltamivir‐resistant influenza virus. Nevertheless, national surveillance data of antiviral resistance by Taiwan Centers for Disease Control showed that only 0·7% of pH1N1 influenza virus in Taiwan was resistant to oseltamivir during the study period. 22 At present, although the relationship between viral shedding measured by a quantitative RT‐PCR and viral transmission has not been clearly demonstrated, our observations of prolonged virus detection in the upper respiratory tract, several days after antiviral treatment, may have implications for infection control. We recommend further clinical investigation into the effects of prolonged use of oseltamivir in immunocompromised or critical patients.
Consistent with previous reports, our results showed that younger adults (<65 years old) might also be hospitalized for severe pH1N1 influenza, and the risk for complications among adult patients increased with age. 13 , 23 , 24 However, none of the complicated cases in our cohort were morbidly obese. Other non‐Western studies also suggest that morbid obesity is not a significant risk factor for severe case. 25 , 26 , 27 Racial differences in the prevalence of obesity should be taken into account when interpreting potential risk factors for complications among patients with pH1N1 influenza.
There were limitations to the present study. First, as a result of limited sample size, the importance of predictors might be underestimated. Second, as we did not perform sampling of lower respiratory specimens simultaneously, which usually requires invasive procedures, we were unable to demonstrate the differences of viral load between the upper and lower respiratory tracts among patients with and without complications. Nevertheless, our observation that severe cases are more likely to have lower initial viral load in upper respiratory tract justifies careful interpretation of negative RIDT or PCR results among these patients. Third, although serial specimens were obtained for analysis, different numbers of specimens were included at different intervals because the point at which the first specimen could be obtained varied in each patient, and a small number of patients refused sampling during the follow‐up period. Lastly, the relatively high positive rate of RIDTs among our study population was probably attributed to sampling bias because those who tested positive were more willing to be enrolled in our study.
In conclusion, patients with complicated pH1N1 infection have prolonged viral shedding but not a higher initial viral load in the upper respiratory tract on presentation when compared with patients with mild illness. This partially accounted for a relatively lower sensitivity of RIDTs among complicated patients. More vigilant surveillance of the lower airway and timely antiviral therapy are required for severe cases with suspected pH1N1 infection but negative RIDTs results. In addition, further research to compare the viral loads in upper and lower respiratory tracts among patients with severe pH1N1 infection is warranted.
Conflict of interest
None to declare.
Acknowledgements
This study was partially funded by the National Science Council of Taiwan (95‐2745‐B‐002‐007, 96‐2321‐B‐002‐027, 97‐2321‐B‐002‐017) and the Department of Health, Taiwan (DOH99‐TD‐B‐111‐001). Preliminary results of this work were presented at the 48th Annual Meeting of IDSA [poster 760; session 72], October 21–24, 2010, Vancouver, Canada.
References
- 1. World Health Organization . Influenza update – 119. Available at: http://www.who.int/csr/disease/influenza/2010_10_20_GIP_surveillance/en/index.html 2010 (Accessed 20 October 2010).
- 2. de Jong MD, Simmons CP, Thanh TT et al. Fatal outcome of human influenza A (H5N1) is associated with high viral load and hypercytokinemia. Nat Med 2006; 12:1203–1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Meunier I, Pillet S, Simonsen JN, von Messling V. Influenza pathogenesis: lessons learned from animal studies with H5N1, H1N1 Spanish, and pandemic H1N1 2009 influenza. Crit Care Med 2010; 4(Suppl):e21–e29. [DOI] [PubMed] [Google Scholar]
- 4. Drexler JF, Helmer A, Kirberg H et al. Poor clinical sensitivity of rapid antigen test for influenza A pandemic (H1N1) 2009 virus. Emerg Infect Dis 2009; 15:1662–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kao TM, Wu UI, Chen YC. Rapid diagnostic tests and severity of illness in pandemic (H1N1) 2009, Taiwan. Emerg Infect Dis 2010; 16:1181–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Singh K, Vasoo S, Stevens J, Schreckenberger P, Trenholme G. Pitfalls in diagnosis of pandemic (novel) A/H1N1 2009 influenza. J Clin Microbiol 2010; 48:1501–1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Blyth CC, Iredell JR, Dwyer DE. Rapid‐test sensitivity for novel swine‐origin influenza A (H1N1) virus in humans. N Engl J Med 2009; 361:2493. [DOI] [PubMed] [Google Scholar]
- 8. Louie JK, Guevara H, Boston E et al. Rapid influenza antigen test for diagnosis of pandemic (H1N1) 2009. Emerg Infect Dis 2010; 16:824–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cheng CK, Cowling BJ, Chan KH et al. Factors affecting QuickVue Influenza A + B rapid test performance in the community setting. Diagn Microbiol Infect Dis 2009; 65:35–41. [DOI] [PubMed] [Google Scholar]
- 10. World Health Organization . CDC protocol for realtime RT‐PCR for influenza A (H1N1). Available at: http://www.who.int/csr/resources/publications/swineflu/CDCRealtimeRTPCR_SwineH1Assay‐2009_20090430.pdf 2009 (Accessed 6 October 2009).
- 11. Jernigan DB, Lindstrom SL, Johnson JR et al. Detecting 2009 pandemic influenza A (H1N1) virus infection: availability of diagnostic testing led to rapid pandemic response. Clin Infect Dis 2011; 52(Suppl 1):S36–S43. [DOI] [PubMed] [Google Scholar]
- 12. Campbell A, Rodin R, Kropp R et al. Risk of severe outcomes among patients admitted to hospital with pandemic (H1N1) influenza. CMAJ 2010; 182:349–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. To KK, Hung IF, Li IW et al. Delayed clearance of viral load and marked cytokine activation in severe cases of pandemic H1N1 2009 influenza virus infection. Clin Infect Dis 2010; 50:850–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yu H, Feng Z, Uyeki TM et al. Risk factors for severe illness with 2009 pandemic influenza A (H1N1) virus infection in China. Clin Infect Dis 2011; 52:457–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Guarner J, Falcon‐Escobedo R. Comparison of the pathology caused by H1N1, H5N1, and H3N2 influenza viruses. Arch Med Res 2009; 40:655–661. [DOI] [PubMed] [Google Scholar]
- 16. Munster VJ, de Wit E, van den Brand JM et al. Pathogenesis and transmission of swine‐origin 2009 A(H1N1) influenza virus in ferrets. Science 2009; 325:481–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Itoh Y, Shinya K, Kiso M et al. In vitro and in vivo characterization of new swine‐origin H1N1 influenza viruses. Nature 2009; 460:1021–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yeh E, Luo RF, Dyner L et al. Preferential lower respiratory tract infection in swine‐origin 2009 A(H1N1) influenza. Clin Infect Dis 2010; 50:391–394. [DOI] [PubMed] [Google Scholar]
- 19. Childs RA, Palma AS, Wharton S et al. Receptor‐binding specificity of pandemic influenza A (H1N1) 2009 virus determined by carbohydrate microarray. Nat Biotechnol 2009; 27:797–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Harper SA, Fukuda K, Uyeki TM, Cox NJ, Bridges CB. Prevention and control of influenza. Recommendations of the Advisory Committee on Immunization Practices (ACIP). MMWR Recomm Rep 2005; 8:1–40. [PubMed] [Google Scholar]
- 21. Ling LM, Chow AL, Lye DC et al. Effects of early oseltamivir therapy on viral shedding in 2009 pandemic influenza A (H1N1) virus infection. Clin Infect Dis 2010; 50:963–969. [DOI] [PubMed] [Google Scholar]
- 22. Taiwan Centers for Disease Control . Taiwan Influenza Express. Available at: http://flu.cdc.gov.tw/public/Data/0841052071.pdf 2010 (Accessed 8 September 2011).
- 23. Lee N, Chan PK, Lui GC et al. Complications and outcomes of pandemic 2009 Influenza A (H1N1) virus infection in hospitalized adults: how do they differ from those in seasonal influenza? J Infect Dis 2011; 203:1739–1747. [DOI] [PubMed] [Google Scholar]
- 24. Shiley KT, Nadolski G, Mickus T, Fishman NO, Lautenbach E. Differences in the epidemiological characteristics and clinical outcomes of pandemic (H1N1) 2009 influenza, compared with seasonal influenza. Infect Control Hosp Epidemiol 2010; 31:676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chien YS, Su CP, Tsai HT et al. Predictors and outcomes of respiratory failure among hospitalized pneumonia patients with 2009 H1N1 influenza in Taiwan. J Infect 2010; 60:168–174. [DOI] [PubMed] [Google Scholar]
- 26. Diaz E, Rodriguez A, Martin‐Loeches I et al. Impact of obesity in patients infected with 2009 influenza A(H1N1). Chest 2011; 139:382–386. [DOI] [PubMed] [Google Scholar]
- 27. Yang P, Deng Y, Pang X et al. Severe, critical and fatal cases of 2009 H1N1 influenza in China. J Infect 2010; 61:277–283. [DOI] [PubMed] [Google Scholar]


