Abstract
We describe the identification and characterization of the H9N2 influenza subtype reported in Egyptian broiler and broiler breeder farms for the first time. Circulation of this subtype in a highly pathogenic H5N1 influenza virus endemic population provides an opportunity for genetic reassortment and emergence of novel viruses.
Keywords: Avian influenza, Egypt, H9N2, phylogeny
The H9N2 subtype virus is a notable member of influenza A genus as it can infect not only avian species but also, although sporadically, mammals such as pigs and humans. 1 , 2 The identification of this subtype in multiple host species combined with its cocirculation with other type A influenza viruses has provided the conditions for H9N2 viruses to accumulate mutations and generate novel variants, thus increasing the probability for this subtype to evolve into a pandemic strain. 3 Since the mid‐1990s, H9N2 avian influenza has established in poultry both in Asia and in the Middle East, giving rise in Middle Eastern countries to four distinct and cocirculating genetic groups provisionally named A, B, C, and D, all belonging to the G1 lineage. 4 Despite H9N2 endemicity in poultry population in the Middle Eastern and Gulf areas, 4 , 5 , 6 at the time of writing detection of this subtype in Egyptian poultry has been reported in a commercial quail farm only 7 and there has been no evidence of circulation of this virus in other commercial poultry sectors. 8 In this study, identification and characterization of the H9N2 subtype in Egyptian commercial broiler and broiler breeder farms are described for the first time.
Cloacal, oropharyngeal swabs and organ samples collected in seven distinct poultry flocks (six broiler farms and one broiler breeder farm), in the governorates of Giza, Behera, Dakahlia, and Sharkia between February 2010 and August 2011, tested positive for H9N2 subtype by real‐time reverse transcription–PCR (rRT–PCR) 9 and virus isolation. 10 At the time of sampling, affected birds showed respiratory signs and depression with mortality rate within the flocks ranging between 7% and 12·8%. At postmortem examination, tracheitis, air sacculitis, and pneumonia were identified. Considering that highly pathogenic avian influenza H5N1 endemically circulates in the Egyptian poultry population, 11 samples were tested for this subtype as well providing negative results. Interestingly, in the H9N2‐positive farms located in Giza, Behera and Sharkia positivity to H5N1 had been detected in the rearing cycles immediately preceding this last survey.
The complete genome of seven representative H9N2 viruses isolated in Egypt between December 2010 and August 2011 was sequenced, as previously described. 4 The nucleotide sequences obtained in this study are made available in the GISAID database under following accession numbers: EPI355083–EPI355122 and EPI355384–EPI355399. Each gene segment was then aligned and compared with the H9N2 sequences of viruses from Central Asian and Middle Eastern countries available in GenBank. Maximum likelihood (ML) trees were estimated using the best‐fit general time reversible (GTR) + I + Γ4 model of nucleotide substitution using PhyML. A bootstrap re‐sampling process (1000 replications) using the neighbor‐joining method was used to assess the robustness of individual nodes of the phylogeny, incorporating the ML substitution model defined above.
Analysis of the eight ML phylogenetic trees reveals that the seven Egyptian viruses cluster with a group of reassortant viruses collected between 2006 and 2010 in Israel and Jordan 4 (Figure 1). In particular, these intersubtype reassortant strains contain four gene segments (HA, NA, NS, and M) belonging to cluster B and the remaining segments from cluster A.
Figure 1.
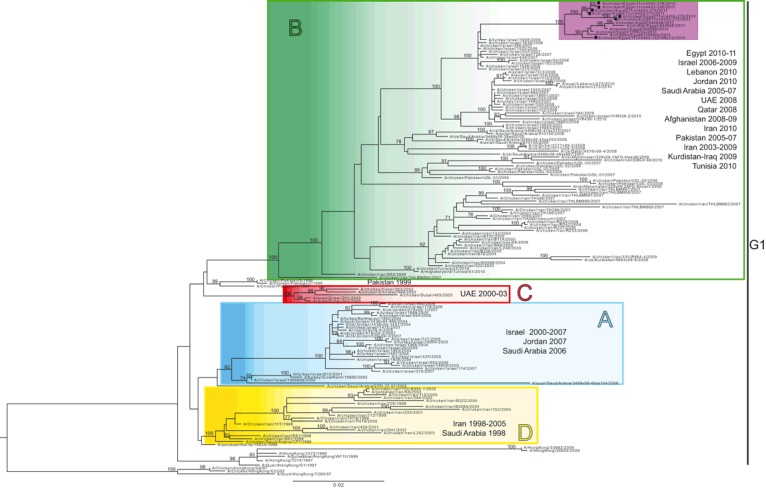
Maximum likelihood phylogenetic tree for the HA gene segment of H9N2 avian influenza subtype. Representative H9N2 sequences of viruses from Central Asian, Middle Eastern, and African countries available in GenBank are included in the phylogeny. Sequences of H9N2 viruses from Egypt are highlighted in pink. Square indicates viruses sequenced for this study. Genetic groups are colored as follows: group A: blue, group B: green, group C: red, and group D: yellow. The numbers at the nodes represent bootstrap values. The year of isolation and the geographical origin of the virus sequences included in the tree are indicated for each genetic group.
Our analysis shows that the H9N2 virus population circulating in the countries bordering Egypt may be considered the main source for the viruses detected in the country; it is however difficult to determine the time and the mechanism of introduction of this subtype in Northern Africa. Indeed, the Egyptian samples form a distinct group, as defined by high bootstrap values (>97%) and long branches in all the phylogenies, and they can be distinguished from the Israeli isolates by a total of 50 key amino acid differences found in the eleven viral proteins with the HA and PB2 genes possessing the majority of these substitutions (7 and 12, respectively). The number of mutations and the long branch observed in the phylogenetic tree may suggest a rather long evolution period during which these strains have been circulating undetected in the poultry population. Although H9N2 Egyptian viruses sequenced in this study are closely related, they show a nucleotide similarity ranging between 98% and 100%, and in the phylogenetic trees generated for the eight gene segments, they are distributed in at least three distinct clusters – one including the isolates A/chicken/Egypt/11vir4453‐273/2011 and A/chicken/Egypt/11vir4453‐272/2011, the other containing the A/chicken/Egypt/11vir4453‐274/2011, A/chicken/Egypt/11vir4453‐275/2011, A/chicken/Egypt/11vir4453‐276/2010, and A/chicken/Egypt/11vir4453‐280/2011 viruses, and the remaining one presenting the A/chicken/Egypt/11vir4453‐132/2011.
Interestingly, analysis of amino acid sequences showed that six of the seven viruses contain the amino acid leucine at position 226 (H3 numbering) at the receptor binding site of the HA, responsible for human virus‐like receptor specificity and critical for replication and direct transmission of H9N2 viruses in ferrets. 12 Only one isolate collected in August 2011 in Dakahlia from a broiler breeder farm (A/chicken/Egypt/11vir4453‐132/2011) presents the amino acid glutamine (Q) at this position which is associated with avian‐like receptor specificity. 13
Although the presence of H9N2 in Northern Africa is an expected event, because of its endemic circulation in the neighboring Near and Middle Eastern countries, up to date little evidence for the circulation of this virus subtype in the region has been reported. 7 , 14 Although vaccination campaigns were implemented in several Asian countries, to date H9N2 is still evolving, spreading, and causing severe problems to the poultry industry. Besides the negative economic drawback, the identification of this subtype in a country that is endemic for highly pathogenic H5N1 avian influenza raises concerns on its control and on the public health implications of such cocirculation. The coexistence of these avian influenza subtypes in the same susceptible poultry population may result in the emergence of natural reassortants, similarly to what has occurred in Pakistan and Southern China in the recent past. 15 , 16 These potentially emerging viruses may possess completely novel genotypes and phenotypes, and thus, their pathogenicity for human beings is difficult to predict. It is clear that H9N2 circulation should be limited and progressively eradicated, although the recent experience in Egypt with H5N1 has shown how an enormous challenge this could be. Certainly, what is to be avoided is to favor virus spread and antigenic evolution by applying uncontrolled vaccination. Continuous virological and serological monitoring have to be implemented to assess the extent of H9N2 circulation in the country and to guarantee the early detection of H9N2/H5N1 reassortants. According to the current international legislation, H9N2 infection in animals is not notifiable. Therefore, the real spread and impact of this virus is likely to be under reported and its circulation difficult to trace. Novel approaches of disease reporting, monitoring, and collaboration between veterinary and public health communities should be implemented to reduce the risk of the emergence of a novel chimeric virus with unpredictable biological properties.
Addendum
Isabella Monne and Hussein A. Hussein: acquired and analyzed the data, interpreted the results, and drafted the article; Alice Fusaro and Viviana Valastro: analyzed the data, interpreted the results, and drafted the article; Mohamed M. Hamoud, Rabab A. Khalefa, Shahin N. Dardir, and Moustafa I. Radwan: collected samples and related information, acquired and analyzed the data, and made a critical revision of the article; Ilaria Capua and Giovanni Cattoli: critically revised the article and provided the final approval.
Acknowledgements
This work was financially supported by the Italian Ministry of Health through the RC IZS VE 14/09. We acknowledge Cairo University, GOVS, and Ministry of Agriculture of Egypt for continuous supports. IFT Corporation, St No 9 Block No 6103 Mokattam, Cairo, Egypt, is acknowledged for the assistance in sample shipment and for partially financing the present study.
References
- 1. Butt KM, Smith GJ, Chen H et al. Human infection with an avian H9N2 influenza A virus in Hong Kong in 2003. J Clin Microbiol 2005; 43:5760–5767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Peiris JS, Guan Y, Markwell D, Ghose P, Webster RG, Shortridge KF. Cocirculation of avian H9N2 and contemporary “human” H3N2 influenza A viruses in pigs in southeastern China: potential for genetic reassortment? J Virol 2001; 75:9679–9686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Alexander PE, De P, Rave S. Is H9N2 avian influenza virus a pandemic potential? Can J Infect Dis Med Microbiol 2009; 20:e35–e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Fusaro A, Monne I, Salviato A et al. Phylogeography and evolutionary history of reassortant H9N2 viruses with potential human health implications. J Virol 2011; 85:8413–8421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Aamir UB, Wernery U, Ilyushina N, Webster RG. Characterization of avian H9N2 influenza viruses from United Arab Emirates 2000 to 2003. Virology 2007; 361:45–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Perk S, Golender N, Banet‐Noach C, Shihmanter E, Pokamunsky S, Pirak M. Phylogenetic analysis of hemagglutinin, neuraminidase, and nucleoprotein genes of H9N2 avian influenza viruses isolated in Israel during the 2000–2005 epizootic. Comp Immunol Microbiol Infect Dis 2009; 32:221–238. [DOI] [PubMed] [Google Scholar]
- 7. El‐Zoghby EF, Arafa AS, Hassan MK et al. Isolation of H9N2 avian influenza virus from bobwhite quail (Colinus virginianus) in Egypt. Arch Virol 2012; 157:1167–1172. [DOI] [PubMed] [Google Scholar]
- 8. Kayali G, El‐Shesheny R, Kutkat MA, Kandeil AM, Mostafa A, Ducatez MF. Continuing threat of influenza (H5N1) virus circulation in Egypt. Emerg Infect Dis 2011; 17:2306–2308. Doi: 10.3201/eid1712.110683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Monne I, Ormelli S, Salviato A et al. Development and validation of a one‐step real‐time PCR assay for simultaneous detection of subtype H5, H7, and H9 avian influenza viruses. J Clin Microb 2008; 46:1769–1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. OIE . Manual of Diagnostic Tests and Vaccines for Terrestrial Animals, 2011. Available at http://www.oie.int/fileadmin/Home/eng/Health_standards/tahm/2.03.04_AI.pdf (Accessed 28 February 2012). [Google Scholar]
- 11. Peyre M, Samaha H, Makonnen YJ et al. Avian influenza vaccination in Egypt: limitations of the current strategy. J Mol Genet Med 2009; 3:198–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wan H, Sorrell EM, Song H et al. Replication and transmission of H9N2 influenza viruses in ferrets: evaluation of pandemic potential. PLoS One 2008; 3:e2923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wan H, Perez DR. Amino acid 226 in the hemagglutinin of H9N2 influenza viruses determines cell tropism and replication in human airway epithelial cells. J Virol 2007; 81:5181–5191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tombari W, Nsiri J, Larbi I, Guerin JL, Ghram A. Genetic evolution of low pathogenecity H9N2 avian influenza viruses in Tunisia: acquisition of new mutations. Virol J 2011; 8:467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Iqbal M, Yaqub T, Reddy K, McCauley JW. Novel genotypes of H9N2 influenza A viruses isolated from poultry in Pakistan containing NS genes similar to highly pathogenic H7N3 and H5N1 viruses. PLoS One 2009; 4:e5788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dong G, Xu C, Wang C et al. Reassortant H9N2 influenza viruses containing H5N1‐like PB1 genes isolated from black‐billedmagpies in Southern China. PLoS One 2011; 6:e25808. [DOI] [PMC free article] [PubMed] [Google Scholar]


