Abstract
Skeletal stem cells (SSCs) are a population of progenitor cells which give rise to postnatal skeletal tissues including bone, cartilage and bone marrow stroma, however not to adipose, haematopoietic or muscle tissue. Growth plate chondrocytes exhibit the ability of continuous proliferation and differentiation, which contributes to the continuous physiological growth. The growth plate has been hypothesized to contain SSCs which exhibit a desirable differentiation capacity to generate bone and cartilage. Due to the heterogeneity of the growth plate chondrocytes, SSCs in the growth plate are not well studied. The present study used cluster of differentiation (CD)146 and CD105 as markers to isolate purified SSCs. CD105+ SSCs and CD146+ SSCs were isolated using a magnetic activated cell sorting method. To quantitatively investigate the proliferation and differentiation ability, the colony-forming efficiency (CFE) and multi-lineage differentiation capacity of CD105+ SSCs and CD146+ SSCs were compared with unsorted cells and adipose-derived stem cells (ASCs). It was revealed that CD105+ and CD146+ subpopulations represented subsets of SSCs which generated chondrocytes and osteocytes, however not adipocytes. Compared with CD105+ subpopulations and ASCs, the CD146+ subpopulation exhibited a greater CFE and continuous high chondrogenic differentiation capacity in vitro. Therefore, the present study suggested that the CD146+ subpopulation represented a chondrolineage-restricted subpopulation of SSCs and may therefore act as a valuable cell source for cartilage regeneration.
Keywords: skeletal stem cells, growth plate, CD105, CD146, adipogenesis, osteogenesis, chondrogenesis
Introduction
Articular cartilage displays a limited capacity of self-renewal when suffered from trauma or degenerative disease. Consequently, stem cell-based tissue engineering is a promising method for cartilage repair (1–4). Mesenchymal stem cells (MSCs) are thought to be a promising cell source for their self-renewal and multi-lineage differentiation abilities, but the utility of MSCs has many unfavorable outcomes including low chondrogenic potential, vascularization and mineralization (5–8). The growth plate chondrocytes have the ability of continuous reproduction and differentiation. Thus it is long thought to be a cell source for cartilage repair strategies. But the growth plate chondrocytes are complicated, ranging from resting cells to proliferative, pre-hypertrophic and ultimately hypertrophic chondrocytes, the research about the growth plate remains elusive (9).
Growth plate chondrocytes are believed to compose progenitors of different cell types (10,11). By investigating the clonal relationship and lineage development of mesenchymal tissues in bone, Chan et al (12) recently isolated a population of postnatal skeletal stem cells (SSCs) which are important for postnatal skeletal development. Unlike mesenchymal stem cells, SSCs seldom diffrentiate into adipocyte (12). Similar research was also conducted by Worthley et al (13), they found that bone morphogenetic protein (BMP) antagonist Gremlin 1 defines a population of osteochondroreticular (OCR) stem cells which mainly concentrated within the metaphysis of long bone and were also thought to be a population of SSCs. These cells could self-renew and generate osteoblasts, chondrocytes and reticular marrow stromal cells, but not adipocytes. They are important for bone development, bone remodeling and fracture repair (13). Like hematopoietic stem cells, SSCs are heterogeneous and contain many lineage-restricted stem cells which lead to unreliable bone and cartilage formation (14). So it is important to isolate purified subpopulation of SSCs which could differentiate along chondrogenic lineage steadily.
Cell surface marker based cell purification is a simple and efficacious cell sorting method. Two alternative markers comprising endoglin (CD105) and melanoma cell adhesion molecule (MCAM; CD146) have been identified on the cell surface of isolated populations of SSCs. These subpopulations of SSCs exhibited different biological characteristics. CD105, a type III receptor for the transforming growth factor β (TGF-β) superfamily, is known as a relatively specific marker for identifying mesenchymal stem cells (15–18). Several lines of evidence showed that CD105 is related to chondrogenic potential of human MSCs or adipose-derived stem cells (ASCs) (4,19–21). Chan et al (12) found that CD105+ subpopulation represented a much more differentiated population of postnatal mouse SSCs compared with CD105− cell population. CD105+ subpopulation is responsible for bone and cartilage regeneration and CD105 is a candidate marker for SSC isolation (12). CD146, a cell adhesion molecule (CAM) that was originally identified as a tumor marker for melanoma (MCAM), has been studied as a putative mesenchymal stem cell marker in human umbilical cord perivascular cells (HUCPVCs) and bone marrow mesenchymal stromal cells (BMSCs) (22–24). Compared with CD146− MSCs, the CD146+ MSCs exhibited a much stronger multi-lineage differentiation potential and capacity of maintaining stemness and phenotype after long cultivation (24,25). There was also evidence showing that CD146 was a marker of cartilage-derived chondroprogenitor cells and CD146+ cartilage subpopulation exhibited greater therapeutic potential in cartilage repair and regeneration (26,27). However, it remains to be ascertained whether CD105 and CD146 could offer improved SSCs isolation in the growth plate.
Based on the aforementioned studies, it was hypothesized that purified SSCs may represent an improved alternative cell source compared with unsorted growth plate chondrocytes and ASCs for cartilage repair and tissue engineering. In the present study, we identified the existence and distribution of CD105+ SSCs and CD146+ SSCs in the growth plate. We then purified SSCs using CD105 and CD146 cell surface markers via magnetic activated cell sorting (MACS) method. Finally, we compared the colony-forming efficiency (CFE) and multi-lineage differentiation capacity of unsorted growth plate chondrocytes, CD105+ SSCs, CD146+ SSCs and ASCs in vitro.
Materials and methods
Isolation and culture of growth plate chondrocytes and ASCs
Growth plate chondrocytes were obtained from 1-week-old Sprague-Dawley rats (Experimental Animal Center of Tongji Hospital, Huazhong University of Science and Technology, Wuhan, China). All animal procedures were approved by the Ethics Committee on Animal Experimentation of Tongji Medical College, Huazhong University of Science and Technology (Wuhan, China).
Growth plate in distal femurs and proximal tibias of SD rats were isolated and subtly cut into pieces. After several rinses in phosphate-buffered saline (PBS), the pieces were subsequently digested with 0.25% trypsin solution (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) for 20 min and 0.1% solution of collagenase type II (Invitrogen; Life Technologies, Carlsbad, CA, USA) at 37°C over night. The cells were then harvested and cultured in Dulbecco's modified Eagles medium/Ham's F-12 (DMEM/F-12) supplemented with 10% fetal bovine serum (FBS) (both from Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) and antibiotics [(100 U/ml penicillin G sodium, 100 µg/ml streptomycin sulfate (Gibco; Thermo Fisher Scientific, Inc.)].
ASCs were isolated from 8-weeks-old Sprague-Dawley rats (Experimental Animal Center of Tongji Hospital). Adipose tissue was digested with collagenase type I (Invitrogen; Life Technologies) for 1 h at 37°C. The cells were then harvested and cultured in the DMEM/F-12 (Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% FBS and antibiotics [(100 U/ml penicillin G sodium, 100 µg/ml streptomycin sulfate (Gibco; Thermo Fisher Scientific, Inc.)]. When reaching 90% confluence, cells were passaged to obtain sufficient number of ASCs.
Flow cytometry analysis of stem cell surface markers on the growth plate chondrocytes
Chondrocytes (1×106) from growth plate (passage 1) were washed in PBS and incubated with corresponding primary antibodies for one hour at 4°C. Primary antibodies used here were conjugated antibodies specific for CD44-FITC (dilution 1:100, #550974; BD Biosciences, San Diego, CA, USA), CD29-PE (dilution 1:80, #48-0291; eBiosciences; Thermo Fisher Scientific, Inc.), CD34-PE (dilution 1:100; #sc-74499 PE; Santa Cruz Biotechnology, Inc., Dallas, TX, USA), CD45-PE (dilution 1:80, #12-0461; eBiosciences; Thermo Fisher Scientific, Inc.) and unconjugated antibodies for CD105 (dilution 1:100, #ab11414) and CD146 (dilution 1:80, #ab75769) (both from Abcam, Cambridge, MA, USA). After washing with PBS, cells stained with CD105 and CD146 were incubated with anti-mouse FITC-conjugated secondary antibody (dilution 1:500, #A0568; Beyotime Institute of Biotechnology, Guangzhou, China) or Alexa Fluor 594 donkey anti-rabbit IgG (dilution 1:500, #A-21207; Invitrogen; Life Technologies) for 30 min at 4°C. The cells were washed twice and then re-suspended in 200 µl PBS for the flow cytometry analysis (FACSort; BD Biosciences).
Immunohistochemistry (IHC) analysis of CD146 and CD105 expression in the growth plate
Proximal tibias of 1-week-old SD rats were isolated and fixed with 4% formaldehyde. The samples were decalcificated with 12.5% EDTA solution, dehydrated, embedded into paraffin wax and sectioned at 5 mm thickness. Primary mouse anti-CD105 antibody (dilution 1:100) and rabbit anti-CD146 antibody (dilution 1:250) (both from Abcam) were used for immune labeling according to manufacturer's instructions. Then sections were incubated with biotinylated goat anti-mouse or goat anti-rabbit secondary antibodies (#BA1001 or #BA1003, dilution 1:2,000; Boster Biological Technology, Wuhan, China) at RT for 30 min. Reactivity was detected with a diaminobenzidine tetrahydrochloride (DAB) substrate kit (Beyotime Institute of Biotechnology), with hematoxylin as the counterstain. Images were captured by microscopy (FV500; Olympus Corporation, Tokyo, Japan).
Cell sorting by MACS
About 5×106 growth plate chondrocytes were harvested for magnetic activated cell sorting (Miltenyi, Teterow, Germany) according to manufacturer's instructions. Briefly, single-cell suspensions were incubated with mouse anti-CD105 antibody (dilution 1:100) or rabbit anti-CD146 antibody (dilution 1:80) (both from Abcam) for 30 min at 37°C. After centrifugated at 300 × g for 10 min and washed by PBS twice, the cells were then incubated with anti-rabbit IgG microbeads or anti-mouse IgG microbeads for 15 min at 37°C. The cells were then washed and resuspended with washing buffer and added into the MS column, the CD146+ SSCs and CD105+ SSCs were obtained following the instructions. The sorted cells were cultured in growth medium to obtain sufficient quantities for further research.
Immunofluorescence assay
To confirm the expression of CD105 and CD146 in sorted cells, immunofluorescence staining was performed. Cells were grown to sub confluence on coverslips in 12-wells plate. Cells were then fixed with 4% paraformaldehyde for 20 min at room temperature and washed 3 times with PBS. After that, the cells were incubated with 0.1% Triton X-100 for 15 min at room temperature (RT) and blocked with 1% bovine serum albumin (Biosharp, Hefei, China) for 1 h at room temperature. Then the cells were incubated with mouse anti-CD105 antibody (dilution 1:100) or rabbit anti-CD146 antibody (dilution 1:80) (both from Abcam) overnight at 4°C. After washing with PBS, cells stained with CD105 or CD146 were incubated with anti-mouse FITC-conjugated secondary antibody (dilution 1:500, #A0568; Beyotime Institute of Biotechnology) or Alexa Fluor 594 donkey anti-rabbit IgG (dilusion 1:1,000, #A-21207; Invitrogen; Life Technologies) for 1 h at room temperature. The nuclei were then labeled with DAPI (50 µg/ml; Sigma-Aldrich; Merck KGaA) for 5 min. Fluorescence images were captured by fluorescence microscopy (FV500; Olympus Corporation).
CFE assay
The proliferative induction capability of the cells was evaluated by CFE analysis. Briefly, 100 cells were seeded and cultured in 60-mm dishes. Two weeks later, colonies formed from single cell were fixed in 4% paraformaldehyde for 20 min at RT. After washing with PBS for three times, Giemsa (#ab150670; Abcam) staining was performed according to manufacturer's instructions. Colonies with >50 cells were counted and recorded.
Multi-lineage differentiation assays
Cells at passage 3 were initially seeded in a 6-well plate at the density of 3×104 cells/cm2. When they reached 90% confluence, cells were washed with PBS and transferred to commercially available adipogenic or osteogenic medium (#RASMD-90031 or #RASMD-90021; Cyagen Biosciences Inc., Santa Clara, CA, USA). After 3 weeks, cells were fixed by 4% paraformaldehyde and washed 3 times with PBS. For adipogenic assay, cells were incubated with Oil red O (Cyagen Biosciences Inc.) for 30 min to detect the intra-cellular lipid droplets. For quantitative evaluation, dye content was extracted using isopropanol and the absorbance at a wavelength of 540 nm was detected. For osteogenic assay, cells were stained with Alizarin Red (Cyagen Biosciences Inc.) for 10 min at room temperature.
A pellet culture system was used for chondrogenic differentiation. Briefly, 1×106 cells were placed and centrifugated at 300 × g for 15 min in a 15 ml polypropylene tube to form a pellet. Then pellets were cultured in the chondrogenic medium (#RASMD-90041; Cyagen Biosciences Inc.), which was changed every 3 days. After induction for 3 weeks, the cell pellets were fixed by 4% paraformaldehyde, dehydrated and then embedded into paraffin wax. Tissue blocks were cut into 5 mm sections and were placed on slides and dried overnight at 37°C. Chondrogenic differentiation was finally assessed by Alician blue staining (Cyagen Biosciences Inc.) and IHC using antibodies against aggrecan (dilution 1:50, #ab36861) and type II collagen (dilution 1:100, #ab185430) (both from Abcam) according to the manufacturer's protocols.
Real-time quantitative polymerase chain reaction (RT-qPCR) analysis
Total mRNA was extracted using TRIzol reagent (Invitrogen; Life Technologies) following the manufacturer's instructions. Complementary DNA (cDNA) was obtained from total RNA using ReverTra Ace qPCR RT kit (Toyobo Co., Ltd., Osaka, Japan) according to the manufacturer's protocols. Then the cDNA was amplified by SYBR Green Real-Time PCR Master Mix (Toyobo Co., Ltd.) with following cycling conditions: 30 sec of polymerase activation at 95°C, followed by 40 cycles of 95°C for 5 sec and 60°C for 30 sec. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as an internal control. The relative expression levels of each gene were calculated using the comparative 2−ΔΔCt method (28). Sequences of primers for interested genes are listed in Table I.
Table I.
Primers used for RT-qPCR.
| Gene | Accession no. | Primer sequence (5′-3′) |
|---|---|---|
| PPAR-γ | NM_001145366 | F: CGGTTGATTTCTCCAGCATT |
| R: TCGCACTTTGGTATTCTTGG | ||
| LPL | NM_012598 | F: AACATTGGAGAAGCCATTCG |
| R: TTCATTCAGCAGGGAGTCAA | ||
| AP2 | NM_053365 | F: ATGTGTCATGAAAGGCGTGA |
| R: AAACCACCAAATCCCATCAA | ||
| RUNX2 | NM_001278483 | F: CTACTCTGCCGAGCTACGAAAT |
| R: TCTGTCTGTGCCTTCTTGGTTC | ||
| ALP | NM_013059 | F: CCTGGACCTCATCAGCATTT |
| R: AGGGAAGGGTCAGTCAGGTT | ||
| OPN | NM_012881 | F: CAAGGACCAACTACAACCA |
| R: GGAGACAGGAGGCAAGG | ||
| SOX9 | NM_080403 | F: GTGGGAGCGACAACTTTACC |
| R: GCGAGCACTTAGCAGAGGC | ||
| Aggrecan | NM_022190 | F: CAAACAGCAGAAACAGCCAAGT |
| R: GAAGGCATAAGCATGTGAAAGTG | ||
| COL2 | NM_012929 | F: CACCCAGAGTGGAAGAGCG |
| R: TCAGTGGACAGTAGACGGAGGA |
F, forward; R, reverse; PPAR-γ, peroxisome proliferators-activated receptor-γ; LPL, lipoprteinlipase; AP2, adipocyte fatty acid-binding protein 2; RUNX2, runt-related transcription factor 2; ALP, alkaline phosphatase; OPN, osteopontin; SOX9, sex-determining region Y-box containing gene 9; AGG, aggrecan; COL2, type II collegen; RT-qPCR, reverse transcription-quantitative polymerase chain reaction.
Statistical analyses
All experiments were performed in triplicate, and one representative set was chosen to be shown. All data are presented as mean and standard deviation (SD) and analyzed with GraphPad Prism 6.0. The significance of difference was determined using the Student's t-test and analysis of variance (ANOVA); differences with P<0.05 were considered significant.
Results
Surface antigen profiles
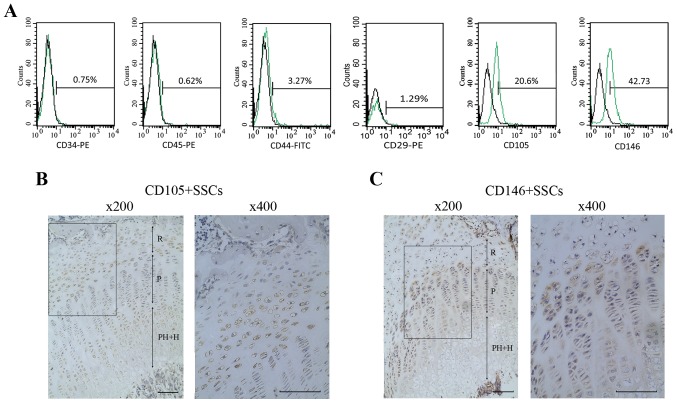
Flow cytometry analyses were performed to assess the phenotype of the growth plate chondrocytes. Results showed that 42.73% of the cells were positive for CD146 and 20.6% of the cells were positive for CD105. Meanwhile, growth plate chondrocytes were negative for the hematopoietic markers (CD34 and CD45) and MSC-associated surface markers (CD29 and CD44) (Fig. 1A).
Figure 1.
Surface markers of growth plate chondrocytes and distribution of CD146+ SSCs and CD105+ SSCs in the growth plate. (A) Flow cytometry analysis of growth plate chondrocytes. The growth plate chondrocytes were positive for CD105 and CD146 while negative for CD34, CD45, CD29 and CD44. (B) Immunohistochemical staining of CD105+ SSCs and CD146+ SSCs. The CD105+ SSCs mainly located at the resting zone and the hypertrophy zone of growth plate. (C) The CD146+ SSCs mainly located at the resting zone and the proliferating zone. Different zones of the growth plate were marked. Scar bar, 100 µm. SSCs, skeletal stem cells; R, resting zone; P, proliferative zone; PH+H, prehypertrophic and hypertrophic zone; CD, cluster of differentiation.
Distribution of CD146+ SSCs and CD105+ SSCs in growth plate
The growth plate was divided into resting zone, proliferative zone, prehypertrophic and hypertrophic zone. We then identified the distribution of CD146+ SSCs and CD105+ SSCs in growth plate using IHC. As shown in Fig. 1B and C, CD146+ SSCs mainly located at the resting zone, partly located at the proliferating zone of the growth plate, while the CD105+ SSCs mainly located at the resting zone and the hypertrophic zone of growth plate.
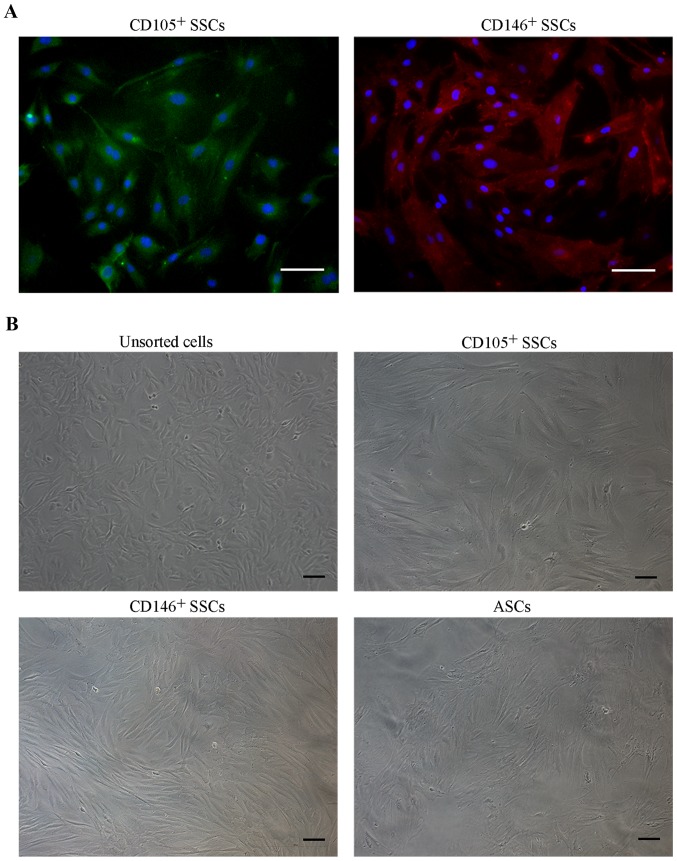
Isolation and cell morphology of CD146+ SSCs and CD105+ SSCs
We then isolated CD146+ SSCs and CD105+ SSCs using MACS method. Immunofluorescence staining of the isolated cells confirmed that the CD146+ SSCs and CD105+ SSCs were successfully isolated and cultured respectively (Fig. 2A). For cell morphology (Fig. 2B), unsorted growth plate chondrocytes generally showed heterogeneous populations containing triangle and fibroblast-like cells. Isolated CD146+ SSCs and CD105+ SSCs cells were more fibroblast-like. Obvious differences in morphology were noted between unsorted and MACS-sorted CD146+ and CD105+ subpopulations.
Figure 2.
Cell morphology of isolated stem cells. (A) Immunofluorescence staining was performed on CD105+ SSCs and CD146+ SSCs, the nuclei were counterstained with DAPI. (B) Morphology of unsorted growth plate cells, CD105+ SSCs, CD146+ SSCs and ASCs. Scar bar, 100 µm. ASCs, adipose-derived stem cells; SSCs, skeletal stem cells; CD, cluster of differentiation.
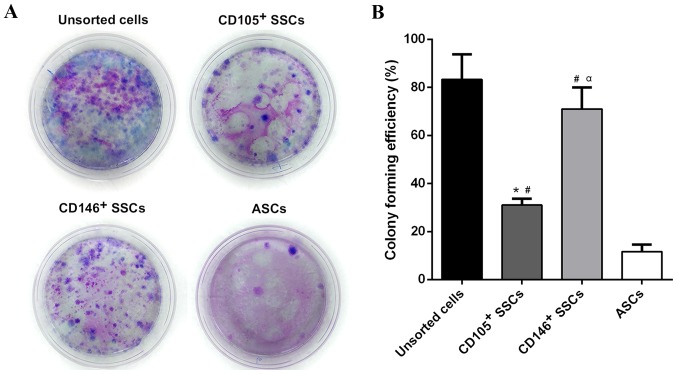
The CFE of CD146+ SSCs and CD105+ SSCs compared with unsorted cells and ASCs
After 14 days of incubation, colony-forming unites were observed in all four groups. The unsorted growth plate chondrocytes yielded the highest number of colonies followed by CD146+ SSCs and CD105+ SSCs. The CFE of CD146+ SSCs was 2.2 fold stronger than CD105+ SSCs. The ASCs showed a much weaker CFE compared with the unsorted growth plate chondrocytes, CD105+ SSCs and CD146+ SSCs (Fig. 3).
Figure 3.
Comparison of cell proliferation capacity. (A) The CFE of unsorted growth plate cells, CD105+ SSCs, CD146+ SSCs and ASCs was detected respectively. (B) Numbers of colony unites were counted. All data are from 3 independent experiments and are presented as means ± SD. *P<0.05 vs. unsorted cells; #P<0.05 vs. ASCs; αP<0.05 vs. CD105+ SSCs. CFE, colony-forming efficiency; ASCs, adipose-derived stem cells; SSCs, skeletal stem cells; CD, cluster of differentiation.
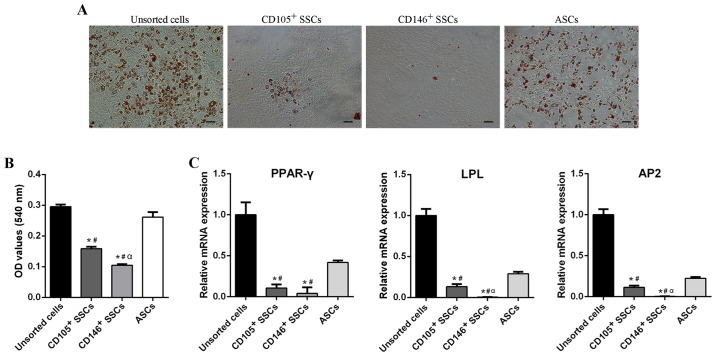
Adipogenic differentiation potential of CD146+ SSCs and CD105+ SSCs compared with unsorted cells and ASCs
The adipogenic differentiation capacity of CD146+ SSCs and CD105+ SSCs was measured and compared with unsorted cells and ASCs. After 21 days of adipogenic induction, a large amount of oil-red positive lipid droplets were found in unsorted cells and ASCs, while little was detected in CD105+ SSCs or CD146+ SSCs (Fig. 4A and B).
Figure 4.
Comparison of adipogenic differentiation potential. (A) Oil Red O staining of unsorted cells, CD105+ SSCs, CD146+ SSCs and ASCs. Scar bar, 200 µm. (B) Quantitative measurement of lipid droplets in all four groups. (C) The mRNA levels of PPAR-γ, LPL and AP2 were detected using reverse transcription-quantitative polymerase chain reaction. All data are from 3 independent experiments and are presented as means ± SD. *P<0.05 vs. unsorted cells; #P<0.05 vs. ASCs; αP<0.05 vs. CD105+ SSCs. PPAR-γ, peroxisome proliferators-activated receptor-γ; LPL, lipoprteinlipase; AP2, adipocyte fatty acid-binding protein 2; ASCs, adipose-derived stem cells; SSCs, skeletal stem cells; CD, cluster of differentiation.
RT-qPCR showed similar results. After 2 week of adipogenic induction, CD105+ and CD146+ enriched populations exhibited decreased peroxisome proliferators-activated receptor-γ (PPAR-γ), lipoprteinlipase (LPL) and adipocyte fatty acid-binding protein 2 (AP2) expression compared with unsorted cells and ASCs. The PPAR-γ expression of unsorted cells was 9-fold over the CD105+ SSCs, 24-fold over the CD146+ SSCs, and 2-fold over ASCs. Similar trends were also observed in the LPL and AP2 expression (Fig. 4C). These results suggested that CD105+ SSCs and CD146+ SSCs could hardly differentiate into adipocytes.
Osteogenic differentiation potential of CD146+ SSCs and CD105+ SSCs compared with unsorted cells and ASCs
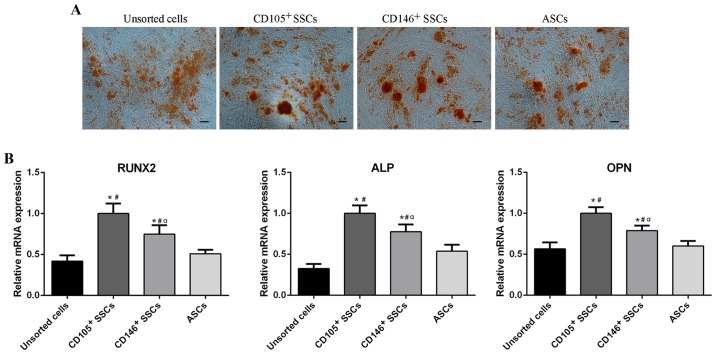
To assess the osteogenic differentiation potential of CD146+ SSCs and CD105+ SSCs, cells were cultured in osteogenic medium for 21 days. All four groups showed formation of red calcium deposits. The CD146+ SSCs and CD105+ SSCs exhibited similar amounts of calcium-containing mineralized nodules compared with unsorted cells and ASCs (Fig. 5A).
Figure 5.
Comparison of osteogenic differentiation potential. (A) Alizarin red staining of unsorted cells, CD105+ SSCs, CD146+ SSCs and ASCs. Scar bar, 100 µm. (B) The mRNA levels of Runx2, ALP and OPN were detected using reverse transcription-quantitative polymerase chain reaction. All data are from 3 independent experiments and are presented as means ± SD. *P<0.05 vs. unsorted cells; #P<0.05 vs. ASCs, αP<0.05 vs. CD105+ SSCs. RUNX2, runt-related transcription factor 2; ALP, alkaline phosphatase; OPN, osteopontin; ASCs, adipose-derived stem cells; SSCs, skeletal stem cells; CD, cluster of differentiation.
Total RNA was extracted from the cells respectively after cultivation in osteogenic medium for 2 weeks. RT-qPCR was then conducted to assess the expression of osteogenic related genes including runt-related transcription factor 2 (RUNX2), alkaline phosphatase (ALP) and osteopontin (OPN). After 2 weeks of osteogenic induction, the CD105+ SSCs showed higher expression of RUNX2, ALP and OPN than the CD146+ SSCs. The RUNX2, ALP and OPN levels in CD105+ SSCs and CD146+ SSCs were higher than unsorted cells and ASCs (Fig. 5B).
Chondrogenic differentiation potential of CD146+ and CD105+ SSCs compared with unsorted cells and ASCs
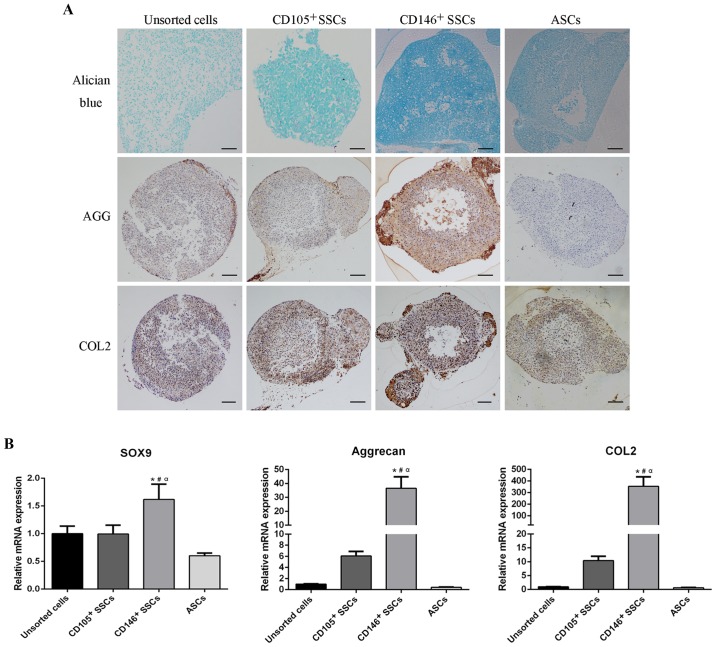
To assess the chondrogenic differentiation potential of CD146+ SSCs and CD105+ SSCs, micromass pellets were cultured in chondrogenic medium for 21 days. Alician blue staining was performed to characterize the proteoglycan formation. Differentiated chondrocytes surface markers, aggrecan and type II collagen, were assessed using IHC. Alician blue staining revealed a higher content of proteoglycan in CD146+ pellets. Consistently, CD146+ pellets showed much higher contents of aggrecan and type II collagen compared with pellets in other three groups (Fig. 6A).
Figure 6.
Comparison of chondrogenic differentiation potential. (A) Pellets derived from unsorted cells, CD105+ SSCs, CD146+ SSCs and ASCs were stained with Alician blue or immunohistochemically stained with aggrecan or type II collagen respectively. Scar bar, 100 µm. (B) The mRNA levels of SOX9, aggrecan and type II collagen were detected using RT-qPCR. All data are from 3 independent experiments and are presented as means ± SD. *P<0.05 vs. unsorted cells; #P<0.05 vs. ASCs; αP<0.05 vs. CD105+ SSCs. SOX9, SRY-box containing gene 9; AGG, aggrecan; COL2, type II collegen; ASCs, adipose-derived stem cells; SSCs, skeletal stem cells.
After cultivation in chondrogenic medium for 2 weeks, total RNA was extracted and subjected to RT-qPCR. The mRNA levels of chondrogenic markers including SRY-box containing gene 9 (SOX9), type II collagen and aggrecan were evaluated. As shown in Fig. 6B, CD146+ SSCs exhibited higher mRNA level of SOX9 compaired with unsorted cells, CD105+ SSCs and ASCs. Furthermore, extraordinarily high levels of aggrecan and type II collagen were observed in the CD146+ SSCs. The aggrecan expression level in CD146+ SSCs was 36-fold over unsorted cells, 6-fold over CD105+ SSCs, and 91.4-fold over ASCs. Consistently, the expression level of type II collegen in CD146+ SSCs was 353-fold over unsorted cells, 34-fold over CD105+ SSCs and 548-fold over ASCs. These results suggested that the CD146+ SSCs exhibited a strong and steady chondrogenic differentiation capacity in vitro.
Discussion
The mesenchymal stem cells represent a powerful tool for cartilage tissue regeneration, but their utility is limited by low chondrogenic potential, vascularization and mineralization (29). So it is important to isolate a stem cell which could differentiate strictly along chondrogenic lineage. The regenerative capacity of bone indicates the presence of skeletal stem cell in bone. However, while this regenerative ability has long been recognized, the identity of the responsible cell population in vivo has only recently been confirmed. Recent researches demonstrated the existence of SSCs at the end of long bones. SSCs could self-renew and generate bone and cartilage, but not adipocytes (12). Like hematopoietic stem cells, SSCs are diverse, with distinct cell-surface marker profiles and distinct fates (14). Theoretically, SSCs with appropriate cell surface markers could offer an ideal cell source for cartilage tissue engineering. In the present study, we successfully isolated SSCs from growth plate using cell surface markers CD105 and CD146. These subpopulations of SSCs could self-renew and differentiate into osteoblasts and chondrocytes but not adipocytes.
The growth plate contains three zones that include chondrocytes at different stages of differentiation (30–32). The zone closest to the epiphysis is termed the resting zone. The resting zone is thought to contain chondrocytes that serve as progenitor cells, which can generate new clones of rapidly proliferating chondrocytes (9,33). In the present study, flow cytometry showed that the growth plate chondrocytes are positive for CD105 and CD146. IHC revealed that the CD105+ SSCs mainly located in the resting zone and hypertrophic zone while the CD146+ SSCs mainly located in the resting zone and proliferating zone. Many researches demonstrated the existence of CD146+ subpopulation in osteoarthritis cartilage chondrocytes and indicated that CD146 is a chondroprogenitor-associated marker (34,35). CD146 was reported to be involved in cell-cell, cell-matrix interactions and cell migration (36–38). There are evidences identifying that CD146+ subpopulation has a high migration abilitiy and is responsible for tumor metastasis as well as tissue repair (39,40). A previous study suggested that CD146 was also expressed in bone marrow derived mesenchymal stem cells and had been used for the isolation of this cell population (41). Thus we postulated that SSCs mainly located in the resting zone of the growth plate and could reproduce and migrate to the place where needed.
SSCs were thought to contribute to the postnatal growth of the long bone and bone fracture repair (42–44). SSCs contain different types of progenitors, which are responsible for bone and cartilage formation, but not for adipose tissue or muscle tissue formation. The CD105 was demonstrated as a marker of SSCs (12). In this study, we found that the CD146+ cells exhibited high osteogenic, chondrogenic differentiation capacity and extremely low adipogeneic capacity in vitro, suggesting that CD146+ cells are also a subset of SSCs. Furthermore, results of RT-qPCR showed that the expression of chondrogenic related genes, such as aggrecan and type II collagen, in the CD146+ SSCs were extraordinarily higher than CD105+ SSCs and ASCs. These results were corroborated by IHC. Taken together, our data implicated that the CD146+ subpopulation represents a subset of SSCs which is much differentiated and prone to differentiate into chondrocytes. The CD146+ subpopulation is a chondrogenic lineage restricted stem cells, or precartilaginous stem cell. It has the ability of efficiently differentiating into chondrocytes with extensive extracellular matrix (ECM) formation.
Both CD105+ and CD146+ SSCs were discussed in the present study. But there are also evidences showing that CD105+ subpopulation is responsible for endochondral ossification and has lost the chondrogenesis potential (6,45). Consistently, we found that CD105+ subpopulation expressed higher levels of osteogenetic genes. Taking into account that CD146+ MSCs derived from bone marrow were prone to differentiate into chondrocytes (46) and that CD146+ chondroprogenitor cells with multi-lineage differentiation abilities were found in articular cartilage (35), CD146 might be regarded as a marker for chondroprogenitor subpopulation. Our data also revealed that CD146+ SSCs isolated from the growth plate exhibited superior chondrogenic differentiation capacity compared CD105+ SSCs. Taken together, these data suggest that CD105+ subpopulations and CD146+ subpopulations may have different cell fate and that CD105+ SSCs may be used to enhance endochondral ossification while CD146+ SSCs are more appropriate as a stem cell source for cartilage repair.
The proliferative capacity of stem cells is important with regard to their application in cell therapy. In the present study, the proliferative capacity of the CD105+ and the CD146+ subpopulations were compared with that of unsorted cells and ASCs using CFE assay. Of note, it was identified that the unsorted growth plate chondrocytes exhibited the highest CFE in vitro. Chan et al (12) suggested that the SSCs are diverse, similar to the diverse hematopoietic progenitor cells that generate various differentiated blood cells. This may imply that the unsorted growth plate chondrocytes contain subpopulations which are more primitive and have higher capacity of self-renewal. However, the oil-red staining results showed that they were prone to differentiate into adipocytes. The results of RT-qPCR identified that the unsorted growth plate chondrocytes expressed much higher level of adipogenic differentiation related genes, including PPAR-γ, LPL and AP2. PPAR-γ is a key transcriptional regulator of adipogenesis. Wang et al (47) found that PPAR-γ is expressed in growth plate chondrocytes and PPAR-γ is able to promote adipogenic differentiation in growth plate chondrocytes, while negatively regulate chondrogenic differentiation and terminal differentiation.
In the present study, we isolated purified subpopulations of SSCs using MACS method. We found that both CD105+ and CD146+ subpopulations had a higher CFE than ASCs. Moreover, compared with the CD105+ subpopulation, the CFE of the CD146+ subpopulation was much higher. Consistently, CD146+ subpopulations mainly located in the resting zone and proliferative zone of the growth plate, which suggested that CD146+ subpopulations are progenitor cells which can generate new clones of rapidly proliferating chondrocytes.
ASCs are attractive stem cell source for tissue engineering (48–50). These cells can be obtained by simple liposuction and had ability of multi-lineage differentiation. Under appropriate culture conditions, they can differentiate along osteogenic lineage (50–52), chondrogenic lineage (53), and adipogenic lineage. Our results showed that CD146+ SSCs exhibited higher colony forming capacity compared compared with ASCs. Most importantly, the CD146+ SSCs showed a steady and high chondrogenic related gene expression and ECM formation in vitro after a long period of cultivation. These results also implied that compared with ASCs the CD146+ SSCs could be a much more appropriate stem cell source for cartilage tissue engineering.
In summary, we identified the existence of SSCs in the growth plate and the CD105+ and the CD146+ subpopulations represented subsets of SSCs which could generate chondrocytes and osteocytes, but not adipocytes. Compared with CD105+ subpopulations and ASCs, the CD146+ subpopulation exhibited higher colony forming capacity and continuous high chondrogenic differentiation capacity in vitro. Thus we propose that CD146+ subpopulation is chondrogenic lineage restricted SSCs and it could provide cell candidates for cell-based cartilage regeneration.
Acknowledgements
This work was supported by grants from the National Natural Science Foundation of China (no. 81371915 and 81572094).
References
- 1.Blunk T, Sieminski AL, Gooch KJ, Courter DL, Hollander AP, Nahir AM, Langer R, Vunjak-Novakovic G, Freed LE. Differential effects of growth factors on tissue-engineered cartilage. Tissue Eng. 2002;8:73–84. doi: 10.1089/107632702753503072. [DOI] [PubMed] [Google Scholar]
- 2.Ishimura D, Yamamoto N, Tajima K, Ohno A, Yamamoto Y, Washimi O, Yamada H. Differentiation of adipose-derived stromal vascular fraction culture cells into chondrocytes using the method of cell sorting with a mesenchymal stem cell marker. Tohoku J Exp Med. 2008;216:149–156. doi: 10.1620/tjem.216.149. [DOI] [PubMed] [Google Scholar]
- 3.Jo CH, Lee YG, Shin WH, Kim H, Chai JW, Jeong EC, Kim JE, Shim H, Shin JS, Shin IS, et al. Intra-articular injection of mesenchymal stem cells for the treatment of osteoarthritis of the knee: A proof-of-concept clinical trial. Stem cells. 2014;32:1254–1266. doi: 10.1002/stem.1634. [DOI] [PubMed] [Google Scholar]
- 4.Jiang T, Liu W, Lv X, Sun H, Zhang L, Liu Y, Zhang WJ, Cao Y, Zhou G. Potent in vitro chondrogenesis of CD105 enriched human adipose-derived stem cells. Biomaterials. 2010;31:3564–3571. doi: 10.1016/j.biomaterials.2010.01.050. [DOI] [PubMed] [Google Scholar]
- 5.Kobayashi S, Takebe T, Inui M, Iwai S, Kan H, Zheng YW, Maegawa J, Taniguchi H. Reconstruction of human elastic cartilage by a CD44+ CD90+ stem cell in the ear perichondrium; Proc Natl Acad Sci USA; 2011; pp. 14479–14484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Crisan M, Yap S, Casteilla L, Chen CW, Corselli M, Park TS, Andriolo G, Sun B, Zheng B, Zhang L, et al. A perivascular origin for mesenchymal stem cells in multiple human organs. Cell Stem Cell. 2008;3:301–313. doi: 10.1016/j.stem.2008.07.003. [DOI] [PubMed] [Google Scholar]
- 7.Bhumiratana S, Eton RE, Oungoulian SR, Wan LQ, Ateshian GA, Vunjak-Novakovic G. Large, stratified, and mechanically functional human cartilage grown in vitro by mesenchymal condensation; Proc Natl Acad Sci USA; 2014; pp. 6940–6945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Oldershaw RA. Cell sources for the regeneration of articular cartilage: The past, the horizon and the future. Int J Exp Pathol. 2012;93:389–400. doi: 10.1111/j.1365-2613.2012.00837.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Belluoccio D, Bernardo BC, Rowley L, Bateman JF. A microarray approach for comparative expression profiling of the discrete maturation zones of mouse growth plate cartilage. Biochim Biophys Acta. 2008;1779:330–340. doi: 10.1016/j.bbagrm.2008.02.010. [DOI] [PubMed] [Google Scholar]
- 10.Pichler K, Schmidt B, Fischerauer EE, Rinner B, Dohr G, Leithner A, Weinberg AM. Behaviour of human physeal chondro-progenitorcells in early growth plate injury response in vitro. Int Orthop. 2012;36:1961–1966. doi: 10.1007/s00264-012-1578-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gothard D, Cheung K, Kanczler JM, Wilson DI, Oreffo RO. Regionally-derived cell populations and skeletal stem cells from human foetal femora exhibit specific osteochondral and multi-lineage differentiation capacity in vitro and ex vivo. Stem Cell Res Ther. 2015;6:251. doi: 10.1186/s13287-015-0247-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chan CK, Seo EY, Chen JY, Lo D, McArdle A, Sinha R, Tevlin R, Seita J, Vincent-Tompkins J, Wearda T, et al. Identification and specification of the mouse skeletal stem cell. Cell. 2015;160:285–298. doi: 10.1016/j.cell.2014.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Worthley DL, Churchill M, Compton JT, Tailor Y, Rao M, Si Y, Levin D, Schwartz MG, Uygur A, Hayakawa Y, et al. Gremlin 1 identifies a skeletal stem cell with bone, cartilage and reticular stromal potential. Cell. 2015;160:269–284. doi: 10.1016/j.cell.2014.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chan CK, Lindau P, Jiang W, Chen JY, Zhang LF, Chen CC, Seita J, Sahoo D, Kim JB, Lee A, et al. Clonal precursor of bone, cartilage, and hematopoietic niche stromal cells; Proc Natl Acad Sci USA; 2013; pp. 12643–12648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chiba H, Ishii G, Ito TK, Aoyagi K, Sasaki H, Nagai K, Ochiai A. CD105-positive cells in pulmonary arterial blood of adult human lung cancer patients include mesenchymal progenitors. Stem cells. 2008;26:2523–2530. doi: 10.1634/stemcells.2008-0037. [DOI] [PubMed] [Google Scholar]
- 16.Salamon A, Jonitz-Heincke A, Adam S, Rychly J, Müller-Hilke B, Bader R, Lochner K, Peters K. Articular cartilage-derived cells hold a strong osteogenic differentiation potential in comparison to mesenchymal stem cells in vitro. Exp Cell Res. 2013;319:2856–2865. doi: 10.1016/j.yexcr.2013.09.008. [DOI] [PubMed] [Google Scholar]
- 17.Amiri F, Halabian R, Harati M Dehgan, Bahadori M, Mehdipour A, Roushandeh A Mohammadi, Roudkenar M Habibi. Positive selection of Wharton's jelly-derived CD105(+) cells by MACS technique and their subsequent cultivation under suspension culture condition: A simple, versatile culturing method to enhance the multipotentiality of mesenchymal stem cells. Hematology. 2015;20:208–216. doi: 10.1179/1607845414Y.0000000185. [DOI] [PubMed] [Google Scholar]
- 18.Odabas S, Sayar F, Güven G, Yanikkaya-Demirel G, Pişkin E. Separation of mesenchymal stem cells with magnetic nanosorbents carrying CD105 and CD73 antibodies in flow-through and batch systems. J Chromatogr B Analyt Technol Biomed Life Sci. 2008;861:74–80. doi: 10.1016/j.jchromb.2007.11.017. [DOI] [PubMed] [Google Scholar]
- 19.Qi J, Chen A, You H, Li K, Zhang D, Guo F. Proliferation and chondrogenic differentiation of CD105-positive enriched rat synovium-derived mesenchymal stem cells in three-dimensional porous scaffolds. Biomed Mater. 2011;6:015006. doi: 10.1088/1748-6041/6/1/015006. [DOI] [PubMed] [Google Scholar]
- 20.Chan CK, Chen CC, Luppen CA, Kim JB, DeBoer AT, Wei K, Helms JA, Kuo CJ, Kraft DL, Weissman IL. Endochondral ossification is required for haematopoietic stem-cell niche formation. Nature. 2009;457:490–494. doi: 10.1038/nature07547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gothard D, Greenhough J, Ralph E, Oreffo RO. Prospective isolation of human bone marrow stromal cell subsets: A comparative study between Stro-1-, CD146- and CD105-enriched populations. J Tissue Eng. 2014;5:2041731414551763. doi: 10.1177/2041731414551763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bakopoulou A, Leyhausen G, Volk J, Koidis P, Geurtsen W. Comparative characterization of STRO-1(neg)/CD146(pos) and STRO-1(pos)/CD146(pos) apical papilla stem cells enriched with flow cytometry. Arch Oral Biol. 2013;58:1556–1568. doi: 10.1016/j.archoralbio.2013.06.018. [DOI] [PubMed] [Google Scholar]
- 23.Middleton J, Americh L, Gayon R, Julien D, Mansat M, Mansat P, Anract P, Cantagrel A, Cattan P, Reimund JM, et al. A comparative study of endothelial cell markers expressed in chronically inflamed human tissues: MECA-79, Duffy antigen receptor for chemokines, von Willebrand factor, CD31, CD34, CD105 and CD146. J Pathol. 2005;206:260–268. doi: 10.1002/path.1788. [DOI] [PubMed] [Google Scholar]
- 24.Schugar RC, Chirieleison SM, Wescoe KE, Schmidt BT, Askew Y, Nance JJ, Evron JM, Peault B, Deasy BM. High harvest yield, high expansion, and phenotype stability of CD146 mesenchymal stromal cells from whole primitive human umbilical cord tissue. J Biomed Biotechnol. 2009;2009:789526. doi: 10.1155/2009/789526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tsang WP, Shu Y, Kwok PL, Zhang F, Lee KK, Tang MK, Li G, Chan KM, Chan WY, Wan C. CD146+ human umbilical cord perivascular cells maintain stemness under hypoxia and as a cell source for skeletal regeneration. PLoS One. 2013;8:e76153. doi: 10.1371/journal.pone.0076153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu CC, Liu FL, Sytwu HK, Tsai CY, Chang DM. CD146+ mesenchymal stem cells display greater therapeutic potential than CD146-cells for treating collagen-induced arthritis in mice. Stem Cell Res Ther. 2016;7:23. doi: 10.1186/s13287-016-0285-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jiang Y, Cai Y, Zhang W, Yin Z, Hu C, Tong T, Lu P, Zhang S, Neculai D, Tuan RS, Ouyang HW. Human cartilage-derived progenitor cells from committed chondrocytes for efficient cartilage repair and regeneration. Stem Cells Transl Med. 2016;5:733–744. doi: 10.5966/sctm.2015-0192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wong HL, Siu WS, Fung CH, Zhang C, Shum WT, Zhou XL, Lau CB, Zhang JF, Leung PC, Fu WM, Ko CH. Characteristics of stem cells derived from rat fascia: In vitro proliferative and multilineage potential assessment. Mol Med Rep. 2015;11:1982–1990. doi: 10.3892/mmr.2014.2967. [DOI] [PubMed] [Google Scholar]
- 29.Qi Y, Du Y, Li W, Dai X, Zhao T, Yan W. Cartilage repair using mesenchymal stem cell (MSC) sheet and MSCs-loaded bilayer PLGA scaffold in a rabbit model. Knee Surg Sports Traumatol Arthrosc. 2014;22:1424–1433. doi: 10.1007/s00167-012-2256-3. [DOI] [PubMed] [Google Scholar]
- 30.Villemure I, Stokes IA. Growth plate mechanics and mechanobiology. A survey of present understanding. J Biomech. 2009;42:1793–1803. doi: 10.1016/j.jbiomech.2009.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lui JC, Nilsson O, Baron J. Recent research on the growth plate: Recent insights into the regulation of the growth plate. J Mol Endocrinol. 2014;53:T1–T9. doi: 10.1530/JME-14-0165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chagin AS, Kronenberg HM. Role of G-proteins in the differentiation of epiphyseal chondrocytes. J Mol Endocrinol. 2014;53:R39–R45. doi: 10.1530/JME-14-0093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Belluoccio D, Etich J, Rosenbaum S, Frie C, Grskovic I, Stermann J, Ehlen H, Vogel S, Zaucke F, von der Mark K, et al. Sorting of growth plate chondrocytes allows the isolation and characterization of cells of a defined differentiation status. J Bone Miner Res. 2010;25:1267–1281. doi: 10.1002/jbmr.30. [DOI] [PubMed] [Google Scholar]
- 34.Xu J, Wang W, Kapila Y, Lotz J, Kapila S. Multiple differentiation capacity of STRO-1+/CD146+ PDL mesenchymal progenitor cells. Stem Cells Dev. 2009;18:487–496. doi: 10.1089/scd.2008.0113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Su X, Zuo W, Wu Z, Chen J, Wu N, Ma P, Xia Z, Jiang C, Ye Z, Liu S, et al. CD146 as a new marker for an increased chondroprogenitor cell sub-population in the later stages of osteoarthritis. J Orthop Res. 2015;33:84–91. doi: 10.1002/jor.22731. [DOI] [PubMed] [Google Scholar]
- 36.Wang Z, Yan X. CD146, a multi-functional molecule beyond adhesion. Cancer Lett. 2013;330:150–162. doi: 10.1016/j.canlet.2012.11.049. [DOI] [PubMed] [Google Scholar]
- 37.Guezguez B, Vigneron P, Lamerant N, Kieda C, Jaffredo T, Dunon D. Dual role of melanoma cell adhesion molecule (MCAM)/CD146 in lymphocyte endothelium interaction: MCAM/CD146 promotes rolling via microvilli induction in lymphocyte and is an endothelial adhesion receptor. J Immunol. 2007;179:6673–6685. doi: 10.4049/jimmunol.179.10.6673. [DOI] [PubMed] [Google Scholar]
- 38.Jin HJ, Kwon JH, Kim M, Bae YK, Choi SJ, Oh W, Yang YS, Jeon HB. Downregulation of melanoma cell adhesion molecule (MCAM/CD146) accelerates cellular senescence in human umbilical cord blood-derived mesenchymal stem cells. Stem Cells Transl Med. 2016;5:427–439. doi: 10.5966/sctm.2015-0109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Qian YN, Luo YT, Duan HX, Feng LQ, Bi Q, Wang YJ, Yan XY. Adhesion molecule CD146 and its soluble form correlate well with carotid atherosclerosis and plaque instability. CNS Neurosci Ther. 2014;20:438–445. doi: 10.1111/cns.12234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bu P, Zhuang J, Feng J, Yang D, Shen X, Yan X. Visualization of CD146 dimerization and its regulation in living cells. Biochim Biophys Acta. 2007;1773:513–520. doi: 10.1016/j.bbamcr.2007.01.009. [DOI] [PubMed] [Google Scholar]
- 41.Sorrentino A, Ferracin M, Castelli G, Biffoni M, Tomaselli G, Baiocchi M, Fatica A, Negrini M, Peschle C, Valtieri M. Isolation and characterization of CD146+ multipotent mesenchymal stromal cells. Exp Hematol. 2008;36:1035–1046. doi: 10.1016/j.exphem.2008.03.004. [DOI] [PubMed] [Google Scholar]
- 42.Balakumaran A, Mishra PJ, Pawelczyk E, Yoshizawa S, Sworder BJ, Cherman N, Kuznetsov SA, Bianco P, Giri N, Savage SA, et al. Bone marrow skeletal stem/progenitor cell defects in dyskeratosis congenita and telomere biology disorders. Blood. 2015;125:793–802. doi: 10.1182/blood-2014-06-566810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dawson JI, Kanczler J, Tare R, Kassem M, Oreffo RO. Concise review: Bridging the gap: Bone regeneration using skeletal stem cell-based strategies-where are we now? Stem cells. 2014;32:35–44. doi: 10.1002/stem.1559. [DOI] [PubMed] [Google Scholar]
- 44.Agarwal S, Loder SJ, Sorkin M, Li S, Shrestha S, Zhao B, Mishina Y, James AW, Levi B. Analysis of bone-cartilage-stromal progenitor populations in trauma induced and genetic models of heterotopic ossification. Stem cells. 2016;34:1692–1701. doi: 10.1002/stem.2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Aslan H, Zilberman Y, Kandel L, Liebergall M, Oskouian RJ, Gazit D, Gazit Z. Osteogenic differentiation of noncultured immunoisolated bone marrow-derived CD105+ cells. Stem cells. 2006;24:1728–1737. doi: 10.1634/stemcells.2005-0546. [DOI] [PubMed] [Google Scholar]
- 46.Aicher WK, Bühring HJ, Hart M, Rolauffs B, Badke A, Klein G. Regeneration of cartilage and bone by defined subsets of mesenchymal stromal cells-potential and pitfalls. Adv Drug Deliv Rev. 2011;63:342–351. doi: 10.1016/j.addr.2010.12.004. [DOI] [PubMed] [Google Scholar]
- 47.Wang L, Shao YY, Ballock RT. Peroxisome proliferator-activated receptor-gamma promotes adipogenic changes in growth plate chondrocytes in vitro. PPAR Res. 2006;2006:67297. doi: 10.1155/PPAR/2006/67297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ferrer-Lorente R, Bejar MT, Tous M, Vilahur G, Badimon L. Systems biology approach to identify alterations in the stem cell reservoir of subcutaneous adipose tissue in a rat model of diabetes: Effects on differentiation potential and function. Diabetologia. 2014;57:246–256. doi: 10.1007/s00125-013-3081-z. [DOI] [PubMed] [Google Scholar]
- 49.Ma T, Liu H, Chen W, Xia X, Bai X, Liang L, Zhang Y, Liang T. Implanted adipose-derived stem cells attenuate small-for-size liver graft injury by secretion of VEGF in rats. Am J Transplant. 2012;12:620–629. doi: 10.1111/j.1600-6143.2011.03870.x. [DOI] [PubMed] [Google Scholar]
- 50.Erdman CP, Dosier CR, Olivares-Navarrete R, Baile C, Guldberg RE, Schwartz Z, Boyan BD. Effects of resveratrol on enrichment of adipose-derived stem cells and their differentiation to osteoblasts in two-and three-dimensional cultures. J Tissue Eng Regen Med. 2012;6(Suppl 3):S34–S46. doi: 10.1002/term.513. [DOI] [PubMed] [Google Scholar]
- 51.Ye Y, Du Y, Guo F, Gong C, Yang K, Qin L. Comparative study of the osteogenic differentiation capacity of human bone marrow- and human adipose-derived stem cells under cyclic tensile stretch using quantitative analysis. Int J Mol Med. 2012;30:1327–1334. doi: 10.3892/ijmm.2012.1123. [DOI] [PubMed] [Google Scholar]
- 52.Quarto N, Senarath-Yapa K, Renda A, Longaker MT. TWIST1 silencing enhances in vitro and in vivo osteogenic differentiation of human adipose-derived stem cells by triggering activation of BMP-ERK/FGF signaling and TAZ upregulation. Stem cells. 2015;33:833–847. doi: 10.1002/stem.1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hamid AA, Idrus RB, Saim AB, Sathappan S, Chua KH. Characterization of human adipose-derived stem cells and expression of chondrogenic genes during induction of cartilage differentiation. Clinics (Sao Paulo) 2012;67:99–106. doi: 10.6061/clinics/2012(02)03. [DOI] [PMC free article] [PubMed] [Google Scholar]