Abstract
Breast cancer is the most prevalent cancer and the leading cause of cancer-associated mortalities among women worldwide today. Accumulating evidence suggested that miR-372 may serve important roles in the initiation and development of various human cancers. However, the role of miR-372 in breast cancer remains unknown. The present study demonstrated that the expression level of miR-372 in human breast cancer tissues and cell lines is significantly reduced compared with normal breast tissues cell lines. Furthermore, results of functional assays indicated that miR-372 inhibits cell proliferation and induces apoptosis in the MCF-7 human breast cancer cell line. E2F1 was identified as a direct functional target of miR-372 in breast cancer. In conclusion, the findings revealed that miR-372 may have the potential to act as a novel molecule for the diagnosis and therapy of patients with breast cancer.
Keywords: miR-372, E2F1, breast cancer, proliferation, apoptosis
Introduction
Breast cancer remains the most prevalent cancer in females around the world today (1). In 2012, breast cancer was ranked first in cancer incidence and was the leading cause of cancer-associated mortalities among females worldwide, with an estimated 1.7 million newly diagnosed cases and 521,900 mortalities worldwide (2). Despite the advances made in breast cancer management, including earlier detection and more effective treatments strategies over the past few decades, ~5 to 10% of females have metastatic disease when first diagnosed with breast cancer, of which only one-fifth survive 5 years (3,4). Therefore, identifying novel molecular correlates with tumorigenesis and development of breast cancer may be helpful for its earlier diagnosis and treatment, so as to improve the prognosis of breast cancer patients.
MicroRNAs (miRNAs/miRs) are small, endogenous, noncoding RNAs that are ~22 nucleotides in length, and regulate gene expression by destabilizing target mRNAs as well as inhibiting their translation (5,6). Mechanistically, miRNAs function by directly binding to the 3′ untranslated region (UTR) of its target mRNA (7). An increasing number of studies have demonstrated that miRNAs may serve as oncogenes or tumor suppressors in human cancers, and aberrant expression of miRNAs has been observed in various human cancers, including lung (8), gastric (9) and prostate (10). Similarly, the role of miRNAs in the initiation and development of breast cancer has been prevalently investigated, including proliferation (11), apoptosis (12), migration and invasion (13). Therefore, various miRNAs may have the potential to serve as diagnostic markers, prognosis predictors and therapeutic targets in breast cancer patients.
It was reported that miR-372 belongs to the Mir-371-372 gene cluster, which is located on chromosome 19q13.42 (14). Accumulating evidence has suggested that miR-372 may regulate cell proliferation, apoptosis, migration and invasion in many types of human cancers, and it may serve oncogenic or suppressive roles in different human cancers. For example, Cho et al (15) demonstrated the oncogenic role of miR-372 in gastric cancer by controlling cell growth, the cell cycle and apoptosis through downregulation of a tumor suppressor gene, large tumor suppressor kinase 2. Chen et al (16) reported that miR-372 may function as an oncogenic miRNA by directly targeting the 3′-UTR of PH domain and leucine rich repeat protein phosphatase 2 in glioma. On the contrary, Huang et al (17) found that miRNA-372 was downregulated in renal cell carcinoma cell lines and tissue specimens, and its over-expression inhibited cell proliferation and invasion by suppressing IGF2BP1. Furthermore, Liu et al (18) demonstrated that miR-372 suppressed cell proliferation, migration, and invasion, and promoted the apoptosis of endometrial carcinoma cells through downregulating RhoC. However, the role of miR-372 in breast cancer remains unknown.
To the best of our knowledge, the present study was the first to investigate the expression level of miR-372 and its role in breast cancer. Firstly, miR-372 expression levels in human breast cancer tissues and cell lines were detected by reverse transcription-quantitative polymerase chain reaction (RT-qPCR). Next, in vitro assays were conducted to investigate the effect of miR-372 on breast cancer cell proliferation and apoptosis. Finally, E2F1 was identified as a direct target of miR-372 for its tumor suppressive role in breast cancer. These results demonstrated that miR-372 inhibits proliferation and induces apoptosis in breast cancer by directly targeting E2F1, and may serve as a therapeutic target for the treatment of breast cancer patients.
Materials and methods
Tissue specimens
A total of 20 paired clinical tissue specimens (tumor and adjacent non-tumor tissues) were collected from patients who were diagnosed with primary breast cancer and underwent surgery in the Department of General Surgery, The Second Affiliated Hospital and Yuying Children's Hospital of Wenzhou Medical University (Wenzhou, China) between May and October 2015. All tissue specimens were frozen in liquid nitrogen immediately and stored at −80°C until use. Both tumor and non-tumor tissues were confirmed histologically. No patients underwent radiation therapy or chemotherapy prior to surgery. Written informed consent was obtained from each participant and this study was approved by the Ethics Committees of The Second Affiliated Hospital and Yuying Children's Hospital of Wenzhou Medical University.
Cell lines
The BT-474, MCF-7, MDA-MB-436 and MDA-MB-231 human breast cancer cell lines were purchased from the Shanghai Institute of Cell Biology, Chinese Academy of Sciences (Shanghai, China). The MCF10A healthy breast cell line was purchased from American Type Culture Collection (Manassas, VA, USA). All breast cancer cell lines were cultured in Dulbecco's modified Eagle's medium (DMEM; Gibco; Thermo Fisher Scientific, Waltham, MA, USA) supplemented with 10% fetal bovine serum (FBS, Gibco; Thermo Fisher Scientific, Inc.), 100 µg/ml penicillin and 100 µg/ml streptomycin (Invitrogen; Thermo Fisher Scientific, Inc.). MCF-10A cells were cultured in DMEM/F12 (Gibco; Thermo Fisher Scientific, Inc.) supplemented with 5% horse serum (Gibco; Thermo Fisher Scientific, Inc.), 20 ng/ml EGF (Peprotech, Inc. Rocky Hill, NJ, USA), 0.5 mg/ml hydrocortisone (Stemcell Technologies, Inc., Vancouver, BC, Canada), 10 µg/ml insulin (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) and 100 ng/ml cholera toxin (Sigma-Aldrich; Merck MGaA). Cells were maintained at 37°C in a humidified atmosphere containing 5% CO2.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted from tissue samples or cell lines using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacturer's protocol and then reverse transcribed into cDNA using a reverse transcription kit (Promega Corporation, Madison, WI, USA). For E2F1 mRNA, qPCR was performed using a SYBR Green Master Mix (Takara Bio, Inc., Otsu, Japan) and GAPDH served as an internal control. The RT-qPCR conditions were 95°C for 3 min; 40 cycles of 95°C for 12 sec, and 62°C for 1 min. For miR-372, TaqMan assays (Applied Biosystems; Thermo Fisher Scientific, Inc.) were performed following the manufacturer's protocol and small nuclear U6 RNA served as an internal control. The relative expression levels were normalized to internal controls using the comparative 2−ΔΔCq method. (19) Primers for miR-372 were as follows: Forward, 5′-ACACTCCAGCTGGGAAAGTGCTGCGACATTT-3′ and reverse, 5′-GTGCAGGGTCCGAGGT-3′. Primers for E2F1 were as follows: Forward, 5′-CCCATCCCAGGAGGTCACTT-3′ and reverse, 5′-CTGCAGGCTCACTGCTCTC-3′. All experiments were conducted in triplicate.
Cell transfection
All transfections were performed using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacture's protocol. miR-372 mimics (miR-372), miR-372 inhibitors (anti-miR-383) and their negative controls (miR-NC and anti-miR-NC) were purchased from Guangzhou RiboBio Co., Ltd. (Guangzhou, China). Small interfering (si)RNA targeting human E2F1 mRNA (si-E2F1) and the scramble (si-NC) were designed by Shanghai GenePharma Co., Ltd. (Shanghai, China). At 48 h after transfection, cells were collected and RT-qPCR was conducted to confirm the transfection efficiency. All assays were conducted in triplicate.
MTT assay
Cell proliferation was analyzed by MTT assay according to the manufacturer's protocol. Briefly, cells were seeded into 96-well plates at a density of 1×104 cells per well and incubated for 0, 24, 48, 72 or 96 h. Following this, 100 µl MTT (5 mg/ml) was added to each well and incubated for 4 h. Next, 150 µl dimethyl sulfoxide was added to halt the reaction. A microplate was used to determine the absorbance at a wavelength of 490 nm. Each assay was performed in triplicate and repeated three times.
Flow cytometry
Cell apoptosis was analyzed by an Annexin V-fluorescein isothiocyanate (FITC)/propidium iodide (PI) apoptosis detection kit (Nanjing KeyGen Biotech, Co., Ltd., Nanjing, China) following the manufacturer's protocol. After transfection for 48 h, cells were harvested by 0.25% trypsin and centrifuged at 251 × g for 5 min at room temperature. Subsequently, cells were washed twice with cold PBS and resuspended in 250 µl binding buffer (Nanjing KeyGen Biotech, Co., Ltd.). FITC Annexin V FITC (5 µl) and 1 µl PI solution were added and cells were incubated for 15 min at room temperature. The apoptosis rate was detected using a Calibur Flow Cytometer (BD Biosciences, San Jose, CA, USA). Three independent experiments were repeated.
Luciferase report assay
TargetScan version 6.2 (http://www.targetscan.org) was used to investigate the potential targets of miR-372 and then it was confirmed by western blotting and a luciferase reporter assay. For the luciferase reporter assay, cells were seeded into 24-well plates 24 h prior to transfection. Cells were co-transfected with the E2F1 wild-type (Wt) or mutant (Mut) 3′UTR reporter vector and miR-372 mimics or NC using Lipofectamine 2000 according to the manufacturer's protocol. After transfection for 48 h, cells were lysed and luciferase activity against Renilla luciferase activity was measured using a Dual Luciferase Reporter Assay System (Promega Corporation) according to the manufacturer's protocol. Experiments were independently repeated three times.
Western blot analysis
Proteins were lysed from treated cells using radioimmunoprecipitation assay buffer containing protease inhibitor (Sigma-Aldrich; Merck KGaA) and collected by centrifugation (16,000 × g at 4°C) for 15 min. The protein concentration was determined using a bicinchoninic Acid Protein Assay kit (Sigma-Aldrich; Merck KGaA). Total protein (40 µg) was separated via 10% SDS-PAGE, transferred to a polyvinylidene difluoride membrane (GE Healthcare Life Sciences, Chalfont, UK) and blocked with 5% non-fat milk for 1 h at room temperature. The membranes were probed with primary antibodies against E2F1 (Abcam, Cambridge UK; cat. no. ab112580; 1:1,000) and β-actin (Santa Cruz Biotechnology, Inc., Dallas, TX, USA; cat. no. sc-130300; 1:2,000) overnight at 4°C, and then incubated with horseradish peroxidase conjugated secondary antibodies (Santa Cruz Biotechnology, Inc.; cat. no. goat anti-mouse sc-2005 and goat anti-rabbit sc-2004; 1:5,000) for 1 h at room temperature. Finally, signals were detected using an enhanced chemiluminescence system with the enhanced chemiluminescense kit (EMD Millipore, Billerica, MA, USA). Each reaction was performed in triplicate.
Statistical analysis
All data are presented as the mean ± standard deviation and were analyzed using SPSS version 17.0 (SPSS Inc., Chicago, IL, USA) and GraphPad Prism software (version 5.0; GraphPad Software, Inc., La Jolla, CA, USA). The difference between groups was analyzed by Student's t test when comparing only two groups and one-way analysis or post hoc test when comparing more than two groups. P<0.05 was considered to indicate a statistically significant difference.
Results
miR-372 is downregulated in breast cancer specimens and cell lines
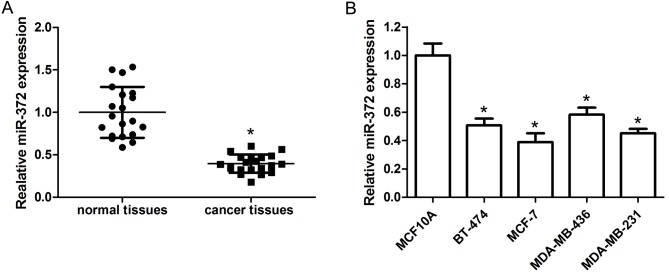
The expression level of miR-372 in human breast cancer tissues and adjacent non-tumor tissues was detected by RT-qPCR. The results demonstrated that miR-372 expression levels were obviously reduced in breast cancer tissue specimens compared with adjacent non-tumor tissues (Fig. 1A). The level of miR-372 in the BT-474, MCF-7, MDA-MB-436 and MDA-MB-231 human breast cancer cell lines and the MCF10A normal breast cell line was further examined. The data indicated that miR-372 was downregulated in breast cancer cell lines compared with MCF10A cells (Fig. 1B).
Figure 1.
miR-372 is downregulated in breast cancer specimens and cell lines. (A) Relative miR-372 expression in breast cancer tissues and adjacent non-tumor tissues, as detected by RT-qPCR. (B) Relative miR-372 expression in the BT-474, MCF-7, MDA-MB-436 and MDA-MB-231 human breast cancer cell lines and the MCF10A normal breast cell line, as detected by RT-qPCR. Data are presented as the mean ± standard deviation of three independent experiments. *P<0.05. RT-qPCR, reverse transcription-quantitative polymerase chain reaction; miR-372, microRNA-372.
miR-372 inhibits cell proliferation and induces apoptosis in MCF-7 cells
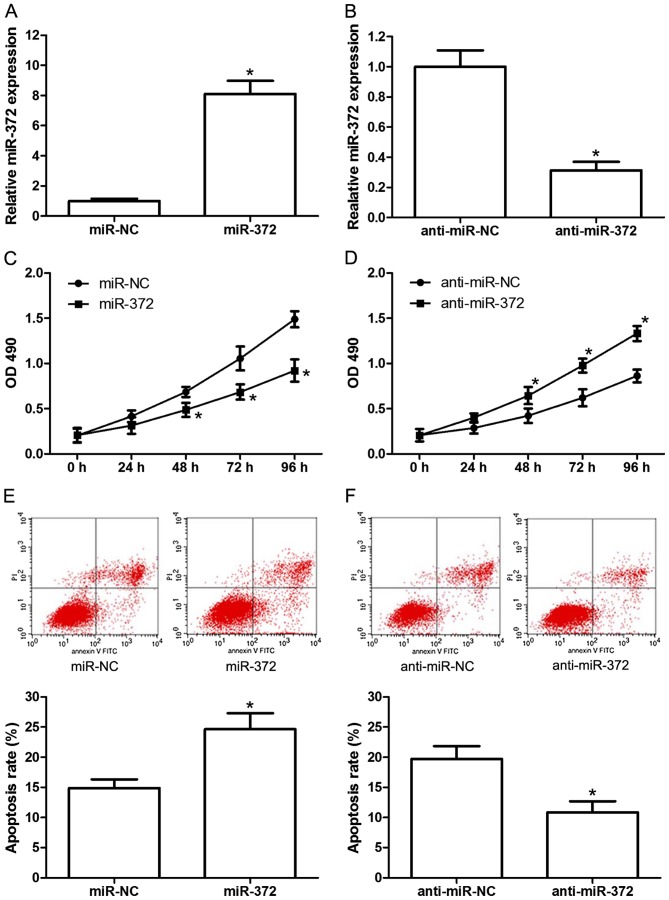
To investigate the effects of miR-372 on the proliferation and apoptosis of breast cancer cells, miR-372 mimics (miR-372), miR-372 inhibitors (anti-miR-372) or their negative controls (miR-NC and anti-miR-NC) were transfected into the MCF-7 human breast cancer cell line. The transfection efficiency was confirmed by RT-qPCR (Fig. 2A and B). Subsequently, cell proliferation and apoptosis were analyzed by MTT assay and flow cytometry, respectively. The results demonstrated that miR-372 mimics significantly inhibited MCF-7 cell proliferation (Fig. 2C), and miR-372 inhibitors significantly increased proliferation (Fig. 2D), compared with the negative control. Additionally, miR-372 mimics significantly induced apoptosis (Fig. 2E) and miR-372 inhibitors markedly reduced apoptosis (Fig. 2F) compared with the control group. These results revealed the tumor suppressive role of miR-372 in human breast cancer development.
Figure 2.
miR-372 inhibits proliferation and induces apoptosis in MCF-7 cells. Relative miR-372 levels in MCF-7 cells when transfected with (A) miR-372 mimics (miR-372) or (B) miR-372 inhibitors (anti-miR-372), as detected by reverse transcription-quantitative polymerase chain reaction. Proliferation of MCF-7 cells transfected with miR-372 (C) inhibitors and (D) mimics, as measured by MTT assay. Apoptosis rate of transfected MCF-7 cells transfected with miR-372 (E) inhibitors and (F) mimics, as analyzed by flow cytometry. Data are presented as the mean ± standard deviation of three independent experiments. *P<0.05. miR-372, microRNA-372; OD, optical density.
E2F1 is a direct target of miR-372 in breast cancer
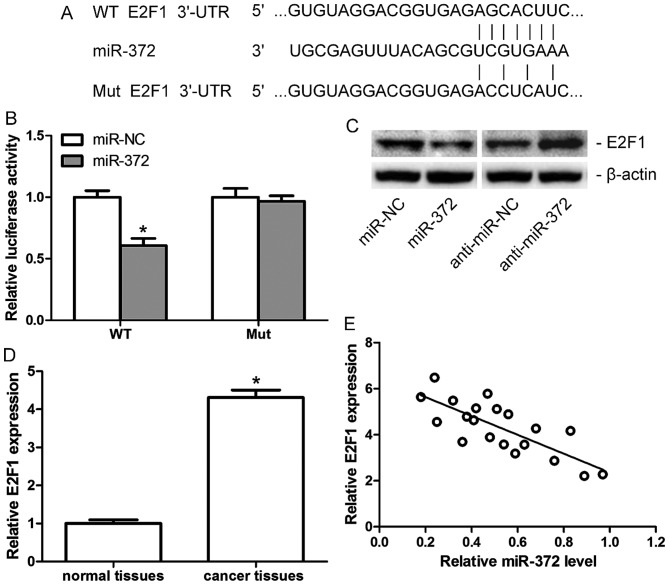
TargetScan 6.2 was used to investigate the functional target of miR-372 in breast cancer, and E2F1 was identified as a candidate target (Fig. 3A). A luciferase reporter assay and western blotting were performed for confirmation. The luciferase reporter assay demonstrated a significant downregulation of luciferase activity in MCF-7 cells when co-transfected with miR-372 mimics (miR-372) and Wt, but not the Mut 3′-UTR of E2F1 (Fig. 3B). The results of western blotting indicated that protein expression levels of E2F1 in MCF-7 cells decreased following transfection with miR-372 mimics (miR-372), but increased when transfected with anti-miR-372 (Fig. 3C). Furthermore, the mRNA expression levels of E2F1 in human breast cancer tissues and adjacent non-tumor tissues were assessed. E2F1 was demonstrated to be upregulated in human breast cancer tissues compared with adjacent normal tissues (Fig. 3D), and its level was negatively correlated with miR-372 levels in breast cancer tissues (Fig. 3E). All the data indicated that E2F1 is a direct target of miR-372 in breast cancer.
Figure 3.
E2F1 is a direct target of miR-372 in breast cancer. (A) The sequences of miR-372 against WT or Mut 3′-UTR of E2F1, as predicted by Targetscan. (B) Relative luciferase activity of MCF-7 cells co-transfected with miR-372 mimics or miR-NC and WT or Mut 3′-UTR of E2F1. (C) E2F1 protein expression levels in MCF-7 cells transfected with miR-372, anti-miR-372 or miR-NC and anti-miR-NC, as assessed by western blotting. (D) Relative mRNA expression levels of E2F1 in breast cancer tissues and adjacent non-tumor tissues, as detected by reverse transcription-quantitative polymerase chain reaction. (E) Correlation between E2F1 expression and miR-372 level in breast cancer tissues. Data are presented as the mean ± standard deviation of three independent experiments. *P<0.05. miR-372, microRNA-372; NC, negative control; UTR, untranslated region; WT, wild-type; Mut, mutant; E2F1, transcription factor E2F1.
Knockdown of E2F1 inhibits proliferation and induces apoptosis in MCF-7 cells
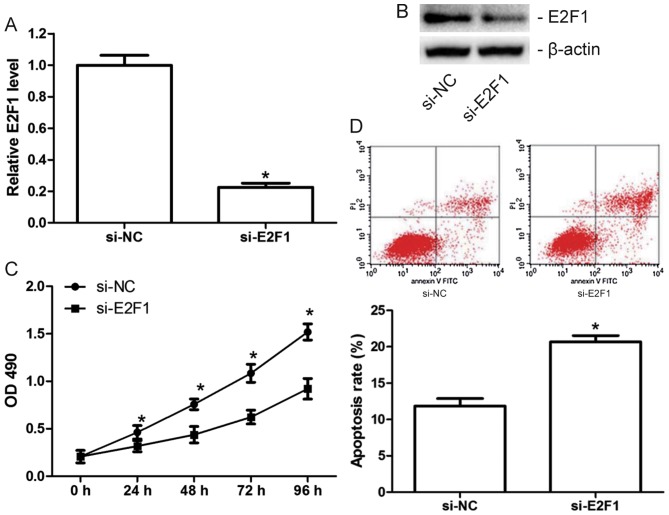
As miR-372 inhibits proliferation and induces apoptosis in MCF-7 cells, and E2F1 is a direct target of miR-372, MCF-7 cells were transfected with si-E2F1 or the scramble (si-NC) to examine the effect of E2F1 on MCF-7 cell proliferation and apoptosis. The transfection efficiency was confirmed by RT-qPCR (Fig. 4A) and western blotting (Fig. 4B). Results of functional experiments demonstrated that knockdown of E2F1 significantly inhibited proliferation (Fig. 4C) and induced apoptosis in MCF-7 cells (Fig. 4D), which suggested that the tumor suppressive role of miR-372 in breast cancer is involved in its suppression of E2F1.
Figure 4.
Knockdown of E2F1 inhibits proliferation and induces apoptosis in MCF-7 cells. Relative miR-372 (A) mRNA and (B) protein expression levels in MCF-7 cells transfected with si-E2F1 or si-NC, as detected by reverse transcription-quantitative polymerase chain reaction and western blotting, respectively. (C) Proliferation of transfected MCF-7 cells measured by MTT assay. (D) Apoptosis of transfected MCF-7 cells, as analyzed by flow cytometry. Data are presented as the mean ± standard deviation of three independent experiments. *P<0.05. si, small interfering; miR-372, microRNA-372; NC, negative control; OD, optical density; E2F1, transcription factor E2F1.
Discussion
Over the past few decades, the detection non-malignant lesions of breast cancer has increased, including atypical ductal hyperplasia, atypical lobular hyperplasia, ductal carcinoma in situ and lobular carcinoma in situ (20). Its early detection is due to an increase in the identification of molecular biomarkers. Apart from established biomarkers, including estrogen receptor, progesterone receptor, receptor tyrosine-protein kinase erbB-2 and Ki67, certain other novel molecular such as bcyclin E, B-Myb, twist, and cyclin D-binding Myb-like Protein 1β have also been identified as novel biomarkers for breast cancer early diagnosis and prognosis prediction (21). A recently published review indicated that miRNAs may serve important roles in initiation, development, metastasis and drug resistance of breast cancer (22). Furthermore, miRNAs are hypothesized to have prognostic, diagnostic and therapeutic potential in breast cancer (23). Thus, identifying novel miRNAs involved in the initiation and development of breast cancer may be helpful for breast cancer patients.
E2F is a family of transcription factors that regulate the expression of genes essential for a wide range of cellular functions, including cell cycle progression, DNA repair, DNA replication, differentiation, proliferation and apoptosis (24). The E2F family is composed of at least eight members that can be divided into distinct subgroups based on their structural and functional similarities (25). E2F1, the most classic member of the E2F family, exhibits dual properties in tumor development by acting as a tumor suppressor or oncogene (26). Accumulating evidence has suggested that E2F1 is regulated by various microRNA in tumor development. For example, Yang et al (27) demonstrated that miR-106a suppresses proliferation and induces apoptosis in human glioma cells, and identified E2F1 as a direct functional target of miR-106a. Dar et al (28) revealed that miR-205 inhibits melanoma cell proliferation and induces apoptosis via regulation of E2F1 protein level. Xiao et al (29) also reported that miR-329 could markedly suppress cell proliferation and colony formation by targeting E2F1 (29).
The present study demonstrated that miR-372 is significantly down-regulated in human breast cancer tissues and cell lines compared with healthy controls, which suggested that miR-372 may serve as a tumor suppressor in breast cancer development. Subsequetly, MCF-7 cells were transfected with miR-372 mimics or inhibitors to investigate its role in breast cancer. The results indicated that miR-372 could inhibit proliferation and induce apoptosis in breast cancer cells. Furthermore, E2F1 was identified as a direct target of miR-372 in the MCF-7 breast cancer cell line, and the mRNA level of E2F1 was negatively correlated with miR-372 levels in breast cancer tissues. Additionally, it was further demonstrated that knockdown of E2F1 inhibited proliferation and induced apoptosis in MCF-7 cells.
In conclusion, to the best of our knowledge, the present study investigated the expression level and roles of miR-372 in breast cancer for the first time. miR-372 was demonstrated to be significantly down-regulated in breast cancer tissues and cell lines compared with normal controls. Furthermore, the results indicated that miR-372 may inhibit proliferation and induce apoptosis in breast cancer cell lines by directly targeting E2F1. These findings suggested that miR-372 may have the potential to serve as a novel molecule for the diagnosis and therapy of patients with breast cancer.
Acknowledgements
The present study was supported by the Foundation of Science and Technology Bureau of Wenzhou (grant nos. Y20100257 and Y20160403), the Foundation of Traditional Chinese Medicine of Zhejiang Province (grant no. 2009C520014) and the Foundation of Public Welfare Technology Application of Zhejiang Province (grant no. 2015C33100).
References
- 1.Cai Y, Yan X, Zhang G, Zhao W, Jiao S. MicroRNA-205 increases the sensitivity of docetaxel in breast cancer. Oncol Lett. 2016;11:1105–1109. doi: 10.3892/ol.2015.4030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 3.Xiao X, Huang X, Ye F, Chen B, Song C, Wen J, Zhang Z, Zheng G, Tang H, Xie X. The miR-34a-LDHA axis regulates glucose metabolism and tumor growth in breast cancer. Sci Rep. 2016;6:21735. doi: 10.1038/srep21735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cardoso F, Harbeck N, Fallowfield L, Kyriakides S, Senkus E, ESMO Guidelines Working Group Locally recurrent or metastatic breast cancer: ESMO clinical practice guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2012;23(Suppl 7):vii11–19. doi: 10.1093/annonc/mds232. [DOI] [PubMed] [Google Scholar]
- 5.Garzon R, Calin GA, Croce CM. MicroRNAs in Cancer. Annu Rev Med. 2009;60:167–179. doi: 10.1146/annurev.med.59.053006.104707. [DOI] [PubMed] [Google Scholar]
- 6.Wang Y, Blelloch R. Cell cycle regulation by MicroRNAs in embryonic stem cells. Cancer Res. 2009;69:4093–4096. doi: 10.1158/0008-5472.CAN-09-0309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van Kouwenhove M, Kedde M, Agami R. MicroRNA regulation by RNA-binding proteins and its implications for cancer. Nat Rev Cancer. 2011;11:644–656. doi: 10.1038/nrc3107. [DOI] [PubMed] [Google Scholar]
- 8.Zhang T, Hu Y, Ju J, Hou L, Li Z, Xiao D, Li Y, Yao J, Wang C, Zhang Y, et al. Downregulation of miR-522 suppresses proliferation and metastasis of non-small cell lung cancer cells by directly targeting DENN/MADD domain containing 2D. Sci Rep. 2016;6:19346. doi: 10.1038/srep19346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhao Y, Lu G, Ke X, Lu X, Wang X, Li H, Ren M, He S. miR-488 acts as a tumor suppressor gene in gastric cancer. Tumour Biol. 2016;37:8691–8698. doi: 10.1007/s13277-015-4645-y. [DOI] [PubMed] [Google Scholar]
- 10.Wang L, Song G, Tan W, Qi M, Zhang L, Chan J, Yu J, Han J, Han B. MiR-573 inhibits prostate cancer metastasis by regulating epithelial-mesenchymal transition. Oncotarget. 2015;6:35978–35990. doi: 10.18632/oncotarget.5427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Song C, Liu LZ, Pei XQ, Liu X, Yang L, Ye F, Xie X, Chen J, Tang H, Xie X. miR-200c inhibits breast cancer proliferation by targeting KRAS. Oncotarget. 2015;6:34968–34978. doi: 10.18632/oncotarget.5198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang L, Li L, Guo R, Li X, Lu Y, Guan X, Gitau SC, Wang L, Xu C, Yang B, Shan H. miR-101 promotes breast cancer cell apoptosis by targeting Janus kinase 2. Cell Physiol Biochem. 2014;34:413–422. doi: 10.1159/000363010. [DOI] [PubMed] [Google Scholar]
- 13.Li G, Yao L, Zhang J, Li X, Dang S, Zeng K, Zhou Y, Gao F. Tumor-suppressive microRNA-34a inhibits breast cancer cell migration and invasion via targeting oncogenic TPD52. Tumour Biol. 2016;37:7481–7491. doi: 10.1007/s13277-015-4623-4. [DOI] [PubMed] [Google Scholar]
- 14.Voorhoeve PM, le Sage C, Schrier M, Gillis AJ, Stoop H, Nagel R, Liu YP, van Duijse J, Drost J, Griekspoor A, et al. A genetic screen implicates miRNA-372 and miRNA-373 as oncogenes in testicular germ cell tumors. Cell. 2006;124:1169–1181. doi: 10.1016/j.cell.2006.02.037. [DOI] [PubMed] [Google Scholar]
- 15.Cho WJ, Shin JM, Kim JS, Lee MR, Hong KS, Lee JH, Koo KH, Park JW, Kim KS. miR-372 regulates cell cycle and apoptosis of ags human gastric cancer cell line through direct regulation of LATS2. Mol Cells. 2009;28:521–527. doi: 10.1007/s10059-009-0158-0. [DOI] [PubMed] [Google Scholar]
- 16.Chen X, Hao B, Han G, Liu Y, Dai D, Li Y, Wu X, Zhou X, Yue Z, Wang L, et al. miR-372 regulates glioma cell proliferation and invasion by directly targeting PHLPP2. J Cell Biochem. 2015;116:225–232. doi: 10.1002/jcb.24949. [DOI] [PubMed] [Google Scholar]
- 17.Huang X, Huang M, Kong L, Li Y. miR-372 suppresses tumour proliferation and invasion by targeting IGF2BP1 in renal cell carcinoma. Cell Prolif. 2015;48:593–599. doi: 10.1111/cpr.12207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu BL, Sun KX, Zong ZH, Chen S, Zhao Y. MicroRNA-372 inhibits endometrial carcinoma development by targeting the expression of the Ras homolog gene family member C (RhoC) Oncotarget. 2016;7:6649–6664. doi: 10.18632/oncotarget.6544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2 (-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 20.Fu SW, Lee W, Coffey C, Lean A, Wu X, Tan X, Man YG, Brem RF. miRNAs as potential biomarkers in early breast cancer detection following mammography. Cell Biosci. 2016;6:6. doi: 10.1186/s13578-016-0071-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Inoue K, Fry EA. Novel molecular markers for breast cancer. Biomark Cancer. 2016;8:25–42. doi: 10.4137/BIC.S38394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Takahashi RU, Miyazaki H, Ochiya T. The Roles of microRNAs in breast cancer. Cancers (Basel) 2015;7:598–616. doi: 10.3390/cancers7020598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mulrane L, Klinger R, McGee SF, Gallagher WM, O'Connor DP. microRNAs: A new class of breast cancer biomarkers. Expert Rev Mol Diagn. 2014;14:347–363. doi: 10.1586/14737159.2014.901153. [DOI] [PubMed] [Google Scholar]
- 24.Zhan L, Zhang Y, Wang W, Song E, Fan Y, Wei B. E2F1: A promising regulator in ovarian carcinoma. Tumour Biol. 2016;37:2823–2831. doi: 10.1007/s13277-015-4770-7. [DOI] [PubMed] [Google Scholar]
- 25.DeGregori J, Johnson DG. Distinct and overlapping roles for E2F family members in transcription, proliferation and apoptosis. Curr Mol Med. 2006;6:739–748. doi: 10.2174/156652406778773484. [DOI] [PubMed] [Google Scholar]
- 26.Knoll S, Emmrich S, Pützer BM. The E2F1-miRNA cancer progression network. Adv Exp Med Biol. 2013;774:135–147. doi: 10.1007/978-94-007-5590-1_8. [DOI] [PubMed] [Google Scholar]
- 27.Yang G, Zhang R, Chen X, Mu Y, Ai J, Shi C, Liu Y, Shi C, Sun L, Rainov NG, et al. MiR-106a inhibits glioma cell growth by targeting E2F1 independent of p53 status. J Mol Med (Berl) 2011;89:1037–1050. doi: 10.1007/s00109-011-0775-x. [DOI] [PubMed] [Google Scholar]
- 28.Dar AA, Majid S, de Semir D, Nosrati M, Bezrookove V, Kashani-Sabet M. miRNA-205 suppresses melanoma cell proliferation and induces senescence via regulation of E2F1 protein. J Biol Chem. 2011;286:16606–16614. doi: 10.1074/jbc.M111.227611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xiao B, Tan L, He B, Liu Z, Xu R. MiRNA-329 targeting E2F1 inhibits cell proliferation in glioma cells. J Transl Med. 2013;11:172. doi: 10.1186/1479-5876-11-172. [DOI] [PMC free article] [PubMed] [Google Scholar]