Abstract
The interaction between airway epithelial progenitor cells and their microenvironment is critical for maintaining lung homeostasis. This microenvironment includes fibroblast cells, which support the growth of airway progenitor cells. However, the mechanism of this support is not fully understood. In the present study, the authors observed that inhibition of transforming growth factor (TGF)-β signal with SB431542 promotes the expression of hepatocyte growth factor (HGF) in fibroblast cells. The HGF receptor, c-Met, is expressed on airway progenitor cells; HGF promotes the colony-forming ability of airway progenitor cells. The deletion of Smad4 in airway progenitor cells increases the colony-forming ability, suggesting that Smad4 plays a negative role in the regulating the proliferation of airway progenitor cells. These data demonstrated that the regulation of airway progenitor cells by TGF-β depends on TGF-βR1/2 on stromal cells, rather than on epithelial progenitor cells. These data suggested a role for the TGF-β-TGF-βR1/2-HGF-Smad4 axis in airway epithelial homeostasis and sheds new light on the interaction between airway progenitor cells and their microenvironment.
Keywords: airway epithelial progenitor cells, TGF-β, HGF, Smad4
Introduction
The airway epithelium is pivotal to host defense against foreign microbial pathogens. There is increasing evidence that epithelial alterations are associated with multiple airway diseases, including asthma, chronic obstructive pulmonary diseases (COPD), obliterative bronchiolitis and cystic fibrosis (1–5). Following injury to the epithelium, airway progenitor cells are a crucial part of the repair process (6). These progenitor cells interact with their microenvironment to determine their proliferation, differentiation and capacity for self-renewal. The growth of these progenitor cells is reported to be supported by the production of growth factors by stromal cells (7). Additionally, the research of Lee et al (8) suggests that endothelial cells direct the specification and differentiation of airway progenitor cells. Furthermore, parabronchial smooth muscle cells activate airway epithelial progenitor cells to undergo a post-injury epithelial to mesenchymal transition (9). Vimentin-positive lung fibroblasts can create a niche for airway progenitor cells (10). Airway progenitor cells cross talk with their microenvironmental elements through direct contact or autocrine and/or paracrine signals. These include Wnt-β-catenin signaling, BMP signaling, Notch signaling and TGF-β signaling (7,11–13).
The TGF-β signal is overactivated in response to pulmonary fibrosis (14,15). Moreover, TGF-β has been reported to be upregulated in COPD and allergic asthma (16). Inhibition of TGF-β signaling with SB431542 promotes the proliferation of airway epithelial progenitor cells (7). TGF-β signal in fibroblasts can act FGF10 to regulate epithelial stem cell growth (7). It was shown that TGF-β inhibits HGF expression in mesenchymal cells through a TGF-β inhibitory element (17). HGF acts as a ligand with its receptor tyrosine kinase, c-Met to fulfill its function (18). HGF modulates the function of Smad4 through activating the Ras/MAPK pathway (19). However, the role of Smad4 in the regulation of airway progenitor cells has not been addressed.
In the present study, the authors adopted an in vitro epithelial-fibroblast co-culture assay developed previously (20). It was observed that TGF-β inhibits the proliferation of airway progenitor cells through the TGF-β receptors on fibroblasts. Inhibition of the TGF-β/TGFR2 pathway alters the secretory properties of fibroblasts, including production of HGF. Deletion of Smad4 resulted in an increase in the colony forming ability of airway progenitor cells.
Materials and methods
Ethics statement
Experimental mice were maintained under pathogen-free conditions in Tianjin Haihe Hospital's animal facility, and the permit number is SYXK (Jin) 2016-0002. Adult mice between the ages of 2–4 months old were sacrificed for experiments according to protocols approved by the Haihe Hospital Animal Care and Use Committee (Tianjin, China). All surgery was performed under 1% sodium pentobarbital 50 mg/kg intraperitoneal injection anesthesia, and all efforts were made to minimize suffering.
Mice
β-actin-GFP, Sftpc-Cre, TGFβR2f/f, and Smad4f/f mice were donated by Stripp B.R. (Cedars-Sinai Medical Center, Los Angeles, CA, USA). The authors crossed Sftpc-Cre mice with TGFβR2f/f mice to generate Sftpc-Cre; TGFβR2f/f mice. Additionally, Sftpc-Cre mice and Smad4f/f mice were crossed to generate Sftpc-Cre; Smad4f/f mice.
Fractionation of airway epithelial progenitor cells
Lung cell suspensions were prepared using an elastase digestion and stained for fluorescence-activated cell sorting (FACS), as previously described (20). Briefly, cells were resuspended in Hanks' balanced saline solution buffer supplemented with 2% fetal bovine serum, 0.1 mM EDTA, 10 mM HEPES, 100 IU/ml penicillin and 100 µg/ml streptomycin (HBSS+). Cells were then stained with the primary antibodies on ice for 45 min. The following antibodies were employed: EpCAM-PE-Cy7 (25-5791-80, 1:100), CD31-Biotin (13-0311-81, 1:40), CD34-Biotin (13-0341-81, 1:10), CD45-Biotin (13-0451-81, 1:100), Sca-1-APC (17-5981-81, 1:100), and CD24-PE (12-0242-81, 1:20) (all from eBioscience, Inc., San Diego, CA, USA). Cells were subsequently stained with the secondary antibody on ice for 40 min using streptavidin-APC-Cy7 (47-4317-82, 1:100; eBioscience, Inc.). Dead cells were identified using 7-aminoactinomycin D staining (BD Biosciences, Franlkin Lakes, NJ, USA).
Cell cultures and treatment
MLg cells (CCL-206; American Type Culture Collection, Manassas, VA, USA) were cultured in Dulbecco's modified Eagle's medium (DMEM; Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA). For the experiments, 6-well culture plates (Thermo Fisher Scientific, Inc.) were seeded with MLg cells at 5,000 cells/well and maintained at 37°C in a humidified atmosphere with 5% CO2. MLg cells were then exposed to 10 µM SB431542 (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) for 48 h. Cultures were visualized under an OLYMPUS IX73 (Olympus Corporation, Tokyo, Japan) inverted microscope before being harvested for reverse transcription-quantitative polymerase chain reaction (RT-qPCR) analysis.
Matrigel cultures of airway progenitor cells
Mouse airway progenitor cells were co-cultured with MLg cells in Matrigel, as described previously (20). In brief, sorted airway progenitor cells were mixed with MLg cells in growth factor-reduced Matrigel (BD Biosciences) and basic medium (BM) at a 1:1 ratio. The basic medium consisted of DMEM/F12 medium (Cellgro, Manassas, VA, USA), 10% FBS (Invitrogen; Thermo Fisher Scientific, Inc.), Insulin-Transferrin-Selenium supplement (Sigma-Aldrich; Merck KGaA), 100 IU/ml penicillin and 100 µg/ml streptomycin. The cell mixture was placed in 24-well Transwell filter inserts (BD Biosciences) in a 24-well flat-bottom culture plate containing culture medium (BM + SB431542). Cultures were incubated at 37°C in a humidified atmosphere with 5% CO2, and medium was replaced every other day. Colony-forming efficiency (CFE) was determined by counting the number of colonies with a diameter of ≥100 µm in each culture and representing this number as a percentage of seeded progenitor cells.
Total RNA isolation and RT-qPCR
Total RNA was extracted from MLg cells with TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.) following the manufacturer's instructions. RNA concentration was assessed and cDNA was synthesized using SuperScript III reagents (Invitrogen; Thermo Fisher Scientific, Inc.) with Oligo-dT and random hexamer priming (Takara Bio, Inc., Otsu, Japan). RT-qPCR was performed using SYBR Green SuperMix (Applied Biosystems; Thermo Fisher Scientific, Inc.) with the Light Cycler 96 Real-Time PCR system (Roche Diagnostics, Indianapolis, IN, USA) under the following reaction conditions: Initial heating cycle of 95°C for 2 min; and 40 cycles of denaturation at 95°C for 25 sec, primer annealing at 60°C for 25 sec and extension at 72°C for 20 sec. Melting curves were used to clarify the identity of amplicons, and the housekeeping gene, β-actin, served as an internal control. The relative mRNA expression levels of targeted genes were calculated using the comparative threshold cycle (CT) method (21) normalized to β-actin mRNA in the same sample. Primers were designed as follows: β-actin forward, 5′-GGCCAACCGTGAAAAGATGA-3′ and reverse, 5′-CAGCCTGGATGGCTACGTACA-3′; Hgf forward, 5′-CCTGGTGTTTCACAAGCAATC-3′ and reverse, 5′-CATGGGACCTCTGTAGCTTTC-3′. β-actin was used as a housekeeping gene.
Microarray analysis
EpCAM+ lung epithelial cells or EpCAM+Sca-1+ airway progenitor cells were sorted and pooled for total RNA extraction using RNeasy Mini kit (Qiagen Sciences, Inc., Gaithersburg, MD, USA). RNA (0.1 µg) was used for Affymetrix microarray analysis using mouse genome 430 2.0 arrays (Affymetrix, Inc., Santa Clara, CA, USA). Data were annotated with Affymetrix Expression Console software (Affymetrix, Inc.). Pathway analysis was performed by online Gather KEGG analysis (Kyoto Encyclopedia of Genes and Genomes; http://gather.genome.duke.edu/). A Bayes factor was also included in the consideration of KEGG pathway association.
Statistical analysis
The data were analyzed using SPSS software (version 13.0; SPSS, Inc., Chicago, IL, USA). Data are expressed as the mean ± standard error of the mean. The significance of the results was assessed using Student's t test between paired groups. P<0.05 was considered to indicate a statistically significant difference.
Results
TGF-β regulates mouse airway progenitor cells through its receptors on MLg cells
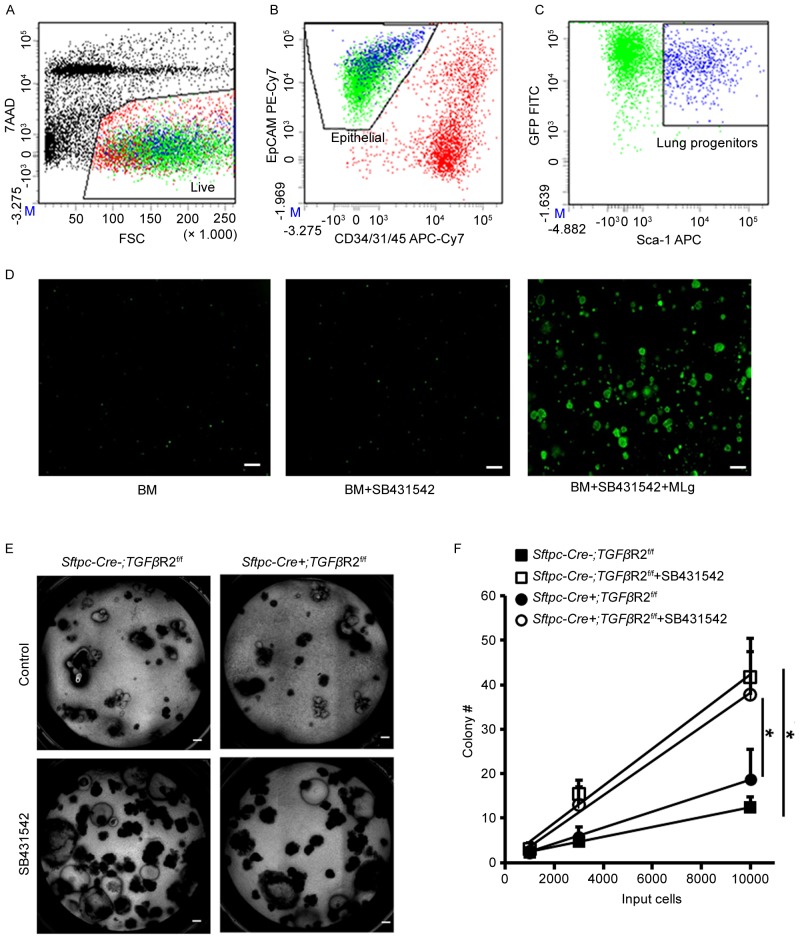
To investigate the role of fibroblasts in the regulation of mouse airway progenitor cells by TGF-β, the authors fractionated mouse airway progenitor cells from β-actin-GFP mice using a FACS-based strategy. Lung cells from β-actin-GFP mice were isolated by elastase digestion. The cells were stained with fluorescent antibodies and a viability dye. Dead cells were detected by 7-AAD staining (Fig. 1A). Endothelial, stromal and hematopoietic cells were excluded by surface staining for CD31, CD34 and CD45 (Fig. 1B). Airway epithelial progenitor cells, also positive for GFP, were further enriched by surface EpCAM and Sca-1 staining (Fig. 1B and C). A significant number of colonies were formed in presence of both SB431542 and MLg cells compared to stromal-free and SB431542 alone cultures (Fig. 1D), which was consistent with the authors' previous findings (20). The authors further examined the TGF-β role in the regulation of airway progenitor cells using Sftpc-Cre+; TGFβR2f/f mice, in which TGFβR2 is absent in their airway progenitor cells. In vitro cultures of airway progenitor cells in presence of MLg cells indicated that the colony-forming ability was comparable between Sftpc-Cre−; TGFβR2f/f and Sftpc-Cre+; TGFβR2f/f (Fig. 1E and F). SB431542 enhanced the colony-forming ability of airway progenitor cells in both Sftpc-Cre−; TGFβR2f/f and Sftpc-Cre+; TGFβR2f/f (Fig. 1E and F). These data suggested that TGF-β exerts its regulatory role in the colony-forming ability of airway progenitor cells by acting on MLg cells, rather than on airway progenitor cells.
Figure 1.
TGF-β regulates mouse airway progenitor cells through its receptors on MLg cells. (A) Exclusion of 7AADpos cells (y-axis). (B) CD34/31/45(Lin) (x-axis) vs. EpCAM (y-axis) analysis of 7AADneg population from A. (C) Sca-1 (x-axis) vs. GFP (y-axis) analysis of Linneg/EpCAMpos population from B represents airway progenitor cells. (D) Sorted progenitor cells were cultured in basic medium, with an addition of 10 µM SB431542, 10 µM SB431542 and MLg cells at day 6. (E) Sorted progenitor cells from Sftpc-Cre−; TGF-β2Rf/f and Sftpc-Cre+; TGF-β2Rf/f mice were cultured under control conditions and with an addition of 10 µM SB431542 for 4 weeks. (F) Quantitative analysis of the colony-forming efficiency in different culture conditions. Data are expressed as the mean ± standard error of the mean. *P<0.05 as indicated. Scale bars, 200 µm. TGF-β, transforming growth factor-β; FSC, forward scatter; GFP, green fluorescent protein; 7-AAD, 7-amino-actinomycin D.
Inhibition of TGF-β signaling promotes the secretion of HGF in fibroblasts
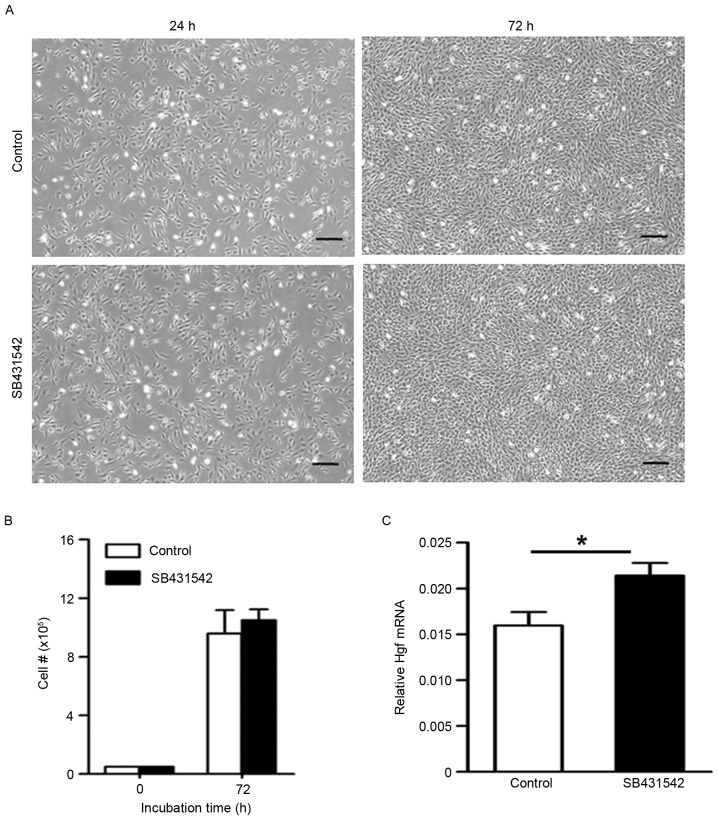
Fibroblasts have been shown to promote airway epithelial progenitor cells by producing growth factors. HGF expression was measured in the absence or presence of SB431542. Within 48 h, the number of MLg cells increased by approximately tenfold in the control group (Fig. 2A). There was no difference in the number of MLg cells between control and SB431542 treatment (Fig. 2B). Additionally, the morphology of MLg cells did not differ between the control and SB431542 treatment (Fig. 2A). Using RT-qPCR, the authors observed that the mRNA expression of Hgf in the SB431542 treatment was higher than that in the control (Fig. 2C). These data suggested that activation of the TGF-β signal pathway inhibits the production of growth factor HGF in fibroblasts.
Figure 2.
Inhibition of transforming growth factor-β signaling promotes the secretion of HGF in fibroblasts. (A) MLg cells were cultured in control condition or with 10 µM SB431542 for 48 h. (B) Quantitative analysis of the number of MLg cells. (C) Reverse transcription-quantitative polymerase chain reaction analysis was performed to quantify Hgf gene expression. Scale bars, 200 µm. Data are expressed as the mean ± standard error of the mean. *P<0.05 as indicated. HGF, hepatocyte growth factor.
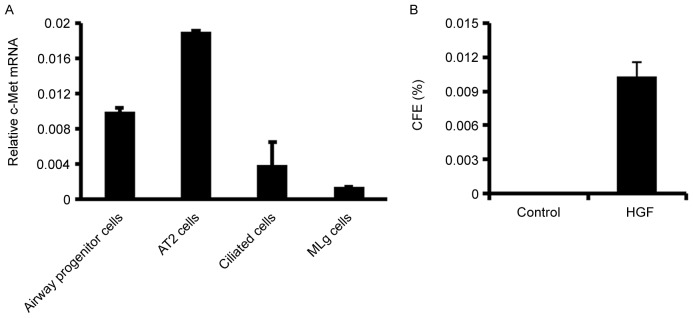
Stromal-derived HGF exerts its function through its specific receptor c-Met. Following this, the c-Met expression was determined in airway progenitor cells, alveolar type 2 (AT2) cells, ciliated cells, and MLg cells using quantitative RT-PCR (Fig. 3A). Mouse AT2 cells and ciliated cells were fractionated as previously (20). c-Met expression was indicated to be more abundant in lung epithelial cells than in stromal cells (Fig. 3A). AT2 cells express more c-Met than airway progenitor cells and ciliated cells in mouse lung (Fig. 3A). In vitro 3-D Matrigel culture indicated that HGF promotes the growth of airway progenitor cells in absence of MLg cells (Fig. 3B), suggesting that HGF acts directly on airway progenitor cells to promote their growth.
Figure 3.
HGF/c-Met regulates airway progenitor cells. (A) The c-Met expression in airway progenitor cells, alveolar type 2 (AT2) cells, ciliated cells, and MLg cells using quantitative reverse transcription-quantitative polymerase chain reaction. (B) CFE of airway progenitor cells is increased in presence of HGF in stromal free 3D Matrigel culture. Data are expressed as the mean ± standard error of the mean. HGF, hepatocyte growth factor; CFE, colony forming efficiency.
Smad4 regulates the proliferation of airway progenitor cells
Smad elements are implicated in downstream TGF-β signal, and have been shown to be modulated by HGF (22). Smad elements were hypothesized to play a role in the regulation of airway progenitor cells by HGF. Microarray analysis was used to identify enriched transcripts of airway progenitor cells as compared to EpCAM+ lung epithelial cells. The top 20 genes enriched in airway progenitor cells were listed in Table I. Notably, TGF-β transcript is enriched by 3.5 fold over total lung epithelial cells (Table I). In addition, the authors sorted the list of transcripts by their relative expression level, and 1213 transcripts were observed that read 1800 or greater which indicate their potential expression in real samples (Table II). As expected, Scgb1a1, Cyp2f2 and Plunc were presented in the list of highly expressed gene list of airway progenitor cells (Table II). Smad4 was in the list even it did not show up in Table II. Gathered analysis of top genes in airway progenitor cells indicated that Smad4 signaling pathway may participate in the regulation of airway progenitor cells proliferation (Table III). In addition, Xenopus fork head domain factor 3 and direct repeat 4 transcription factors may also serve a role in regulating airway progenitor cells. These data suggested that Smad4 may be a regulator of airway progenitor cells.
Table I.
Top genes expressed in airway progenitor cells compared to total lung epithelial cells.
| Gene ID | Gene name | Gene symbol | Sca-1+/EpCAM |
|---|---|---|---|
| 742 | Proline/serine-rich coiled-coil 1 | Psrc1 | 12.0 |
| 12623 | Carboxylesterase 1 | Ces1 | 8.6 |
| 232400 | cDNA sequence BC048546 | BC048546 | 7.4 |
| 17984 | Necdin | Ndn | 7.4 |
| 67005 | Polymerase (RNA) III (DNA directed) polypeptide K | Polr3k | 4.8 |
| 140474 | Mucin 4 | Muc4 | 4.7 |
| 23886 | Growth differentiation factor 15 | Gdf15 | 4.6 |
| 22270 | Uroplakin 3A | Upk3a | 4.3 |
| 13078 | Cytochrome P450, family 1, subfamily b, polypeptide 1 | Cyp1b1 | 3.7 |
| 11670 | Aldehyde dehydrogenase family 3, subfamily A1 | Aldh3a1 | 3.6 |
| 27280 | Pleckstrin homology-like domain, family A, member 3 | Phlda3 | 3.6 |
| 70337 | Iodotyrosine deiodinase | Iyd | 3.6 |
| 21808 | Transforming growth factor, β | TGF-β | 3.5 |
| 399638 | RIKEN cDNA A030001D16 gene | A030001D16Rik | 3.5 |
| 215446 | Ectonucleoside triphosphate diphosphohydrolase 3 | Entpd3 | 3.4 |
| 18104 | NAD(P)H dehydrogenase, quinone 1 | Nqo1 | 3.4 |
| 217830 | RIKEN cDNA 9030617O03 gene | 9030617O03Rik | 3.4 |
| 77914 | Keratin associated protein 17-1 | Krtap17-1 | 3.4 |
| 70536 | Glutaminyl-peptide cyclotransferase (glutaminyl cyclase) | Qpct | 3.4 |
| 19363 | RAD51-like 1 (S. cerevisiae) | Rad51l1 | 3.3 |
Table II.
Highly expressed genes in airway progenitor cells.
| Entrez gene | Gene name | Gene symbol | Ave.Sca1+ | Ave.EpCAM+ |
|---|---|---|---|---|
| 287 | Secretoglobin, family 1A, member 1 (uteroglobin) | Scgb1a1 | 33358 | 33006 |
| 13107 | Cytochrome P450, family 2, subfamily f, polypeptide 2 | Cyp2f2 | 31168 | 26482 |
| 12409 | Carbonyl reductase 2 | Cbr2 | 28700 | 24926 |
| 117158 | Secretoglobin, family 3A, member 2 | Scgb3a2 | 28154 | 25734 |
| 15439 | Haptoglobin | Hp | 25201 | 19952 |
| 67701 | WAP four-disulfide core domain 2 | Wfdc2 | 24106 | 19162 |
| 22070 | Tumor protein, translationally-controlled 1 | Tpt1 | 23147 | 22975 |
| 13627 | Eukaryotic translation elongation factor 1 alpha 1 | Eef1a1 | 21612 | 21224 |
| 68662 | Secretoglobin, family 3A, member 1 | Scgb3a1 | 21599 | 19061 |
| 56615 | Microsomal glutathione S-transferase 1 | Mgst1 | 21468 | 18682 |
| 19241 | Thymosin, β 4, X chromosome | Tmsb4x | 21214 | 22595 |
| 20387 | Surfactant associated protein A1 | Sftpa1 | 20590 | 27427 |
| 22190 | Ubiquitin C | Ubc | 20572 | 18226 |
| 18843 | Palate, lung, and nasal epithelium associated | Plunc | 20304 | 13823 |
| 20341 | Selenium binding protein 1 | Selenbp1 | 20207 | 15170 |
| 14319 | Ferritin heavy chain 1 | Fth1 | 19942 | 17755 |
| 100042862 | Predicted gene 4076 | Gm4076 | 19843 | 18573 |
| 14281 | FBJ osteosarcoma oncogene | Fos | 19779 | 18595 |
| 65019 | Ribosomal protein L23 | Rpl23 | 19029 | 18042 |
| 14319 | Ferritin heavy chain 1 | Fth1 | 18279 | 15370 |
Table III.
Potential transcription factors active in airway progenitor cells by Gather analysis.
| n | Gene name | Total gene (n) | Bayes factor | P-value | Genes |
|---|---|---|---|---|---|
| 1 | SMAD4 | 88 | 3.9 | 0.0004 | AW210596, Abcc3, Acas2, Acp5, Adhfe1, Afp, Aox1, Arhgap24, Asgr1, Atp2b2, BC004853, BC010843, BC018371, BC024561, Btbd11, Bucs1, C030018L16Rik, Cbs, Ccng1, Ces1, Chn2, Col4a6, Cyp1b1, Cyp2b10, Cyp4f15, D430038H04Rik, Daf1, Dapk2, Dp1l1, Edg7, Efna5, Entpd3, Epdr2, Ephx1, Fmo2, Fmo3, Foxq1, Gabrp, Gdpd2, Gmnn, Gpr120, Gpx2, Gpx3, Grasp, Gss, Gsta3, Gstk1, Gstm2, Gsto1, Hod, Hpgd, Hspa5bp1, Ifit1, Itih5, Kdr, Lnx1, Ltf, Ly6a, Mscp, Nupr1, Oact1, Palmd, Pgam2, Pgd, Pglyrp1, Phlda3 Polr3k, Ppp2r2b, Prdx6, Qpct, Rab6b, Rarb, Rbp4, Samd4, Sema3a Serpinf1, Slc23a1, Stat4, Stxbp6, Sulf2, Tacstd2, Tgfb2, Tmem40, Tnfrsf21, Tnrc9, Trim3, Trim30 U46068 |
| 2 | XFD3 | 7 | 2.3 | 0.002 | Abca4, Ace2, Ces1, Epdr2, Hpgd, Igf1r, Lmyc1 |
| 3 | DR4 | 148 | 1.6 | 0.005 | 1810008K03Rik, 4933428I03Rik, 5430413I02Rik, 9030408N13Rik, A630065K24Rik, AW210596, Abca4, Abcc3, Acas2, Ace2, Acp5, Acpp, Adhfe1, Afp, Aldh3a1, Aqp4, Arhgap24, Armcx2, Asgr1, Atp2b2, Atp6v0d2, Azgp1, BC004853, BC010843, BC018371, BC024561, BC048546, Bax, Btbd11, Bucs1, C030018L16Rik, Cbs, Cckar, Ccng1, Ces1, Chn2, Cldn10, Cldn23, Cnga2, Col23a1, Col4a6 Cyp1b1, Cyp2b10, Cyp4f15, D430038H04Rik, Daf1, Dapk2, Dp1l1, Dtna, Edg7, Efna5, Enah, Entpd3, Epdr2, Ephx1, Fign, Fmo1, Fmo2, Foxq1, Frem1, Gabrp, Gcnt2, Gdf15, Gdpd2, Ggh, Gmnn, Gpr120, Gpx2, Gpx3, Grasp, Gss, Gsta2, Gsta3, Gstk1, Gstm1, Gstm2, Gsto1, Hck, Hmox1, Hod, Hpgd, Hspa5bp1, Igf1r, Il1f9, Il6ra, Itih2, Itih5, Kcnk2, Kdr, Kitl, Ldh2, Lif, Lmyc1, Lnx1, Ltf, Ly6a, MGC25972, Mgat3, Mia1, Mscp, Ndn, Nfe2l2, Notch3, Nqo1, Nrarp, Oact1, Palmd, Pgam2, Pgd, Pglyrp1, Phlda3, Pir, Polr3k, Pparg, Prdx6, Ptger4, Qpct, Rab6b, Rad51l1, Rarb, Rbp4, Rpgrip1, Samd4, Scnn1b, Scnn1g, Sdpr, Sec8l1, Sema3a, Serpinf1, Slc1a1, Slc23a1, Slc38a1, Slc9a9, Sphk1, Stat4, Stxbp6, Sulf2, Tacstd2, Tgfb2, Tmem40, Tmie, Tnfrsf21, Tnrc9, Trim30, Ttpa, U46068, Upk3a, Zfhx1a |
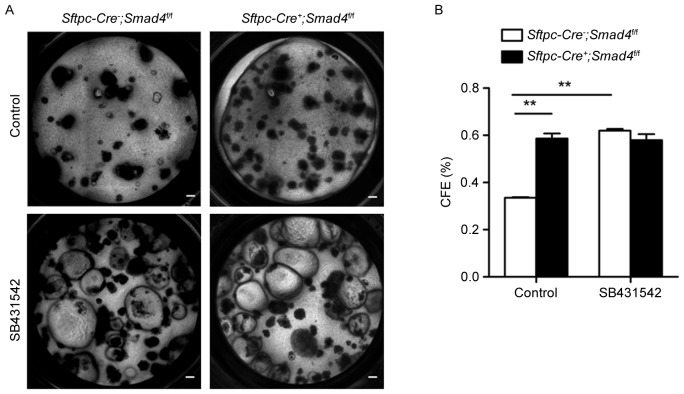
To examine this, the authors used a transgenic mouse model, Sftpc-Cre−; Smad4f/f, in which Smad4 is absent in all lung epithelia. Airway progenitor cells from Sftpc-Cre−; Smad4f/f or Sftpc-Cre+; Smad4f/f mice were sorted and cultured in the presence of MLg cells. More epithelial colonies were observed in the Sftpc-Cre+; Smad4f/f group compared to the Sftpc-Cre−; Smad4f/f control group in the absence of SB431542 (Fig. 4A and B), suggesting that Smad4 plays a negative role in the proliferation of airway progenitor cells. This change was absent, however, between these two groups in the presence of SB431542 (Fig. 4A and B). These data suggest that TGF-β activation is required for the regulation of airway progenitor cells by Smad4.
Figure 4.
Smad4 regulates the proliferation of airway progenitor cells. (A) Airway progenitor cells from Sftpc-Cre-; Smad4flf or Sftpc-Cre+; Smad4f/f mice were cultured in control condition or with the addition of 10 µM SB431542 at week 4. (B) Quantitative analysis of the colony-forming efficiency in different culture conditions. Scale bars, 200 µm. Data are expressed as the mean ± standard error of the mean. **P<0.01 as indicated. CFE, colony forming efficiency.
It was proposed, therefore, that the TGFβ/TGFβR1/2 axis plays an inhibitory role in the expression and/or secretion of growth factors HGF in fibroblasts (Fig. 5). Fibroblast-derived HGF, through its receptor c-Met on airway progenitor cells, supports the proliferation of airway epithelial progenitor cells (Fig. 5). Smad4 may serve as a negative regulator, given its deletion promotes the proliferation of airway progenitor cells (Fig. 5). Epithelial TGFβR1/2 seems not to be involved in the regulation of airway progenitor cell proliferation.
Figure 5.
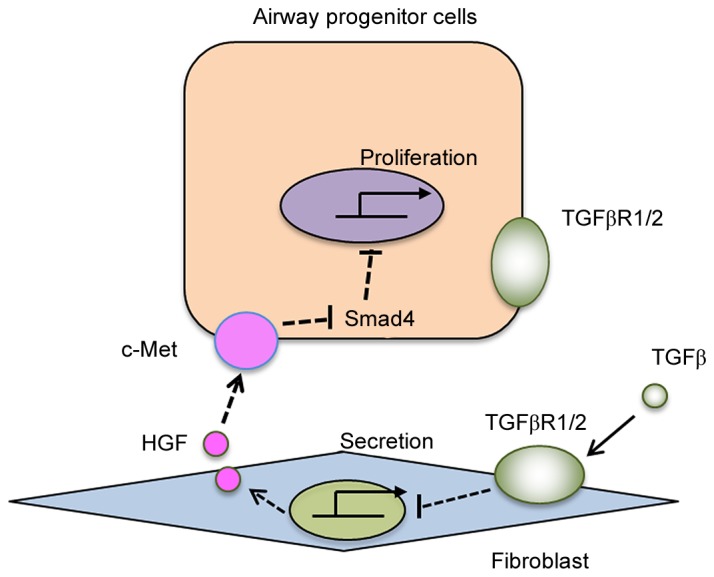
Model for Fibroblasts-airway progenitor cell interaction. The TGF-β signaling inhibits the secretion of HGF in fibroblasts, which binds to its receptor c-Met on airway progenitor cells to promote their growth. Downstream Smad4 may serve as a repressor in the regulating airway progenitor cell function. TGF-β, transforming growth factor-β; HGF, hepatocyte growth factor.
Discussion
In the present study, the authors demonstrated that airway progenitor cell proliferation is not affected by inhibition of the TGF-β signal or deletion of the TGF-β receptor 2 on progenitor cells in particular. Rather, inhibition of TGF-β signaling specifically in fibroblasts contributes to the proliferation of airway progenitor cells, likely due to the inducible role of TGF-β signaling in the secretion of fibroblast growth factors HGF.
TGF-β signal controls lung progenitor cell fate by altering the microenvironment of the lung progenitor cells niche (23). The TGF-β signal has been implicated in the regeneration of airway epithelium following injury. For instance, the upregulation of TGF-β stimulates extracellular matrix secretion and activates the profibrotic signaling pathway in lung fibrosis (24). Li et al (25) demonstrated that deletion of TGF-βR2 on the epithelia prevented mice from bleomycin-induced lung fibrosis. Furthermore, TGF-β signaling promotes differentiation of club cells and alveolar type 2 cells (26–28). These results strongly suggest that TGF-β signaling reduces the proliferation of epithelial progenitor cells and inhibits the regeneration of injured epithelium. In the present study, the authors found that TGF-β indirectly regulates airway progenitor cells. More specifically, TGF-β ligand binds to a cell membrane serine/threonine kinase heteromeric receptor complex, which is composed of type 1 and type 2 receptors. TGFβ1R is known to be inhibited by SB431542 (29). Stimulation of TGFβ1R leads to phosphorylation of Smad2 and Smad3, which combine with Smad4 and translocate into the nucleus (30). Smad plays a critical role in the TGF-β1/Smad signal pathway. Inhibition of TGF-β1/Smad3 signaling induces alveolar repair in the lung of hyperoxic mice (31). It also has been shown that the deletion of Smad4 promotes lung cancer growth and metastasis (32). The authors observed that the proliferation of airway progenitor cells was also promoted in absence of Smad4. Conversely, Smad4 can direct the repair of epithelium under microenvironmental signals.
Growth factors are critical in the regulation of airway epithelial cells by TGF-β. McQualter et al (7) found that inhibition of TGF-β can restore stromal cell epithelial-supportive capacity, and upregulate the expression of FGF10 in stromal cells (7). In addition, colonies of airway progenitor cells were apparent in cultures supplemented with FGF10 and HGF in stromal-free conditions by Mungunsukh and Day (33). Similar to FGF10, HGF expression is suppressed by TGF-β signaling post-transcription in a human adult fibroblast culture. In contrast, HGF expression is upregulated by knocking out TGF-β receptor 2 in mammary fibroblasts (34). Consistent with these data, the authors observed that inhibition of TGF-β promotes HGF expression. These findings suggest that blocking TGF-β activates HGF, after which HGF binds to its receptor c-Met on epithelial and endothelial cells to fulfill its functions.
Theoretically, HGF and FGF have been demonstrated to inhibit Smad4 function through activating the Ras/MAPK pathway (19). In the present study, the authors observed that as compared with fibroblasts, airway progenitor cells express c-Met abundantly. c-Met may mediate HGF's role in regulating airway progenitor cells which may be dependent on Smad4. Therefore, the TGFβ-TGFβR1/2-HGF-Smad4 axis may play a role in airway epithelial homeostasis.
Nonetheless, there are still some limitations of the current study. The relationship between HGF and Smad4 was not detected directly, which can be addressed by creating Sftpc-Cre+; Met f/f mice to understand the interactions between c-Met and Smad4 in the regulation of airway epithelial regeneration.
In conclusion, the study suggests that TGF-β represses the proliferation of airway progenitor cells via the TGF-β receptor on fibroblasts, thereby altering their secretory properties including the secretion of HGF. Additionally, Smad4 may play a mechanistic role in the regulation of airway progenitor cells by HGF.
Acknowledgements
The current work was supported by the National Natural Science Foundation of China (grant nos. 31471121, 81728001 and 81773394) and the Natural Science Foundation of Tianjin City (grant nos. 13JCYBJC40000 and 17JCYBJC24700), and and Key Projects of Health and Family Planning Commission of Tianjin (grant nos. 16KG163 and 16KG164).
References
- 1.O'Koren EG, Hogan BL, Gunn MD. Loss of basal cells precedes bronchiolitis obliterans-like pathological changes in a murine model of chlorine gas inhalation. Am J Respir Cell Mol Biol. 2013;49:788–797. doi: 10.1165/rcmb.2012-0369OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Trinh NT, Bardou O, Privé A, Maillé E, Adam D, Lingée S, Ferraro P, Desrosiers MY, Coraux C, Brochiero E. Improvement of defective cystic fibrosis airway epithelial wound repair after CFTR rescue. Eur Respir J. 2012;40:1390–1400. doi: 10.1183/09031936.00221711. [DOI] [PubMed] [Google Scholar]
- 3.Lambrecht BN, Hammad H. The airway epithelium in asthma. Nat Med. 2012;18:684–692. doi: 10.1038/nm.2737. [DOI] [PubMed] [Google Scholar]
- 4.Proud D, Leigh R. Epithelial cells and airway diseases. Immunol Rev. 2011;242:186–204. doi: 10.1111/j.1600-065X.2011.01033.x. [DOI] [PubMed] [Google Scholar]
- 5.Tzortzaki EG, Siafakas NM. A hypothesis for the initiation of COPD. Eur Respir J. 2009;34:310–315. doi: 10.1183/09031936.00067008. [DOI] [PubMed] [Google Scholar]
- 6.Hogan BL, Barkauskas CE, Chapman HA, Epstein JA, Jain R, Hsia CC, Niklason L, Calle E, Le A, Randell SH, et al. Repair and regeneration of the respiratory system: Complexity, plasticity, and mechanisms of lung stem cell function. Cell Stem Cell. 2014;15:123–138. doi: 10.1016/j.stem.2014.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McQualter JL, McCarty RC, Van der Velden J, O'Donoghue RJ, Asselin-Labat ML, Bozinovski S, Bertoncello I. TGF-β signaling in stromal cells acts upstream of FGF-10 to regulate epithelial stem cell growth in the adult lung. Stem Cell Res. 2013;11:1222–1233. doi: 10.1016/j.scr.2013.08.007. [DOI] [PubMed] [Google Scholar]
- 8.Lee JH, Bhang DH, Beede A, Huang TL, Stripp BR, Bloch KD, Wagers AJ, Tseng YH, Ryeom S, Kim CF. Lung stem cell differentiation in mice directed by endothelial cells via a BMP4-NFATc1-thrombospondin-1 axis. Cell. 2014;156:440–455. doi: 10.1016/j.cell.2013.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Volckaert T, Dill E, Campbell A, Tiozzo C, Majka S, Bellusci S, De Langhe SP. Parabronchial smooth muscle constitutes an airway epithelial stem cell niche in the mouse lung after injury. J Clin Invest. 2011;121:4409–4419. doi: 10.1172/JCI58097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ruiz EJ, Oeztuerk-Winder F, Ventura JJ. A paracrine network regulates the cross-talk between human lung stem cells and the stroma. Nat Commun. 2014;5:3175. doi: 10.1038/ncomms4175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tadokoro T, Gao X, Hong CC, Hotten D, Hogan BL. BMP signaling and cellular dynamics during regeneration of airway epithelium from basal progenitors. Development. 2016;143:764–773. doi: 10.1242/dev.126656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lynch TJ, Anderson PJ, Xie W, Crooke AK, Liu X, Tyler SR, Luo M, Kusner DM, Zhang Y, Neff T, et al. Wnt signaling regulates airway epithelial stem cells in adult murine submucosal glands. Stem Cells. 2016;34:2758–2771. doi: 10.1002/stem.2443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rock JR, Gao X, Xue Y, Randell SH, Kong YY, Hogan BL. Notch-dependent differentiation of adult airway basal stem cells. Cell Stem Cell. 2011;8:639–648. doi: 10.1016/j.stem.2011.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim KK, Kugler MC, Wolters PJ, Robillard L, Galvez MG, Brumwell AN, Sheppard D, Chapman HA. Alveolar epithelial cell mesenchymal transition develops in vivo during pulmonary fibrosis and is regulated by the extracellular matrix; Proc Natl Acad Sci USA; 2006; pp. 13180–13185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Larson-Casey JL, Deshane JS, Ryan AJ, Thannickal VJ, Carter AB. Macrophage Akt1 kinase-mediated mitophagy modulates apoptosis resistance and pulmonary fibrosis. Immunity. 2016;44:582–596. doi: 10.1016/j.immuni.2016.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang YC, Zhang N, Van Crombruggen K, Hu GH, Hong SL, Bachert C. Transforming growth factor-beta1 in inflammatory airway disease: A key for understanding inflammation and remodeling. Allergy. 2012;67:1193–1202. doi: 10.1111/j.1398-9995.2012.02880.x. [DOI] [PubMed] [Google Scholar]
- 17.Plaschke-Schlütter A, Behrens J, Gherardi E, Birchmeier W. Characterization of the scatter factor/hepatocyte growth factor gene promoter. Positive and negative regulatory elements direct gene expression to mesenchymal cells. J Biol Chem. 1995;270:830–836. doi: 10.1074/jbc.270.2.830. [DOI] [PubMed] [Google Scholar]
- 18.Hoot KE, Oka M, Han G, Bottinger E, Zhang Q, Wang XJ. HGF upregulation contributes to angiogenesis in mice with keratinocyte-specific Smad2 deletion. J Clin Invest. 2010;120:3606–3616. doi: 10.1172/JCI43304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Korc M. Smad4: Gatekeeper gene in head and neck squamous cell carcinoma. J Clin Invest. 2009;119:3208–3211. doi: 10.1172/JCI41230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Teisanu RM, Chen H, Matsumoto K, McQualter JL, Potts E, Foster WM, Bertoncello I, Stripp BR. Functional analysis of two distinct bronchiolar progenitors during lung injury and repair. Am J Respir Cell Mol Biol. 2011;44:794–803. doi: 10.1165/rcmb.2010-0098OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 22.Shukla MN, Rose JL, Ray R, Lathrop KL, Ray A, Ray P. Hepatocyte growth factor inhibits epithelial to myofibroblast transition in lung cells via Smad7. Am J Respir Cell Mol Biol. 2009;40:643–653. doi: 10.1165/rcmb.2008-0217OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Quantius J, Schmoldt C, Vazquez-Armendariz AI, Becker C, El Agha E, Wilhelm J, Morty RE, Vadász I, Mayer K, Gattenloehner S, et al. Influenza virus infects epithelial stem/progenitor cells of the distal lung: Impact on Fgfr2b-driven epithelial repair. PLoS Pathog. 2016;12:e1005544. doi: 10.1371/journal.ppat.1005544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gauldie J, Kolb M, Ask K, Martin G, Bonniaud P, Warburton D. Smad3 signaling involved in pulmonary fibrosis and emphysema; Proc Am Thorac Soc; 2006; pp. 696–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li M, Krishnaveni MS, Li C, Zhou B, Xing Y, Banfalvi A, Li A, Lombardi V, Akbari O, Borok Z, Minoo P. Epithelium-specific deletion of TGF-β receptor type II protects mice from bleomycin-induced pulmonary fibrosis. J Clin Invest. 2011;121:277–287. doi: 10.1172/JCI42090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jain R, Barkauskas CE, Takeda N, Bowie EJ, Aghajanian H, Wang Q, Padmanabhan A, Manderfield LJ, Gupta M, Li D, et al. Plasticity of Hopx(+) type I alveolar cells to regenerate type II cells in the lung. Nat Commun. 2015;6:6727. doi: 10.1038/ncomms7727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhao L, Yee M, O'Reilly MA. Transdifferentiation of alveolar epithelial type II to type I cells is controlled by opposing TGF-β and BMP signaling. Am J Physiol Lung Cell Mol Physiol. 2013;305:L409–L418. doi: 10.1152/ajplung.00032.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xing Y, Li C, Li A, Sridurongrit S, Tiozzo C, Bellusci S, Borok Z, Kaartinen V, Minoo P. Signaling via Alk5 controls the ontogeny of lung clara cells. Development. 2010;137:825–833. doi: 10.1242/dev.040535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Prime SS, Pring M, Davies M, Paterson IC. TGF-beta signal transduction in oro-facial health and non-malignant disease (part I) Crit Rev Oral Biol Med. 2004;15:324–336. doi: 10.1177/154411130401500602. [DOI] [PubMed] [Google Scholar]
- 30.Mori S, Matsuzaki K, Yoshida K, Furukawa F, Tahashi Y, Yamagata H, Sekimoto G, Seki T, Matsui H, Nishizawa M, et al. TGF-beta and HGF transmit the signals through JNK-dependent Smad2/3 phosphorylation at the linker regions. Oncogene. 2004;23:7416–7429. doi: 10.1038/sj.onc.1207981. [DOI] [PubMed] [Google Scholar]
- 31.Kayalar O, Oztay F. Retinoic acid induced repair in the lung of adult hyperoxic mice, reducing transforming growth factor-β1 (TGF-β1) mediated abnormal alterations. Acta Histochem. 2014;116:810–819. doi: 10.1016/j.acthis.2014.01.009. [DOI] [PubMed] [Google Scholar]
- 32.Liu J, Cho SN, Akkanti B, Jin N, Mao J, Long W, Chen T, Zhang Y, Tang X, Wistub II, et al. ErbB2 Pathway Activation upon Smad4 loss promotes lung tumor growth and metastasis. Cell Rep. 2015;pii:S2211–S1247. doi: 10.1016/j.celrep.2015.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mungunsukh O, Day RM. Transforming growth factor-β1 selectively inhibits hepatocyte growth factor expression via a micro-RNA-199-dependent posttranscriptional mechanism. Mol Biol Cell. 2013;24:2088–2097. doi: 10.1091/mbc.E13-01-0017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cheng N, Chytil A, Shyr Y, Joly A, Moses HL. Transforming growth factor-beta signaling-deficient fibroblasts enhance hepatocyte growth factor signaling in mammary carcinoma cells to promote scattering and invasion. Mol Cancer Res. 2008;6:1521–1533. doi: 10.1158/1541-7786.MCR-07-2203. [DOI] [PMC free article] [PubMed] [Google Scholar]