Abstract
Long noncoding RNAs (lncRNAs) have been regarded as important regulators in numerous biological processes during cell development. However, the holistic lncRNA expression pattern and potential functions during osteoblast differentiation under simulated microgravity remain unknown. In the present study, a high throughput microarray assay was performed to detect lncRNA and mRNA expression profiles during MC3TC-E1 pre-osteoblast cell osteo-differentiation under simulated microgravity. The expression of 857 lncRNAs and 2,264 mRNAs was significantly altered when MC3T3-E1 cells were exposed to simulated microgravity. A relatively consistent distribution pattern on the chromosome and a co-expression network were observed between the differentially-expressed lncRNAs and mRNAs. Genomic context analysis further identified 132 differentially-expressed lncRNAs and nearby coding gene pairs. Subsequently, 3 lncRNAs were screened out for their possible function in osteoblast differentiation, based on their co-expression association and potential cis-acting regulatory pattern with the deregulated mRNAs. The present study aimed to provide a comprehensive understanding of and a foundation for future studies into lncRNA function in mechanical signal-mediated osteoblast differentiation.
Keywords: osteoblast differentiation, long noncoding RNA, simulated microgravity, bioinformatic analysis
Introduction
Studies have demonstrated that mechanical unloading may result in severe bone loss, as observed in astronauts undergoing long-term space flight or in patients subjected to long-duration immobility or bedrest (1,2). Decreased bone formation caused by abnormal osteoblast function is considered to be one of the primary causes of bone loss (3). Studies have demonstrated that real or simulated microgravity may affect the processes of osteoblast proliferation, differentiation and mineralization (4–6). Genomic analysis revealed that the expression of hundreds of genes in osteoblasts is altered upon exposure to microgravity (7,8). Efforts have been made to identify and characterize the key regulators or genes that control alterations in osteoblasts. However, existing studies have not completely clarified the reason for the delay in osteoblast development induced by microgravity; it is therefore necessary to further examine more regulatory mechanisms, in order to obtain a deep insight into this area and to develop effective preventative measures.
Previously, high throughput genomic analysis and transcriptome sequencing demonstrated that much of the genome is transcribed to noncoding RNAs (ncRNAs) (9). Increasing numbers of ncRNAs, which were previously considered to be transcriptional ‘noise’, have been observed to be important regulators of gene expression in development, physiology and disease (10,11). A subset of these ncRNAs whose strand length exceeds 200 nucleotides are termed long noncoding RNAs (lncRNAs) (12). Studies have demonstrated that lncRNAs are involved in a range of important cellular processes, including chromatin modification, RNA processing and gene transcription (11). ncRNAs are able to regulate gene expression in close genomic proximity (cis-acting) or target distant transcriptional activators or inhibitors (trans-acting) via diverse mechanisms (13). Such regulatory effects have been additionally observed in the osteogenic differentiation of mesenchymal stem cells (MSCs). lncRNA-maternally expressed 3, which is located near the bone morphogenetic protein 4 (BMP4) gene locus, is able to dissociate the repressor transcription factor SOX-2 from the BMP4 promoter and thereby activate the transcription of BMP4, resulting in the osteogenic differentiation of bone marrow MSCs (14). These previous studies suggested that lncRNAs serve an important role upstream of gene expression during normal osteoblast development. However, there are no reports of lncRNA expression patterns in the microgravity-induced inhibition of osteogenic development, and whether mechanical signal-mediated osteoblast differentiation relies on the modulation of lncRNA expression remains unclear.
The present study screened for differentially-expressed lncRNAs and mRNAs in the MC3T3-E1 pre-osteoblast cells, prior to and following exposure to simulated microgravity, using microarray profiling. A series of bioinformatic analyses, including Gene Ontology (GO) analysis, pathway analysis, genomic context analysis and co-expression analysis, were used to predict potential functional lncRNAs. The results of the present study suggested that lncRNAs may serve important roles during osteoblast differentiation under simulated microgravity conditions.
Materials and methods
Cell culture and osteogenic differentiation
Mouse MC3T3-E1 pre-osteoblast cells were obtained from the Chinese Academy of Sciences Cell Bank (Shanghai, China) and cultured in Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal bovine serum (FBS) (both from Hyclone; GE Healthcare Life Sciences, Logan, UT, USA), 100 U/ml penicillin G and 100 mg/ml streptomycin, at 37°C in a humidified atmosphere of 5% CO2 in air. To induce osteoblast differentiation, the DMEM was supplemented with 10% FBS, 0.1 mM dexamethasone, 10 mM β-glycerophosphate, and 50 µg/ml ascorbic acid, and was changed every 2–3 days.
mRNA extraction and reverse transcription-quantitative polymerase chain reaction (RT-qPCR) analysis
Total mRNA was extracted using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA), according to the manufacturer's instructions. The RNA concentration was quantified using a NanoDrop 1000 Spectrophotometer (Thermo Fisher Scientific, Inc.). RNA quality was tested using an Agilent 2100 bioanalyzer (Agilent Technologies, Inc., Santa Clara, CA, USA) and measured using the RNA integrity number (RIN). RNA samples were submitted for further analysis if the RIN score was >5.0.
For RT, first-strand cDNA was synthesized using the PrimeScript® RT reagent kit (cat. no. DRR037; Takara Bio, Inc., Otsu, Japan). The expression levels of target genes were determined quantitatively using a CFX96 qPCR system (Bio-Rad Laboratories, Inc., Hercules, CA, USA) using SYBR® Premix Ex Taq™ II (cat. no. DRR820A; Takara Bio, Inc.), according to the manufacturer's instructions. The primer pairs were as follows: Runt-related transcription factor 2 (Runx2; GenBank accession no. NM_053470) forward, 5′-CCATAACGGTCTTCACAAATCC-3′ and reverse, 5′-GCGGGACACCTACTCTCATACT-3′; transcription factor Sp7 (Osx; accession no. NM_001037632) forward, 5′-CAGTAATCTTCGTGCCAGACC-3′ and reverse, 5′-CTTCTTTGTGCCTCCTTTTCC-3′; polypyrimidine tract-binding protein 2 (Ptbp2; accession no. NM_001310711) forward, 5′-AAAGTCGCTCTGAGTTGTTAT-3′ and reverse, 5′-GCGAAGAGTTTGTCCTCAACC-3′; transportin-1 (Tnpo1; accession no. NM_001048267) forward, 5′-AATTCGCGGTGACTCAGTCTGG-3′ and reverse, 5′-TCCATCTTGGTTTGCGAGGC-3′; exostosin-1 (Ext1; accession no. NM_010162) forward, 5′-GAAGAGCACAGTGGTCGGAA-3′ and reverse, 5′-CTCGATGGCCGCTAGAATGT-3′; NONMMUT044983 (NONCODE gene ID NONMMUG027774.1) forward, 5′-CGGCAGGCCTAGTCTTGTAT-3′ and reverse, 5′-ACAGCAGAGAGAGCCAAGGA-3′; NONMMUT023474 (gene ID NONMMUG014520.1) forward, 5′-TCTCGAACCCTAGGAGAGCA-3′ and reverse, 5′-GGGACAAGGTAAATGGCTCA-3′; NONMMUT018832 (gene ID NONMMUG011720.1) forward, 5′-AACATCTGAGGCTTGGCACT-3′ and reverse, 5′-TCATGGTACTGGCATCTCCA-3′; and GAPDH (accession no. NM_008084) forward, 5′-CAGTGCCAGCCTCGTCTCAT-3′ and reverse, 5′-AGGGGCCATCCACAGTCTTC-3′. GAPDH was used as an internal control. The qPCR thermal cycling was performed as follows: Initial incubation for 15 sec at 95°C, followed by denaturing for 40 cycles at 95°C for 5 sec and annealing for 31 sec at 60°C. The relative expression of target genes was calculated using the 2−ΔΔCq method (15).
Alkaline phosphatase (ALP) activity assay
Cells were seeded at 1×106 cells/well in 6-well plates (Corning Inc., Corning, NY, USA) and cultured for 24 h. When ALP activity was determined, confluent cell layers were washed with PBS, lysed with 0.1 mol/l M-PER mammalian protein extraction reagent (Pierce; Thermo Fisher Scientific, Inc.) for 15–30 min, and centrifuged at 12,000 × g for 15 min at room temperature, according to the manufacturer's instructions. The supernatants were collected for determining ALP activity using an ALP assay kit (Nanjing Jiancheng Bioengineering Institute, Nanjing, China). Protein concentrations were measured using a Bicinchoninic Acid Protein Assay kit (Pierce; Thermo Fisher Scientific, Inc.). ALP activity (IU/l) was defined as the release of 1 nmol p-nitrophenol/min/µg total cellular protein.
Simulated microgravity exposure
The Rotating Wall Vessel Bioreactor (RWVB) clinostat is an effective, ground-based device which is used to simulate microgravity (7). Microgravity is achieved by keeping cells rotating uniformly around a horizontal axis. Therefore, there is a vector-averaged reduction in the apparent gravity acting on the cells when rotated by 360°. A 2D-RWVB (China Astronaut Research and Training Center, Beijing, China) was used in the present study, as previously described (16). MC3T3-E1 cells were seeded on coverslips and incubated until cell confluence reached 70%. The coverslips were subsequently fixed in the bioreactor and placed 12.5 mm away from the rotational axis. The bioreactor was completely filled with culture medium. Gentle aspiration was performed to clear away air bubbles in order to avoid shear stress during rotation. The bioreactor was fixed onto the clinostat and rotated around a horizontal axis at 24 rpm. The group rotating around a vertical axis was regarded as the control. The entire system was placed in a humidified incubator at 37°C under 5% CO2.
Microarray profiling
An Affymetrix GeneChip® Mouse Transcriptome assay (version 1.0; Affymetrix Inc., Santa Clara, CA, USA) provided global profiling of transcripts in the mouse genome, which were selected from the most authoritative databases, including NONCODE (www.noncode.org/index.php) and ENSEMBL (www.ensembl.org/index.html). In the microarray, every transcript was detected with 10 specific probes in order to improve the confidence of the statistical results. Protein-coding and non-coding genes were represented on a separate array to supply coincident hybridization. RNA labeling and array hybridization were performed according to the manufacturer's protocol. The microarray work was performed by Shanghai Ming Information Technology Co., Ltd. (Shanghai, China).
GO and pathway analysis
GO analysis was applied to analyze the primary function of the differentially-expressed genes according to the principles of Gene Ontology, which is able to organize genes into hierarchical categories and reveal the gene regulatory network on the basis of biological process and molecular function (17). Pathway analysis was additionally performed to examine the significant pathways of the differentially-expressed genes according to the Kyoto Encyclopedia of Genes and Genomes, Biocarta and Reatome databases (18). An online Bioinformatics enrichment tool (DAVID; david.ncifcrf.gov) was used to perform the GO and pathway analysis in the present study (19). Biological processes of GO terms were illustrated in the GO analysis. The P-value indicated the significance of GO term and pathway term enrichment in the differentially-expressed mRNA list (P<0.05 was considered to be statistically significant).
Construction of the lncRNA-mRNA co-expression network
A co-expression regulatory network is an undirected graph, where each node corresponds to coding or ncRNAs, and a pair of nodes is connected by an edge if there is a significant co-expression association between them (20). lncRNA-mRNA networks were built to identify the interactions between the differentially-expressed genes and lncRNAs, according to the normalized signal intensity of specific expression in the gene and lncRNA. For lncRNA-gene pair, the Pearson correlation was calculated and the significant correlation pairs with which to construct the network were selected (21). In network analysis, degree centrality, which is defined as the number of connections between one node and another, is the simplest and most important measure of the centrality of a gene or lncRNA within a network which determines its relative importance (22).
Statistical analysis
Experiments were repeated a minimum of three times. Data were statistically analyzed using SPSS 19.0 software and expressed as the mean ± standard deviation. A one-way repeated measures analysis of variance followed by Dunnett's post hoc test was used to compare the time course-dependent variables. Two group comparisons were performed using a two-tailed t-test. P<0.05 was considered to indicate a statistically significant difference.
Results
Differentially-expressed lncRNAs and mRNAs in MC3T3-E1 cells exposed to simulated microgravity
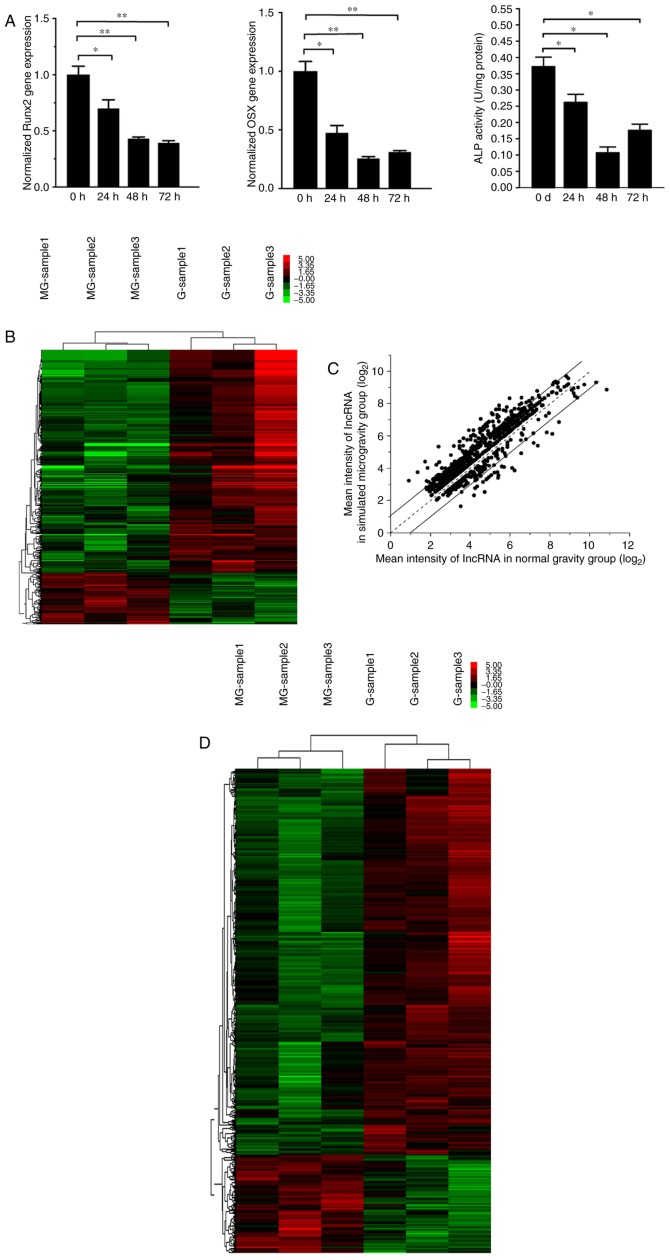
MC3T3-E1 cells were cultured in an RWVB clinostat and tested following rotation for 72 h. The effect of simulated microgravity on osteoblasts was assessed by observing the gene expression of Runx2 and Osx, in addition to the activity of ALP, which were considered to be important transcription factors for osteoblast differentiation. The data demonstrated that the gene expression of Runx2 and Osx and the protein activity of ALP were significantly decreased, of which, Osx and ALP reached lowest levels at 48 h and Runx2 decreased slightly further at 72 h (Fig. 1A). The results suggested that osteo-differentiation process of MC3T3-E1 cells was inhibited by simulated microgravity.
Figure 1.
Expression profile of lncRNAs and mRNAs in MC3T3-E1 cells exposed to simulated microgravity. (A) Osteogenic differentiation was assessed by reverse transcription-quantitative polymerase chain reaction analysis of osteoblast marker genes and activity analysis of ALP. n=3. *P<0.05, **P<0.01. (B) Expression profiles of ncRNAs in MC3T3-E1 cells when exposed to simulated microgravity. Green represents downregulated ncRNAs, and red represents upregulated ncRNAs. n=3. (C) The deregulated lncRNAs with a length exceeding 200 nucleotides were screened out and exhibited on a scatter plot. (D) Expression profile of mRNAs in MC3T3-E1 cells when exposed to simulated microgravity. n=3. lncRNA, long noncoding RNA; ALP, alkaline phosphatase; Runx2, runt-related transcription factor 2; OSX, transcription factor Sp7.
In order to detect the differentially-expressed lncRNAs and mRNAs in MC3T3-E1 when exposed to simulated microgravity, a transcriptome assay was performed to compare expression profiles between the normal gravity and simulated microgravity groups. The expression levels of 1,481 ncRNAs were observed to be significantly altered, which is presented in the hierarchical clustering analysis heat map (Fig. 1B). Among them, 857 lncRNAs whose length exceeded 200 nucleotides, including 168 upregulated and 689 downregulated, were screened out and are exhibited on a scatter plot (Fig. 1C). In addition, 2,264 mRNAs were demonstrated to be differentially-expressed, including 459 upregulated mRNAs and 1,805 downregulated mRNAs, presented on a clustering heat map (Fig. 1D).
Construction of the co-expression network between differentially-expressed lncRNAs and mRNAs
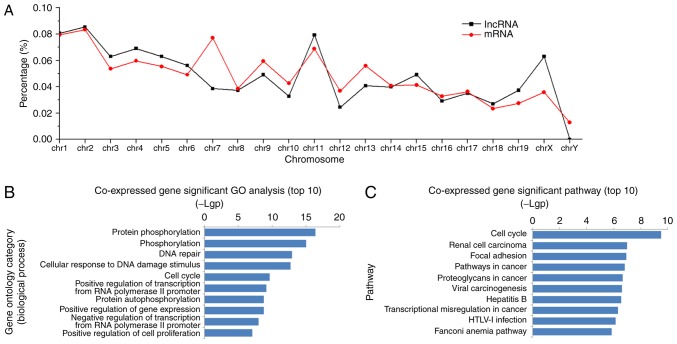
Locational distributions of the aberrantly-expressed lncRNAs and mRNAs were analyzed synchronously. Significantly altered lncRNAs and mRNAs were spread across the chromosomes, with the largest amounts on chromosomes 1, 2 and 11. A relatively consistent variation pattern was observed between aberrant lncRNA and mRNA species, except for on chromosome 7 (Fig. 2A). In order to elucidate the detailed association, a large and complex lncRNA-mRNA co-expression network was generated according to the normalized signal intensity of differentially-expressed lncRNAs and mRNAs. This co-expression network consisted of 916 nodes and 4,813 connections between 354 lncRNAs and 562 mRNAs. Within this network, there were 3,798 pairs presenting as positive regulatory associations and 1,015 pairs as negative associations. Subsequently, mRNAs in the network were submitted for GO enrichment analysis. The results demonstrated that the aberrant mRNAs were frequently enriched in such biological process as phosphorylation, response to DNA damage, the cell cycle, regulation of gene expression and cell proliferation (Fig. 2B), and were involved in the cell signaling pathways of the cell cycle, focal adhesion, cancer and viral infection (Fig. 2C).
Figure 2.
Chromosome location, GO and pathway analyses of deregulated lncRNAs and mRNAs. (A) Location distributions of deregulated lncRNAs and mRNAs on chromosomes. The co-expressed genes of deregulated lncRNAs were submitted for GO and pathway enrichment analyses. (B) The top 10 enriched GO terms (biological process) and (C) signal pathways are presented in bar diagrams. The value of -log10 (P-value) was calculated to reflect the significance of enrichment. GO, Gene Ontology; lncRNA, long noncoding RNA.
Genomic context analysis of the aberrantly-expressed lncRNAs
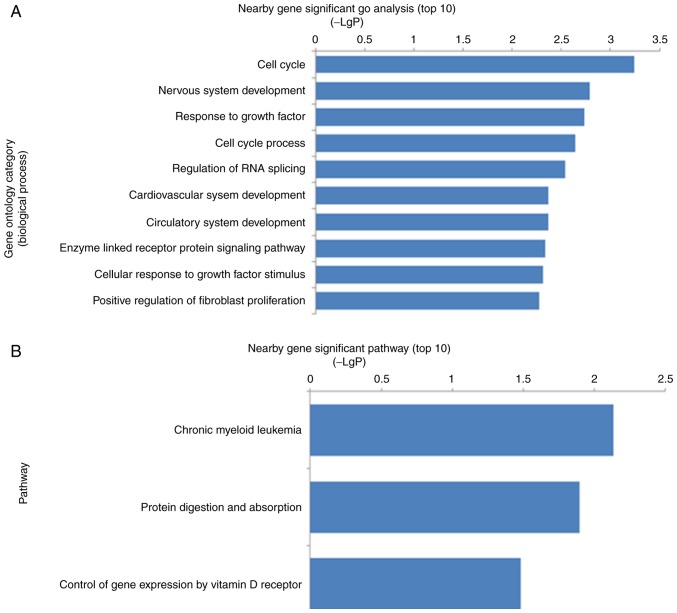
In order to further elicit information from the co-expression network and examine the possible functional lncRNAs, the nearby mRNAs located within 100 kb up or downstream of aberrantly-expressed lncRNAs were identified and matched with the aberrantly-expressed mRNAs in the microarray data. Among them, 132 differentially-expressed lncRNAs and nearby mRNAs pairs were identified (data not shown). GO analysis indicated that the nearby genes were involved in the significant biological processes, including the cell cycle, nervous system development, response to growth factors, regulation of RNA splicing, circulatory system development, enzyme-linked receptor protein signaling pathway and positive regulation of fibroblast proliferation (Fig. 3A). Pathway analysis demonstrated that the most significant pathways were chronic myeloid leukemia, protein digestion and absorption, and control of gene expression by vitamin D receptor (Fig. 3B). A literature review further demonstrated that ≥15 mRNAs included in 17 lncRNA-mRNA pairs have been reported to be involved in the regulation of osteoblast proliferation, differentiation and mineralization (Table I).
Figure 3.
GO and pathway analyses of genes located near the deregulated long noncoding RNAs. (A) The top 10 enriched GO terms (biological process). (B) The top 10 enriched signal pathways. GO, Gene Ontology.
Table I.
Osteoblast function-associated genes located near to deregulated lncRNAs.
| Author, year | Deregulated lncRNAa | lncRNA fold change | Nearby gene | Expression styleb | Location of nearby gene | Osteoblast-associated function of nearby genec | (Refs.) |
|---|---|---|---|---|---|---|---|
| Hong et al, 2010 | NONMMUT000809 | 0.47 | Col5a2 | Downregulated | Same strand with overlap | Positive on differentiation | (23) |
| Verlinden et al, 2013 | NONMMUT001206 | 0.83 | Nrp2 | Downregulated | Same strand with overlap | Positive on proliferation | (24) |
| Tang et al, 2011 | NONMMUT044983 | 0.54 | Ptbp2 | Downregulated | Same strand with overlap | Positive on development | (25) |
| Lekva et al, 2012 | NONMMUT044301 | 0.81 | Txnip | Downregulated | Complementary with overlap | Negative on differentiation | (26) |
| Ogasawara et al, 2004 | KnowTID_00005517 | 0.83 | Cdk6 | Downregulated | Upstream, 10,000 bp | Negative on differentiation | (27) |
| Fan et al, 2016 | NONMMUT054273 | 0.80 | Ptpn11 | Downregulated | Same strand with overlap | Positive on differentiation | (28) |
| Sun et al, 2015 | NONMMUT058601 | 0.51 | Cacna1c | Downregulated | Same strand with overlap | Positive on proliferation | (29) |
| Chang et al, 2009 | KnowTID_00006206 | 0.82 | Cdkn1b | Downregulated | Upstream, 10,000 bp | Positive on differentiation | (30) |
| Yano et al, 2014 | NONMMUT055574 | 0.54 | Col1a2 | Downregulated | Same strand with overlap | Positive on differentiation | (31) |
| Haasper et al, 2008 | KnowTID_00006709 | 3.21 | Fosb | Upregulated | Upstream, 10,000 bp | Positive on differentiation | (32) |
| Choi et al, 2015 | KnowTID_00007668 | 0.66 | Cbl | Downregulated | Upstream, 10,000 bp | Negative on differentiation | (33) |
| Yi et al, 2017 | NONMMUT069918 | 0.42 | Tcf12 | Downregulated | Same strand with overlap | Negative on differentiation | (34) |
| Di et al, 2014 | NONMMUT018832 | 0.68 | Tnpo1 | Downregulated | Complementary strand with overlap | Positive on differentiation | (35) |
| Matsumoto et al, 2010 | NONMMUT023472 | 0.55 | Ext1 | Downregulated | Same strand overlap | Positive on development | (36) |
| Matsumoto et al, 2010 | NONMMUT023474 | 0.50 | Ext1 | Downregulated | Same strand with overlap | Positive on development | (36) |
| Matsumoto et al, 2010 | NONMMUT023475 | 0.55 | Ext1 | Downregulated | Same strand with overlap | Positive on development | (36) |
| Lu et al, 2016 | NONMMUT025233 | 0.65 | Crebbp | Downregulated | Upstream, 10,000 bp | Positive on differentiation | (37) |
The transcript ID of lncRNAs in the NONCODE database is listed to instead of the gene symbol due to the lack of unified nomenclature.
The expression style of the lncRNAs and their nearby genes are coincident according to the microarray data.
The direct or indirect osteoblast-associated functions of the nearby genes are summarized with one representative supporting reference.
Bioinformatics screening of potential functional lncRNAs
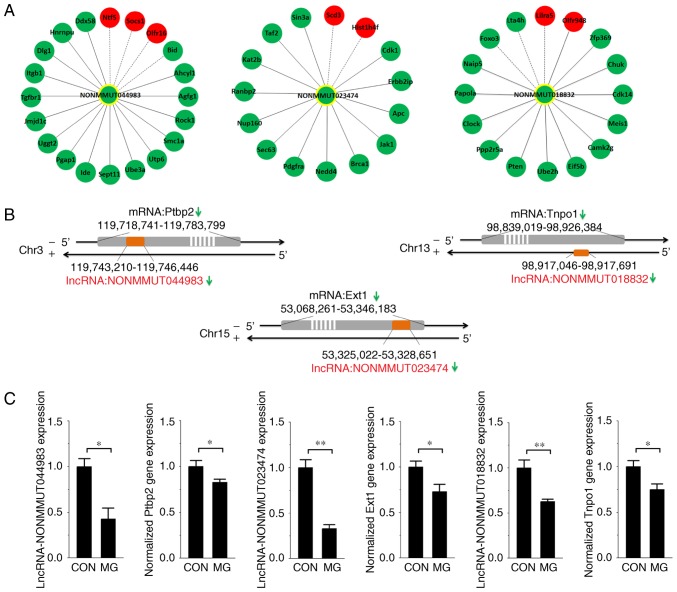
In the co-expression network, degree centrality was considered to be the simplest and most important measure by which to estimate the core regulatory factors. In order to obtain more accurate candidate lncRNAs, the degree centrality of the 17 lncRNAs whose nearby genes had been reported to be associated with osteoblast function was calculated. The lncRNAs NONMMUT044983, NONMMUT023474 and NONMMUT018832 were screened out via the degree centrality analysis (degree >15). Their associations with deregulated mRNAs were obtained from the overall co-expression network and rebuilt (Fig. 4A). The locations of the lncRNAs compared with mRNAs on the chromosome are exhibited in Fig. 4B. In addition, RT-qPCR analysis was performed to determine the expression of the three lncRNAs and their nearby genes in MC3T3-E1 cells exposed to the RWVB clinostat for 48 h. The results of the present study demonstrated that the expression of each lncRNA and mRNA was significantly decreased under simulated microgravity (Fig. 4C), consistent with the microarray data.
Figure 4.
Association between target lncRNAs and co-expressed or nearby genes. (A) The co-expression network was rebuilt between the target lncRNAs and their co-expressed deregulated mRNAs. Red nodes represent upregulated genes and green nodes represent downregulated genes. A solid line between two nodes represents a positive association while a dotted line represents a negative association. (B) The positional association on the chromosome of the target lncRNAs and their adjacent protein-coding genes. (C) Reverse transcription-quantitative polymerase chain reaction analysis of lncRNAs and their nearby genes in MC3T3-E1 cells exposed to simulated microgravity for 48 h. n=3. *P<0.05, **P<0.01. lncRNA, long noncoding RNA; Ptbp2, polypyrimidine tract-binding protein 2; Ext1, exostosin-1; Tnpo1, transportin-1; CON, control; MG, microgravity.
Discussion
Osteoblasts, the main functional cells of bone formation, have been reported to be associated with the bone loss induced by microgravity (38,39). A number of important regulators or genes have been studied to examine how microgravity may affect osteoblast function. In the present study, global genome analysis was performed and it was observed that 857 lncRNAs and 2,264 mRNAs were significantly altered in MC3T3-E1 cells exposed to simulated microgravity. Bioinformatic analyses were performed to identify potential functional lncRNAs. To the best of our knowledge, the present study is the first to suggest that lncRNAs may serve important roles in osteoblast differentiation under simulated microgravity conditions and provide a novel perspective on the effects of microgravity on osteoblast function.
Previous studies have revealed that a number of lncRNAs are not transcriptional noise and serve important regulatory roles (11,13). However, unlike the relatively definite regulatory mechanisms of miRNAs, lncRNA function may not be accurately predicted from the sequence or structure. A previous study demonstrated that the potential function of lncRNA may be inferred from their co-expressed genes (40). In the present study, a relatively consistent variation pattern, except for on chromosome 7, was observed between the differentially-expressed lncRNAs and mRNAs, suggesting that they were more co-expressed or inversely-expressed than may be expected by chance. The co-expression regulatory network was built according to the normalized signal intensity of the differentially-expressed lncRNAs and mRNAs. The co-expressed lncRNAs/genes in the network were hypothesized to be controlled by the same transcriptional regulatory pathway, to be functionally similar, or to be members of the same pathway or protein complex (41). Therefore, the functions of the deregulated lncRNAs may cover the biological process of the cell cycle, regulation of gene expression or cell proliferation, which were inferred from functional enrichment analysis of their co-expressed genes.
Apart from expression-based approaches, the genomic context of lncRNAs is frequently studied in order to elucidate their functions. An important localized regulatory mechanism through which lncRNAs may affect the expression of nearby protein-coding genes has been confirmed (42,43). For example, lncRNA-Evf2, generated from the homeobox protein Dlx-5/6 ultra conserved region, is able to act concurrently with the transcription factor Dlx2 to increase the transcriptional activity of the Dlx-5/6 enhancer in a target- and homeodomain-specific manner (43). Studying the nearby coding mRNAs may enhance the understanding of the function and potential regulatory mechanisms of lncRNAs. In the present study, 132 differentially-expressed lncRNAs and their nearby coding mRNA pairs were identified. The 132 differentially-expressed mRNAs were involved in the significant biological processes of the cell cycle, nervous system development and response to growth factors, or in the signaling pathways of chronic myeloid leukemia, protein digestion and absorption, and control of gene expression by vitamin D receptor. In particular, the vitamin D receptor pathway is associated with osteogenic development (44). Certain deregulated lncRNAs were more likely to affect osteoblast function by acting on their nearby osteoblast-associated mRNAs. Notably, 15 of the nearby mRNAs have been previously reported to be involved in osteoblast development. The corresponding 17 lncRNAs are therefore important candidates for examining the function of lncRNAs in osteoblast development.
To identify the most likely functional lncRNAs, degree centrality within the co-expression network was considered. Larger degree centrality values indicate an increased possibility of a regulatory role. Therefore, the lncRNAs NONMMUT044983, NONMMUT018832 and NONMMUT023474 were screened out via degree centrality at the forefront. According to the degree of functional association and similarity between co-expression genes, it may be possible to infer the potential function of these lncRNAs from their co-expressed genes, although GO analysis was not feasible due to the insufficient sample size. Alternative analyses of the functions of the three lncRNAs focused on the nearby genes. Their nearby coding mRNAs exhibited an association with osteoblast development, according to previous literature. Ptbp2 regulates the mutually exclusive exons 8a and 8 in the CaV1.2 calcium channel transcript (25), and the latter serves fundamental roles in cellular responses to external stimuli, including mechanical forces and hormonal signals, in osteoblastic lineage bone cells (45,46). Tnpo1 is able to facilitate the nuclear translocation of oxytocin receptors and result in osteoblast maturation. The knockdown of Tnpo1 may abrogate the oxytocin-induced expression of osteoblast differentiation genes Osx, cyclic AMP-dependent transcription factor Atf4 and osteocalcin (35). In addition, it has been reported that conditional ablation of Ext1 may lead to dysregulation of BMP signaling and severe skeletal defects (36). Microarray and RT-qPCR analyses suggested that the expression of the three lncRNAs and their nearby genes were significantly decreased under simulated microgravity. Therefore, it was hypothesized that downregulation of these lncRNAs may abrogate the expression of their nearby osteoblast-associated genes and result in inhibited osteogenesis under simulated microgravity. Future studies are required to investigate the potential regulatory mechanisms.
In conclusion, 857 differentially-expressed lncRNAs were identified in the present study when the osteo-differentiating MC3T3-E1 cells were exposed to simulated microgravity. A number of potential functional lncRNAs, and the possible regulatory mechanisms through which lncRNAs may control osteoblast differentiation in a microgravity environment were predicted using bioinformatics analysis. Although further experiment studies are required to test this hypothesis, the results of the present study demonstrated that lncRNAs are novel and potent candidates for studies into the effects and mechanism of microgravity in osteoblast function.
Acknowledgements
The present study was supported by grants from the National Natural Science Foundation of China (grant nos. 31570939, 81701856, 81471815 and 31170889).
References
- 1.Bloomfield SA. Disuse osteopenia. Curr Osteoporos Rep. 2010;8:91–97. doi: 10.1007/s11914-010-0013-4. [DOI] [PubMed] [Google Scholar]
- 2.Ohshima H. Secondary osteoporosis UPDATE. Bone loss due to bed rest and human space flight study. Clin Calcium. 2010;20:709–716. (In Japanese) [PubMed] [Google Scholar]
- 3.Morey ER, Baylink DJ. Inhibition of bone formation during space flight. Science. 1978;201:1138–1141. doi: 10.1126/science.150643. [DOI] [PubMed] [Google Scholar]
- 4.Zayzafoon M, Gathings WE, McDonald JM. Modeled microgravity inhibits osteogenic differentiation of human mesenchymal stem cells and increases adipogenesis. Endocrinology. 2004;145:2421–2432. doi: 10.1210/en.2003-1156. [DOI] [PubMed] [Google Scholar]
- 5.Rodionova NV. The dynamics of proliferation and differentiation of osteogenic cells under supportive unloading. Tsitol Genet. 2011;45:22–27. [PubMed] [Google Scholar]
- 6.Hu LF, Li JB, Qian AR, Wang F, Shang P. Mineralization initiation of MC3T3-E1 preosteoblast is suppressed under simulated microgravity condition. Cell Biol Int. 2015;39:364–372. doi: 10.1002/cbin.10391. [DOI] [PubMed] [Google Scholar]
- 7.Capulli M, Rufo A, Teti A, Rucci N. Global transcriptome analysis in mouse calvarial osteoblasts highlights sets of genes regulated by modeled microgravity and identifies a ‘mechanoresponsive osteoblast gene signature’. J Cell Biochem. 2009;107:240–252. doi: 10.1002/jcb.22120. [DOI] [PubMed] [Google Scholar]
- 8.Patel MJ, Liu W, Sykes MC, Ward NE, Risin SA, Risin D, Jo H. Identification of mechanosensitive genes in osteoblasts by comparative microarray studies using the rotating wall vessel and the random positioning machine. J Cell Biochem. 2007;101:587–599. doi: 10.1002/jcb.21218. [DOI] [PubMed] [Google Scholar]
- 9.Claverie JM. Fewer genes, more noncoding RNA. Science. 2005;309:1529–1530. doi: 10.1126/science.1116800. [DOI] [PubMed] [Google Scholar]
- 10.Kapranov P, Cheng J, Dike S, Nix DA, Duttagupta R, Willingham AT, Stadler PF, Hertel J, Hackermüller J, Hofacker IL, et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007;316:1484–1488. doi: 10.1126/science.1138341. [DOI] [PubMed] [Google Scholar]
- 11.Mercer TR, Dinger ME, Mattick JS. Long noncoding RNAs: Insights into functions. Nat Rev Genet. 2009;10:155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 12.Brosnan CA, Voinnet O. The long and the short of noncoding RNAs. Curr Opin Cell Biol. 2009;21:416–425. doi: 10.1016/j.ceb.2009.04.001. [DOI] [PubMed] [Google Scholar]
- 13.Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 14.Zhuang W, Ge X, Yang S, Huang M, Zhuang W, Chen P, Zhang X, Fu J, Qu J, Li B. Upregulation of lncRNA MEG3 promotes osteogenic differentiation of mesenchymal stem cells from multiple myeloma patients by targeting BMP4 transcription. Stem Cells. 2015;33:1985–1997. doi: 10.1002/stem.1989. [DOI] [PubMed] [Google Scholar]
- 15.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 16.Hu Z, Wang Y, Sun Z, Wang H, Zhou H, Zhang L, Zhang S, Cao X. miRNA-132-3p inhibits osteoblast differentiation by targeting Ep300 in simulated microgravity. Sci Rep. 2015;5:18655. doi: 10.1038/srep18655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: Tool for the unification of biology. The gene ontology consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Draghici S, Khatri P, Tarca AL, Amin K, Done A, Voichita C, Georgescu C, Romero R. A systems biology approach for pathway level analysis. Genome Res. 2007;17:1537–1545. doi: 10.1101/gr.6202607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.da W Huang, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 20.Stuart JM, Segal E, Koller D, Kim SK. A gene-coexpression network for global discovery of conserved genetic modules. Science. 2003;302:249–255. doi: 10.1126/science.1087447. [DOI] [PubMed] [Google Scholar]
- 21.Prieto C, Risueño A, Fontanillo C, De las Rivas J. Human gene coexpression landscape: Confident network derived from tissue transcriptomic profiles. PLoS One. 2008;3:e3911. doi: 10.1371/journal.pone.0003911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barabási AL, Oltvai ZN. Network biology: Understanding the cell's functional organization. Nat Rev Genet. 2004;5:101–113. doi: 10.1038/nrg1272. [DOI] [PubMed] [Google Scholar]
- 23.Hong D, Chen HX, Yu HQ, Liang Y, Wang C, Lian QQ, Deng HT, Ge RS. Morphological and proteomic analysis of early stage of osteoblast differentiation in osteoblastic progenitor cells. Exp Cell Res. 2010;316:2291–2300. doi: 10.1016/j.yexcr.2010.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Verlinden L, Kriebitzsch C, Beullens I, Tan BK, Carmeliet G, Verstuyf A. Nrp2 deficiency leads to trabecular bone loss and is accompanied by enhanced osteoclast and reduced osteoblast numbers. Bone. 2013;55:465–475. doi: 10.1016/j.bone.2013.03.023. [DOI] [PubMed] [Google Scholar]
- 25.Tang ZZ, Sharma S, Zheng S, Chawla G, Nikolic J, Black DL. Regulation of the mutually exclusive exons 8a and 8 in the CaV1.2 calcium channel transcript by polypyrimidine tract-binding protein. J Biol Chem. 2011;286:10007–10016. doi: 10.1074/jbc.M110.208116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lekva T, Ueland T, Bøyum H, Evang JA, Godang K, Bollerslev J. TXNIP is highly regulated in bone biopsies from patients with endogenous Cushing's syndrome and related to bone turnover. Eur J Endocrinol. 2012;166:1039–1048. doi: 10.1530/EJE-11-1082. [DOI] [PubMed] [Google Scholar]
- 27.Ogasawara T, Kawaguchi H, Jinno S, Hoshi K, Itaka K, Takato T, Nakamura K, Okayama H. Bone morphogenetic protein 2-induced osteoblast differentiation requires Smad-mediated down-regulation of Cdk6. Mol Cell Biol. 2004;24:6560–6568. doi: 10.1128/MCB.24.15.6560-6568.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fan D, Liu S, Jiang S, Li Z, Mo X, Ruan H, Zou GM, Fan C. The use of SHP-2 gene transduced bone marrow mesenchymal stem cells to promote osteogenic differentiation and bone defect repair in rat. J Biomed Mater Res A. 2016;104:1871–1881. doi: 10.1002/jbm.a.35718. [DOI] [PubMed] [Google Scholar]
- 29.Sun Z, Cao X, Hu Z, Zhang L, Wang H, Zhou H, Li D, Zhang S, Xie M. miR-103 inhibits osteoblast proliferation mainly through suppressing Cav1.2 expression in simulated microgravity. Bone. 2015;76:121–128. doi: 10.1016/j.bone.2015.04.006. [DOI] [PubMed] [Google Scholar]
- 30.Chang SF, Chang TK, Peng HH, Yeh YT, Lee DY, Yeh CR, Zhou J, Cheng CK, Chang CA, Chiu JJ. BMP-4 induction of arrest and differentiation of osteoblast-like cells via p21 CIP1 and p27 KIP1 regulation. Mol Endocrinol. 2009;23:1827–1838. doi: 10.1210/me.2009-0143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yano H, Hamanaka R, Nakamura-Ota M, Adachi S, Zhang JJ, Matsuo N, Yoshioka H. Sp7/Osterix induces the mouse pro-α2 (I) collagen gene (Col1a2) expression via the proximal promoter in osteoblastic cells. Biochem Biophys Res Commun. 2014;452:531–536. doi: 10.1016/j.bbrc.2014.08.100. [DOI] [PubMed] [Google Scholar]
- 32.Haasper C, Jagodzinski M, Drescher M, Meller R, Wehmeier M, Krettek C, Hesse E. Cyclic strain induces FosB and initiates osteogenic differentiation of mesenchymal cells. Exp Toxicol Pathol. 2008;59:355–363. doi: 10.1016/j.etp.2007.11.013. [DOI] [PubMed] [Google Scholar]
- 33.Choi YH, Han Y, Lee SH, Jin YH, Bahn M, Hur KC, Yeo CY, Lee KY. Cbl-b and c-Cbl negatively regulate osteoblast differentiation by enhancing ubiquitination and degradation of Osterix. Bone. 2015;75:201–209. doi: 10.1016/j.bone.2015.02.026. [DOI] [PubMed] [Google Scholar]
- 34.Yi S, Yu M, Yang S, Miron RJ, Zhang Y. Tcf12, A member of basic helix-loop-helix transcription factors, mediates bone marrow mesenchymal stem cell osteogenic differentiation in vitro and in vivo. Stem Cells. 2017;35:386–397. doi: 10.1002/stem.2491. [DOI] [PubMed] [Google Scholar]
- 35.Di Benedetto A, Sun L, Zambonin CG, Tamma R, Nico B, Calvano CD, Colaianni G, Ji Y, Mori G, Grano M, et al. Osteoblast regulation via ligand-activated nuclear trafficking of the oxytocin receptor; Proc Natl Acad Sci USA; 2014; pp. 16502–16507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Matsumoto Y, Matsumoto K, Irie F, Fukushi J, Stallcup WB, Yamaguchi Y. Conditional ablation of the heparan sulfate-synthesizing enzyme Ext1 leads to dysregulation of bone morphogenic protein signaling and severe skeletal defects. J Biol Chem. 2010;285:19227–19234. doi: 10.1074/jbc.M110.105338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lu J, Qu S, Yao B, Xu Y, Jin Y, Shi K, Shui Y, Pan S, Chen L, Ma C. Osterix acetylation at K307 and K312 enhances its transcriptional activity and is required for osteoblast differentiation. Oncotarget. 2016;7:37471–37486. doi: 10.18632/oncotarget.9650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kacena MA, Todd P, Landis WJ. Osteoblasts subjected to spaceflight and simulated space shuttle launch conditions. In Vitro Cell Dev Biol Anim. 2003;39:454–459. doi: 10.1290/1543-706X(2003)039<0454:OSTSAS>2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 39.Bucaro MA, Fertala J, Adams CS, Steinbeck M, Ayyaswamy P, Mukundakrishnan K, Shapiro IM, Risbud MV. Bone cell survival in microgravity: Evidence that modeled microgravity increases osteoblast sensitivity to apoptogens. Ann N Y Acad Sci. 2004;1027:64–73. doi: 10.1196/annals.1324.007. [DOI] [PubMed] [Google Scholar]
- 40.Necsulea A, Soumillon M, Warnefors M, Liechti A, Daish T, Zeller U, Baker JC, Grützner F, Kaessmann H. The evolution of lncRNA repertoires and expression patterns in tetrapods. Nature. 2014;505:635–640. doi: 10.1038/nature12943. [DOI] [PubMed] [Google Scholar]
- 41.Matthew T Weirauch. Gene coexpression networks for the analysis of DNA microarray data. In: Dehmer M, Emmert-Streib F, Graber A, Salvador A, editors. Applied Statistics for Network Biology: Methods in Systems Biology. Wiley-VCH Verlag GmbH & Co. KGaA; Weinheim, Germany: 2011. [Google Scholar]
- 42.Martianov I, Ramadass A, Barros A Serra, Chow N, Akoulitchev A. Repression of the human dihydrofolate reductase gene by a non-coding interfering transcript. Nature. 2007;445:666–670. doi: 10.1038/nature05519. [DOI] [PubMed] [Google Scholar]
- 43.Feng J, Bi C, Clark BS, Mady R, Shah P, Kohtz JD. The Evf-2 noncoding RNA is transcribed from the Dlx-5/6 ultraconserved region and functions as a Dlx-2 transcriptional coactivator. Genes Dev. 2006;20:1470–1484. doi: 10.1101/gad.1416106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Olivares-Navarrete R, Sutha K, Hyzy SL, Hutton DL, Schwartz Z, McDevitt T, Boyan BD. Osteogenic differentiation of stem cells alters vitamin D receptor expression. Stem Cells Dev. 2012;21:1726–1735. doi: 10.1089/scd.2011.0411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Duncan RL, Akanbi KA, Farach-Carson MC. Calcium signals and calcium channels in osteoblastic cells. Semin Nephrol. 1998;18:178–190. [PubMed] [Google Scholar]
- 46.Iqbal J, Zaidi M. Molecular regulation of mechanotransduction. Biochem Biophys Res Commun. 2005;328:751–755. doi: 10.1016/j.bbrc.2004.12.087. [DOI] [PubMed] [Google Scholar]