Abstract
The present study aimed to reveal the potential genes associated with the pathogenesis of intervertebral disc degeneration (IDD) by analyzing microarray data using bioinformatics. Gene expression profiles of two regions of the intervertebral disc were compared between patients with IDD and controls. GSE70362 containing two groups of gene expression profiles, 16 nucleus pulposus (NP) samples from patients with IDD and 8 from controls, and 16 annulus fibrosus (AF) samples from patients with IDD and 8 from controls, was downloaded from the Gene Expression Omnibus database. A total of 93 and 114 differentially expressed genes (DEGs) were identified in NP and AF samples, respectively, using a limma software package for the R programming environment. Gene Ontology (GO) function enrichment analysis was performed to identify the associated biological functions of DEGs in IDD, which indicated that the DEGs may be involved in various processes, including cell adhesion, biological adhesion and extracellular matrix organization. Pathway enrichment analysis using the Kyoto Encyclopedia of Genes and Genomes (KEGG) demonstrated that the identified DEGs were potentially involved in focal adhesion and the p53 signaling pathway. Further analysis revealed that there were 35 common DEGs observed between the two regions (NP and AF), which may be further regulated by 6 clusters of microRNAs (miRNAs) retrieved with WebGestalt. The genes in the DEG-miRNA regulatory network were annotated using GO function and KEGG pathway enrichment analysis, among which extracellular matrix organization was the most significant disrupted biological process and focal adhesion was the most significant dysregulated pathway. In addition, the result of protein-protein interaction network modules demonstrated the involvement of inflammatory cytokine interferon signaling in IDD. These findings may not only advance the understanding of the pathogenesis of IDD, but also identify novel potential biomarkers for this disease.
Keywords: intervertebral disc degeneration, differentially expressed genes, enrichment analysis, microRNA regulatory network
Introduction
Intervertebral disc degeneration (IDD) is characterized by increased extracellular matrix breakdown and abnormal matrix synthesis leading to reduced hydration, loss of disc height, and decreased ability to absorb load (1,2). It is considered to be the primary source of chronic lower back pain and spine-associated disease, which leads to major economic and social burdens that affect millions of individuals globally (3). The major clinical manifestations of IDD are disc herniation, vertebral instability and spinal stenosis. The ability to treat IDD effectively is hindered by an incomplete understanding of the biological processes that control intervertebral disc development, function and disease. At present, IDD continues to be treated with symptomatic interventions, which do not lead to substantially improved outcomes, as no disease-modifying drugs are currently available (4). Consequently, the clinical management of IDD pathologies remains severely limited, with no options at present for early intervention or predictive patient screening. Therefore, an improved understanding of the pathophysiology and molecular mechanisms underlying IDD is essential for diagnosis and the development of novel therapeutic approaches.
Although the etiology of IDD is likely to be multifactorial, genetic factors are considered to be the greatest contributors (5). Recently, the molecular basis of degenerative disc disease has received increased attention in research, which has substantially improved the understanding of the biology underlying this process. Studies that employed classic experimental approaches to investigate the molecular changes associated with the pathophysiology of IDD have established criteria to define degenerative intervertebral discs (5,6). While helpful, these criteria involve relatively few factors. In recent years, there has been an increase in the use of transcriptomic approaches to identify the large spectrum of factors that exhibit altered expression during IDD. For instance, Chen et al (7) identified mitogen-activated protein kinase kinase 6 and Rho-related BTB domain-containing 2 as two specific therapeutic molecular targets in the treatment of IDD. Periostin was proven to be upregulated in the progression of human IDD (8). Furthermore, high-throughput screening of human patient samples may identify potential biomarkers of IDD, leading to more precise diagnostic criteria, classification of disease progression and prognosis (9).
The intervertebral disc is composed of specialized connective tissue structures that link adjacent vertebral bodies along the spine and confer flexibility and mechanical stability to the body trunk during axial compression. There are three morphologically distinct regions in the intervertebral disc; the nucleus pulposus (NP), annulus fibrosis (AF) and cartilaginous endplates (10). Previous microarray analysis of mRNA isolated from AF cells identified differential expression of insulin-like growth factor binding protein 3 and interferon-induced protein with tetratricopeptide repeats 3 in the AF of IDD samples when compared with the control samples (11). However, the results obtained were limited as the study did not contain samples from NP, which is an important region of the human intervertebral disc. Therefore, the reanalysis of the gene expression profile by applying bioinformatics methods remains necessary to identify differentially expressed genes (DEGs) in IDD and further elucidate the potential pathogenesis mechanisms of the disease.
The present study aimed to identify the DEGs and further analyze their functions and pathways associated with the progression of IDD by utilizing a bioinformatics method to analyze microarray expression profiles from the NP and AF, and to obtain additional insights regarding the mechanisms of IDD.
Materials and methods
Microarray data
The gene expression dataset GSE70362 was downloaded from the Gene Expression Omnibus database (http://www.ncbi.nlm.nih.gov/geo) (11). It contained two groups of gene expression profiles, including 16 NP samples from patients with IDD and 8 from controls, and 16 AF samples from patients with IDD and 8 from controls. The platform was GPL17810 [HG-U133_Plus_2] Affymetrix Human Genome U133A Plus 2.0 array (Affymetrix; Thermo Fisher Scientific, Inc., Waltham, MA, USA). Probe annotation files were also acquired.
Preprocessing and differential analysis
Raw data were converted into a recognizable format with the package affy of R (http://bioconductor.org/packages/release/bioc/html/affy.html, version 1.54.0), and missing values were then inferred by a method based on k-nearest neighbors (k-NN). The k-NN-based method selects genes with expression profiles similar to the gene of interest to infer missing values (12). Following background correction and data normalization with the median method (13), differential analysis between degeneration samples and controls was performed using the limma package (version 3.32.5) (14). The design matrix indicates which RNA samples have been applied to each array, and the contrast matrix specifies the comparisons one would like to make between the RNA samples. For statistical analysis and the assessment of differential expression, limma employs an empirical Bayes method to moderate the standard errors of the estimated log-fold changes. The basic statistic used for significance analysis is the moderated t-statistic, which is computed for each probe and for each contrast. Moderated t-statistics lead to P-values in the same way as ordinary t-statistics, except that the degrees of freedom are increased, reflecting the greater reliability associated with the smoothed standard errors. Limma includes the functions top Table and decide Tests, which summarize the results of the linear model, perform hypothesis tests and adjust the P-values for multiple testing. The results obtained include (log) fold changes, standard errors, t-statistics and P-values (15). Log |(fold change)| >1 and P<0.05 were set as the cut-offs to screen out DEGs.
Gene Ontology (GO) functional enrichment analysis of DEGs
In order to identify disturbed biological functions in IDD and to understand the importance of genes, GO classification was performed, which included the following categories: BP_Fat (biological process); CC_Fat (cellular component); and MF_Fat (molecular function). GO functional enrichment analysis was performed for DEGs using the Database for Annotation, Visualization and Integrated Discovery (DAVID; https://david.ncifcrf.gov/) with a threshold of P<0.05 (16). DAVID provides a comprehensive set of functional annotation tools for the investigation of the biological context of large lists of genes.
Pathway enrichment analysis of DEGs
The Kyoto Encyclopedia of Genes and Genomes (KEGG; http://www.genome.jp/kegg) database is a collection of online databases consisting of genomes, enzymatic pathways and biological chemicals (17). The present study performed KEGG pathway enrichment analysis to determine the function of DEGs using KOBAS 2.0 with a threshold of P<0.05. KOBAS 2.0 is a web server that provides a comprehensive functional annotation tool for associating biological pathways with genes based on mapping to genes with established annotations, and also performs statistical testing to identify statistically significantly enriched pathways and diseases (18).
Comparison of DEGs between NP and AF
Common DEGs from two regions of intervertebral disc (NP and AF) were obtained using the package Venn of R verson:1.2 (https://cran.r-project.org/web/packages/venn/index.html). A Venn diagram demonstrated the numbers of genes significant in each comparison and extracted the same genes from them.
Protein-protein interaction network modules construction
Although a number of enrichment analysis tools support protein-protein interaction network-based enrichment analysis, the majority typically rely on gene sets derived from network decomposition at a single level, without considering the hierarchical structure of the network. However, it is established that hierarchical organization is a critical intrinsic property of complex systems such as biological networks (19). The present study employed WebGestalt (http://www.webgestalt.org) (20) to construct hierarchical protein interaction network modules with common DEGs and modules with adjusted P<0.05 were selected.
Establishing interaction network between common DEGs and microRNAs (microRNAs/miRs)
The miRNAs that targeted the common DEGs were retrieved with WebGestalt. WebGestalt is based on an Oracle relational database, GeneKeyDB (http://www.webgestalt.org/option.php). It supports 7 gene identifiers from a number of public databases for human and mouse, including Entrez Gene ID, Gene Symbol, RefSeq for DNA, RefSeq for Protein, Unigene, Ensemble ID and Uniprot ID, and it has added regulatory modules defined as sets of genes sharing common transcription factors or microRNA binding sites, which have been inferred from comparative genomic analysis and made available through MSigDB (19). miRNAs with adjusted P<0.05 were selected. The regulatory network between DEGs and miRNAs, and interactions between DEGs were then visualized with Cytoscape version 3.4.0 (http://www.cytoscape.org).
Results
Identification of DEGs
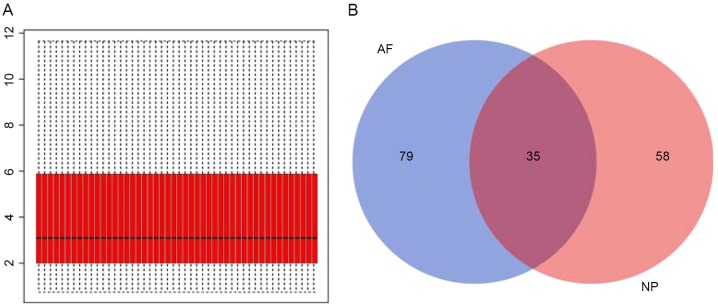
Following gene expression data normalization (Fig. 1A), 93 NP and 114 AF DEGs were identified by comparison between the samples from patients with IDD and controls. As demonstrated in Fig. 1B, 35 common DEGs present in NP and AF of patients with IDD were extracted from these identified DEGs (Table I).
Figure 1.
(A) Box plot for normalized gene expression data. The medians (black lines) are almost at the same level, indicating a good performance of normalization. (B) Venn diagram of differentially expressed genes identified from the NP and AF of degenerative intervertebral discs. NP, nucleus pulposus; AF, annulus fibrosus.
Table I.
The 35 common differentially expressed genes present in the nucleus pulposus and annulus fibrosis of patients with intervertebral disc degeneration.
| Gene symbol | Entrez_gene_ID | Gene name |
|---|---|---|
| COL6A2 | 1292 | Collagen, type VI, α2 |
| LYVE1 | 10894 | Lymphatic vessel endothelial hyaluronan receptor 1 |
| IRX5 | 10265 | Iroquois homeobox 5 |
| SOX4 | 6659 | Sex determining region Y-box 4 |
| GATA6 | 2627 | GATA binding protein 6 |
| IBSP | 3381 | Integrin-binding sialoprotein |
| VAMP8 | 8673 | Vesicle-associated membrane protein 8 (endobrevin) |
| FOXF2 | 2295 | Forkhead box F2 |
| IFIT1 | 3434 | Interferon-induced protein with tetratricopeptide repeats 1 |
| IFIT2 | 3433 | Interferon-induced protein with tetratricopeptide repeats 2 |
| SCGB2A2 | 4250 | Secretoglobin, family 2A, member 2 |
| IGFBP3 | 3486 | Insulin-like growth factor binding protein 3 |
| IFIT3 | 3437 | Interferon-induced protein with tetratricopeptide repeats 3 |
| NANOS1 | 340719 | Nanos homolog 1 (Drosophila) |
| CCND1 | 595 | Cyclin D1 |
| ZFP36L1 | 677 | Zinc finger protein 36, C3H type-like 1 |
| TFPI | 7035 | Tissue factor pathway inhibitor (lipoprotein-associated coagulation inhibitor) |
| RAP1A | 5906 | RAP1A, member of RAS oncogene family |
| ITGBL1 | 9358 | Integrin, β-like 1 (with EGF-like repeat domains) |
| S100A2 | 6273 | S100 calcium binding protein A2 |
| GBP1 | 2633 | Guanylate binding protein 1, interferon-inducible, 67 kDa |
| PTHLH | 5744 | Parathyroid hormone-like hormone |
| GREM1 | 26585 | Gremlin 1, cysteine knot superfamily, homolog (Xenopus laevis) |
| MGST1 | 4257 | Microsomal glutathione S-transferase 1 |
| Hyal1 | 3373 | Hyaluronoglucosaminidase 1 |
| IRX3 | 79191 | Iroquois homeobox 3 |
| C5ORF62 | 85027 | Mstp150 |
| SLITRK4 | 139065 | SLIT and NTRK-like family, member 4 |
| GDF15 | 9518 | Growth differentiation factor 15 |
| SCGB1D2 | 10647 | Secretoglobin, family 1D, member 2 |
| CHST10 | 9486 | Carbohydrate sulfotransferase 10 |
| LY75 | 4065 | CD302 molecule; lymphocyte antigen 75 |
| ZNF185 | 7739 | Zinc finger protein 185 (LIM domain) |
| MT1G | 4495 | Metallothionein 1G (MT1G) |
| CLEC3A | 10143 | C-type lectin domain family 3 member A (CLEC3A) |
EGF, epidermal growth factor; Mstp150, small integral membrane protein 3.
GO functional enrichment and pathway enrichment analysis of DEGs
The present study used function annotation for the DEGs, and the clustering groups were obtained by GO function enrichment analysis. A total of 24 and 50 GO terms were enriched among the DEGs in NP and AF, respectively (Fig. 2), and DEGs from NP and AF appeared to share similar biological processes, including cell adhesion, biological adhesion and extracellular matrix organization.
Figure 2.
Enriched GO terms for differentially expressed genes from the (A) nucleus pulposus and (B) annulus fibrosus. GO, Gene Ontology.
A total of 8 and 12 pathways were disclosed for DEGs by KEGG pathway enrichment analysis in NP and AF, respectively (Fig. 3), and DEGs from NP and AF contained similar pathways, including focal adhesion and the p53 signaling pathway.
Figure 3.
KEGG pathway enrichment analysis for differentially expressed genes from the (A) nucleus pulposus and (B) annulus fibrosus. ECM, extracellular matrix. KEGG, Kyoto Encyclopedia of Genes and Genomes.
Protein-protein interaction network modules
As demonstrated in Fig. 4A, the enriched directly acyclic graph revealed enriched network modules in red and their non-enriched parents in black. Enriched modules were identified at different hierarchical levels and the Hsapiens_module_922 was the most enriched hierarchical network module. To reveal associations among genes within the most enriched network module, the Cytoscape Web plug in was used to visualize in a network graph the input genes (in green) and their direct neighbors (in white). As demonstrated in Fig. 4B, interferon-induced protein with tetratricopeptide repeats (IFIT) 1, IFIT2 and IFIT3 were the hub genes of the network, which indicated that the inflammatory cytokine interferon signaling may serve an important role in degenerative human intervertebral disc.
Figure 4.
(A) Enriched protein interaction network modules in a directly acyclic graph. (B) Visualization of input genes and their direct neighbors in an enriched module using a node-link diagram. adjP, P-value following adjustment with the Benjamini-Hochberg correction method; IFIT, interferon-induced protein with tetratricopeptide repeats.
miRNAs and gene regulatory network
A total of 6 relevant clusters of miRNAs were retrieved with WebGestalt for the common DEGs (Table II). Subsequently, the miRNA-DEG regulatory network and DEG-DEG interaction network were visualized with Cytoscape (Fig. 5). Functional annotation was applied on the genes in the network, in which 11 GO terms and 4 pathways were revealed (Tables III and IV), among which the extracellular matrix organization was the most significant disrupted cellular component and focal adhesion the most significant dysregulated pathway.
Table II.
Relevant miRNAs for the 35 common differentially expressed genes.
| Parameters | |||||
|---|---|---|---|---|---|
| miRNA | DB_ID | O | rawP | adjP | Target genes |
| hsa_GTGCCAA, miR-96 | 821 | 5 | 0.0002 | 0.0052 | FOXF2, CHST10, ZFP36l1, VAMP8, NANOS1 |
| hsa_TTGCCAA, miR-182 | 757 | 4 | 0.0022 | 0.0286 | FOXF2, ZFP36l1, PTHLH, SLITRK4 |
| hsa_ATCTTGC, miR-31 | 870 | 2 | 0.0070 | 0.0385 | SLITRK4, ZFP36l1 |
| hsa_CTCAAGA, miR-526B | 779 | 2 | 0.0072 | 0.0385 | SLITRK4, ZFP36l1 |
| hsa_AAGGGAT, miR-188 | 871 | 2 | 0.0074 | 0.0385 | SLITRK4, SOX4 |
| hsa_TTTGCAC, miR-19A, miR-19B | 696 | 4 | 0.0112 | 0.0485 | FOXF2, SOX4, RAP1A, IGFBP3 |
miRNA, microRNA; O, number of differentially expressed genes regulated by the miRNA; rawP, initial P-value calculated according to the hypergeometric distribution; adjP, P-value following adjustment with the Benjamini-Hochberg correction method; miR, microRNA; FOXF2, forkhead box F2; RAP1A, RAP1A, member of RAS oncogene family.
Figure 5.
The integrated DEG-microRNA regulatory and DEG-DEG interaction network. DEGs, differentially expressed genes; miR, microRNA; COL6A2, collagen type VI α2; RAP1A, RAP1A, member of RAS oncogene family; FOXF2, forkhead box F2.
Table III.
GO functional enrichment analysis for the regulatory network.
| GO term | P-value | Gene sets |
|---|---|---|
| Extracellular matrix organization | 0.012232 | IBSP, FOXF2, COL6A2 |
| Ossification | 0.014810 | IBSP, PTHLH, IGFBP3 |
| Bone development | 0.016820 | IBSP, PTHLH, IGFBP3 |
| Wnt receptor signaling pathway through β-catenin | 0.027305 | CCND1, SOX4 |
| Extracellular structure organization | 0.028464 | IBSP, FOXF2, COL6A2 |
| Endoderm development | 0.028889 | PTHLH, GATA6 |
| Regulation of cellular protein metabolic process | 0.040149 | ZFP36L1, CCND1, NANOS1, IGFBP3 |
| Posttranscriptional regulation of gene expression | 0.045585 | ZFP36L1, NANOS1, SOX4 |
| Negative regulation of cell differentiation | 0.047545 | PTHLH, IRX3, CCND1 |
| Tube development | 0.049135 | PTHLH, GATA6, GREM1 |
| Negative regulation of signal transduction | 0.049536 | CCND1, GREM1, IGFBP3 |
GO, Gene Ontology; FOXF2, forkhead box F2; COL6A2, collagen type VI α2.
Table IV.
KEGG pathway enrichment analysis for the regulatory network.
| KEGG pathway | P-value | Gene sets |
|---|---|---|
| Focal adhesion | 0.0003 | CCND1, COL6A2, RAP1A, IBSP |
| p53 signaling pathway | 0.0055 | CCND1, IGFBP3 |
| ECM-receptor interaction | 0.0084 | COL6A2, IBSP |
| Platelet activation | 0.0453 | VAMP8, RAP1A |
KEGG, Kyoto Encyclopedia of Genes and Genomes; ECM, extracellular matrix; COL6A2, collagen type VI α2; RAP1A, RAP1A, member of RAS oncogene family.
Discussion
Despite years of investigation, the pathogenesis underlying IDD remains poorly understood and continues to require further investigation. The emergence of bioinformatics methods has accelerated the progress of research on the mechanisms of human disease. The present study identified 93 and 114 DEGs in the NP and AF respectively, through the comparative analysis of the transcriptome between degenerative intervertebral disc samples and controls. The analysis identified 35 common DEGs in the two regions, and protein-protein interaction network modules demonstrated that the inflammatory cytokine interferon signaling may serve an important role in human IDD. In addition, a total of 6 associated miRNAs (miR-96, miR-182, miR-31, miR-526B, miR-188 and miR-19) were identified, which targeted these common DEGs.
Alterations in the production of extracellular matrix and inflammatory cytokines by intervertebral discs have an important role in the pathogenesis of IDD (5). In the present study, functional enrichment analysis indicated a number of DEGs involved in biological processes, including cell adhesion, biological adhesion and extracellular matrix organization, in the two regions of the intervertebral disc. Pathway enrichment analysis demonstrated that focal adhesion and the p53 signaling pathway were disrupted in NP and AF. In addition, 3 common DEGs in the regulatory network were enriched in extracellular matrix organization and 4 common DEGs in the regulatory network were enriched in the focal adhesion signaling pathway.
The extracellular matrix is a component of all mammalian tissues, and is a network that consists predominantly of the fibrous proteins collagen, elastin and fibronectin. In addition to a structural function, the extracellular matrix exhibits a number of other roles; as a major component of the cellular microenvironment, it affects various cell behaviors, which include proliferation, adhesion and migration, and also regulates cell differentiation and death (21). Extracellular matrix composition is particularly heterogeneous and dynamic, and abnormal extracellular matrix dynamics may lead to dysregulated cell proliferation, cell death failure and loss of cell differentiation, which subsequently results in congenital defects and pathological processes such as tissue fibrosis and cancer (22). It has been reported that during IDD, the ability of intervertebral disc cells to produce extracellular matrix reduces, however, the production of degradative enzymes does not change; this phenomenon is hypothesized to accelerate the degeneration by degrading the extracellular matrix of the disc, ultimately resulting in the macroscopic changes of the intervertebral disc (6).
Collagen, which gives tissues the ability to recover following stretching, is the most abundant fibrous protein within the extracellular matrix. It has been identified that the integrin-binding sialoprotein (IBSP) interacts with collagen and appears to modulate cell-matrix interactions (23). Cell-matrix adhesions have essential roles in a number of important biological processes, including cell motility, proliferation and differentiation, and the regulation of gene expression and cell survival; at contact points between the cell and extracellular matrix, specialized structures termed focal adhesions are formed (24).
The RAP1A gene encodes a member of the Ras family of small GTPases. Alterations in the conformation and activity of the protein encoded by RAP1A occur depending on whether GTP or GDP is bound to the protein, which are involved in regulating signaling pathways that affect cell proliferation and adhesion (25,26). Therefore, the dysregulation of collagen type VI α2 (COL6A2), IBSP and RAP1A in human intervertebral discs may induce de-adhesion, characterized by disruption of extracellular matrix organization and focal adhesions, which accelerates the degeneration of intervertebral discs.
Since the discovery of interferons, they have been investigated widely in a large number of studies, and considerable progress has been made in describing the nature of the cytokines themselves (27). Originally, interferons were known for their antiviral properties, however, interferons are currently better known for their distinct cellular functions, which include inhibition of proliferation and angiogenesis, induction of differentiation and regulation of the immune system (28). Previous research has demonstrated that IFIT3 may lead to AF cell growth arrest via its antiproliferative activity, which negatively regulates the cell cycle and directly or indirectly induces cell apoptosis (11,29,30). This is similar to the results of the present study. However, the difference and innovation of the present study primarily lies in the different groups included in the differential analysis. The present study included two sets of differential analysis, between the NP and control groups and the AF and control groups, respectively, while in the study by Kazezian et al (11), the differential analysis was only conducted between the NP and AF groups, and so the DEGs were not exactly the same. According to the results of protein-protein interaction network modules presented in the present study, three interferon-induced genes (IFIT1, IFIT2 and IFIT3) were enriched in the degenerative human discs. Therefore, based on the above information, the upregulated IFITs may negatively regulate the cell cycle, and thus reduce the disc cell number, subsequently accelerating degeneration.
miRNAs are considered to serve a crucial role in gene expression, which affects numerous biological processes, including cell differentiation, proliferation, metabolism, apoptosis and tumorigenesis, and have become therapeutic targets for diseases such as IDD (31–33). To investigate the potential molecular targets, miRNAs interacting with the common DEGs were retrieved in the present study and a regulatory network was also constructed. Aberrant expression of miR-96 or miR-182 has been reported in a number of human diseases, including pulmonary arterial hypertension and cancer (34–37). The results of the present study, and of previous studies, have demonstrated that forkhead transcription factor F2 (FOXF2) may be regulated by miR-96 or miR-182 (38,39). FOX is a super family of transcriptional regulators that exhibit numerous functions in human diseases (38–40). The FOXF subfamily consists of two members, FOXF1 and FOXF2. A previous study indicated that FOXF2 promoted extracellular matrix production, and in FOXF2 mutant animals, the extracellular matrix, particularly collagens, was severely reduced, which causes tissue disintegration (41). In addition, FOXF2 may act as a mesenchymal factor that controls cell proliferation and survival (42). Therefore, we hypothesized that FOXF2, miR-96 and miR-182 are worthy of further investigation to determine their specific roles in IDD.
In conclusion, the present study provides integrated network insight into the pathogenesis of IDD and offers potential therapeutic targets for controlling the disease. The dysregulation of COL6A2, IBSP, RAP1A and FOXF2 in NP and AF are associated with IDD progression by disrupting the extracellular matrix organization and focal adhesions pathway. In addition, IFIT1, IFIT2 and IFIT3 may negatively regulate the cell cycle, and thus decrease the number of disc cells, eventually accelerating degeneration of intervertebral discs. However, further experiments, clinical and mechanistic, are required to confirm the results of the present study.
Acknowledgements
The present study was supported by the State Key Program of National Nature Science Foundation of China (grant no. 81330042), the Special Program for Sino-Russian Joint Research Sponsored by the Ministry of Science and Technology, China (grant no. 2014DFR31210) and the Key Program Sponsored by the Tianjin Science and Technology Committee, China (grant nos. 13RCGFSY19000 and 14ZCZDSY00044).
References
- 1.Buckwalter JA. Aging and degeneration of the human intervertebral disc. Spine (Phila Pa 1976) 1995;20:1307–1314. doi: 10.1097/00007632-199506000-00022. [DOI] [PubMed] [Google Scholar]
- 2.Costi JJ, Stokes IA, Gardner-Morse MG, Iatridis JC. Frequency-dependent behavior of the intervertebral disc in response to each of six degree of freedom dynamic loading: Solid phase and fluid phase contributions. Spine (Phila Pa 1976) 2008;33:1731–1738. doi: 10.1097/BRS.0b013e31817bb116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kalichman L, Hunter DJ. The genetics of intervertebral disc degeneration. Familial predisposition and heritability estimation. Joint Bone Spine. 2008;75:383–387. doi: 10.1016/j.jbspin.2007.11.003. [DOI] [PubMed] [Google Scholar]
- 4.Friedman BW, O'Mahony S, Mulvey L, Davitt M, Choi H, Xia S, Esses D, Bijur PE, Gallagher EJ. One-week and 3-month outcomes following an emergency department visit for undifferentiated musculoskeletal low back pain. Ann Emerg Med. 2012;59:128–133. doi: 10.1016/j.annemergmed.2011.09.012. [DOI] [PubMed] [Google Scholar]
- 5.Kepler CK, Ponnappan RK, Tannoury CA, Risbud MV, Anderson DG. The molecular basis of intervertebral disc degeneration. Spine J. 2013;13:318–330. doi: 10.1016/j.spinee.2012.12.003. [DOI] [PubMed] [Google Scholar]
- 6.Gopal D, Ho AL, Shah A, Chi JH. Molecular basis of intervertebral disc degeneration. Adv Exp Med Biol. 2012;760:114–133. doi: 10.1007/978-1-4614-4090-1_8. [DOI] [PubMed] [Google Scholar]
- 7.Chen Y, Chen K, Li M, Li C, Ma H, Bai YS, Zhu XD, Fu Q. Genes associated with disc degeneration identified using microarray gene expression profiling and bioinformatics analysis. Genet Mol Res. 2013;12:1431–1439. doi: 10.4238/2013.April.26.5. [DOI] [PubMed] [Google Scholar]
- 8.Tsai TT, Lai PL, Liao JC, Fu TS, Niu CC, Chen LH, Lee MS, Chen WJ, Fang HC, Ho NY, Pang JH. Increased periostin gene expression in degenerative intervertebral disc cells. Spine J. 2013;13:289–298. doi: 10.1016/j.spinee.2013.01.040. [DOI] [PubMed] [Google Scholar]
- 9.He J, Xue R, Li S, Lv J, Zhang Y, Fan L, Teng Y, Wei H. Identification of the potential molecular targets for human intervertebral disc degeneration based on bioinformatic methods. Int J Mol Med. 2015;36:1593–1600. doi: 10.3892/ijmm.2015.2389. [DOI] [PubMed] [Google Scholar]
- 10.Choi YS. Pathophysiology of degenerative disc disease. Asian Spine J. 2009;3:39–44. doi: 10.4184/asj.2009.3.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kazezian Z, Gawri R, Haglund L, Ouellet J, Mwale F, Tarrant F, O'Gaora P, Pandit A, Alini M, Grad S. Gene expression profiling identifies interferon signalling molecules and IGFBP3 in human degenerative annulus fibrosus. Sci Rep. 2015;5:15662. doi: 10.1038/srep15662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Troyanskaya O, Cantor M, Sherlock G, Brown P, Hastie T, Tibshirani R, Botstein D, Altman RB. Missing value estimation methods for DNA microarrays. Bioinformatics. 2001;17:520–525. doi: 10.1093/bioinformatics/17.6.520. [DOI] [PubMed] [Google Scholar]
- 13.Fujita A, Sato JR, Lde O Rodrigues, Ferreira CE, Sogayar MC. Evaluating different methods of microarray data normalization. BMC Bioinformatics. 2006;7:469. doi: 10.1186/1471-2105-7-469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 3 doi: 10.2202/1544-6115.1027. Article 3, 2004. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- 16.da W Huang, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 17.Kanehisa M, Goto S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xie C, Mao X, Huang J, Ding Y, Wu J, Dong S, Kong L, Gao G, Li CY, Wei L. KOBAS 2.0: A web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 2011;39:W316–W322. doi: 10.1093/nar/gkr483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang J, Duncan D, Shi Z, Zhang B. WEB-based GEne SeT AnaLysis Toolkit (WebGestalt): Update 2013. Nucleic Acids Res. 2013;41:W77–W83. doi: 10.1093/nar/gkt439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang B, Kirov S, Snoddy J. WebGestalt: An integrated system for exploring gene sets in various biological contexts. Nucleic Acids Res. 2005;33:W741–W748. doi: 10.1093/nar/gki475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hynes RO. The extracellular matrix: Not just pretty fibrils. Science. 2009;326:1216–1219. doi: 10.1126/science.1176009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Frantz C, Stewart KM, Weaver VM. The extracellular matrix at a glance. J Cell Sci. 2010;123:4195–4200. doi: 10.1242/jcs.023820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bornstein P. Matricellular proteins: An overview. J Cell Commun Signal. 2009;3:163–165. doi: 10.1007/s12079-009-0069-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mitra SK, Hanson DA, Schlaepfer DD. Focal adhesion kinase: In command and control of cell motility. Nat Rev Mol Cell Biol. 2005;6:56–68. doi: 10.1038/nrm1549. [DOI] [PubMed] [Google Scholar]
- 25.Xiang J, Bian C, Wang H, Huang S, Wu D. miR-203 down-regulates Rap1A and suppresses cell proliferation, adhesion and invasion in prostate cancer. J Exp Clin Cancer Res. 2015;34:8. doi: 10.1186/s13046-015-0125-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mott HR, Owen D. Structures of Ras superfamily effector complexes: What have we learnt in two decades? Crit Rev Biochem Mol Biol. 2015;50:85–133. doi: 10.3109/10409238.2014.999191. [DOI] [PubMed] [Google Scholar]
- 27.Sadler AJ, Williams BR. Interferon-inducible antiviral effectors. Nat Rev Immunol. 2008;8:559–568. doi: 10.1038/nri2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brierley MM, Fish EN. Review: IFN-alpha/beta receptor interactions to biologic outcomes: Understanding the circuitry. J Interferon Cytokine Res. 2002;22:835–845. doi: 10.1089/107999002760274845. [DOI] [PubMed] [Google Scholar]
- 29.Foser S, Redwanz I, Ebeling M, Heizmann CW, Certa U. Interferon-alpha and transforming growth factor-beta co-induce growth inhibition of human tumor cells. Cell Mol Life Sci. 2006;63:2387–2396. doi: 10.1007/s00018-006-6256-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kiepe D, Ulinski T, Powell DR, Durham SK, Mehls O, Tönshoff B. Differential effects of insulin-like growth factor binding proteins-1, −2, −3 and −6 on cultured growth plate chondrocytes. Kidney Int. 2002;62:1591–1600. doi: 10.1046/j.1523-1755.2002.00603.x. [DOI] [PubMed] [Google Scholar]
- 31.Pritchard CC, Cheng HH, Tewari M. MicroRNA profiling: Approaches and considerations. Nat Rev Genet. 2012;13:358–369. doi: 10.1038/nrg3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xu YQ, Zhang ZH, Zheng YF, Feng SQ. Dysregulated miR-133a mediates loss of type II collagen by directly targeting matrix metalloproteinase 9 (MMP9) in human intervertebral disc degeneration. Spine (Phila Pa 1976) 2016;41:E717–E724. doi: 10.1097/BRS.0000000000001375. [DOI] [PubMed] [Google Scholar]
- 33.Ji ML, Zhang XJ, Shi PL, Lu J, Wang SZ, Chang Q, Chen H, Wang C. Downregulation of microRNA-193a-3p is involved in invertebral disc degeneration by targeting MMP14. J Mol Med (Berl) 2016;94:457–468. doi: 10.1007/s00109-015-1371-2. [DOI] [PubMed] [Google Scholar]
- 34.Wallace E, Morrell NW, Yang XD, Long L, Stevens H, Nilsen M, Loughlin L, Mair KM, Baker AH, MacLean MR. A sex-specific microRNA-96/5-hydroxytryptamine 1B axis influences development of pulmonary hypertension. Am J Respir Crit Care Med. 2015;191:1432–1442. doi: 10.1164/rccm.201412-2148OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu L, Pu X, Wang Q, Cao J, Xu F, Xu LI, Li K. miR-96 induces cisplatin chemoresistance in non-small cell lung cancer cells by downregulating SAMD9. Oncol Lett. 2016;11:945–952. doi: 10.3892/ol.2015.4000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yu S, Lu Z, Liu C, Meng Y, Ma Y, Zhao W, Liu J, Yu J, Chen J. miRNA-96 suppresses KRAS and functions as a tumor suppressor gene in pancreatic cancer. Cancer Res. 2010;70:6015–6025. doi: 10.1158/0008-5472.CAN-09-4531. [DOI] [PubMed] [Google Scholar]
- 37.Bian DL, Wang XM, Huang K, Zhai QX, Yu GB, Wu CH. Expression and regulatory effects of microRNA-182 in osteosarcoma cells: A pilot study. Oncol Lett. 2016;11:3040–3048. doi: 10.3892/ol.2016.4375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kundu ST, Byers LA, Peng DH, Roybal JD, Diao L, Wang J, Tong P, Creighton CJ, Gibbons DL. The miR-200 family and the miR-183~96~182 cluster target Foxf2 to inhibit invasion and metastasis in lung cancers. Oncogene. 2016;35:173–186. doi: 10.1038/onc.2015.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang Y, Wang X, Wang Z, Tang H, Fan H, Guo Q. miR-182 promotes cell growth and invasion by targeting forkhead box F2 transcription factor in colorectal cancer. Oncol Rep. 2015;33:2592–2598. doi: 10.3892/or.2015.3833. [DOI] [PubMed] [Google Scholar]
- 40.Kong PZ, Yang F, Li L, Li XQ, Feng YM. Decreased FOXF2 mRNA expression indicates early-onset metastasis and poor prognosis for breast cancer patients with histological grade II tumor. PLoS One. 2013;8:e61591. doi: 10.1371/journal.pone.0061591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Van der Heul-Nieuwenhuijsen L, Dits N, Van Ijcken W, de Lange D, Jenster G. The FOXF2 pathway in the human prostate stroma. Prostate. 2009;69:1538–1547. doi: 10.1002/pros.20996. [DOI] [PubMed] [Google Scholar]
- 42.Ormestad M, Astorga J, Landgren H, Wang T, Johansson BR, Miura N, Carlsson P. Foxf1 and Foxf2 control murine gut development by limiting mesenchymal Wnt signaling and promoting extracellular matrix production. Development. 2006;133:833–843. doi: 10.1242/dev.02252. [DOI] [PubMed] [Google Scholar]