Abstract
Two cases of liveborn unrelated children with developmental delay and overlapping unbalanced translocations der(8)t(8;16)(p23.2;q23.3) and der (8)t(8;16)(p23.1;q23.1), leading to partial monosomy 8p and partial trisomy 16q, are reported in the present study. The first patient was a 10-year-old boy with mild developmental delay and minor congenital anomalies (borderline microcephaly, clinodactyly, hypertelorism, epicanthus, mild systolic murmur and kidney reflux). The second patient was a 3 year-old girl with developmental delay, gross motor milestone delay and dysmorphic features. Array-comparative genomic hybridization analysis revealed that partial chromosome 8p monosomy extended from 8p23.2 to 8pter (4.8 Mb) in Patient 1 and from 8p23.1 to 8pter (9.5 Mb) in Patient 2, and partial chromosome 16 trisomy extended from 16q23.3 to 16qter (5.6 Mb) in Patient 1 and from 16q23.1 to 16qter (11.7 Mb) in Patient 2. The mechanism of appearance of the rearrangement in association with the genes involved and the architecture of the region is discussed.
Keywords: translocation, monosomy 8p, trisomy 16q, array comparative genomic hybridization, developmental delay, congenital abnormalities
Introduction
Monosomy 8p is a rare chromosomal disorder characterized by deletion of a part of the eighth chromosome. The incidence of the 8p23.1 deletion was estimated at 1:18,542 in amniotic fluid samples and 1:5,072 in postnatal samples (1). Since the first report of an 8p23.1 deletion by Fagan and Morris (2), >50 cases have been reported (3). The majority of the cases are not studied with high resolution molecular techniques or characterized at the molecular level (4). Interstitial deletions of the sub-band 8p23.1 have primarily been associated with facial and other phenotypic abnormalities, whereas terminal deletions are associated with heart defects (3,5). Notably, distal deletion of 8p23.2-pter has additionally been observed in apparently healthy individuals (1).
In the majority of cases, monosomy 8p appears to result from de novo errors in early embryonic development that occur for unknown reasons. Associated symptoms and findings differ between cases (6). However, in most cases clinical manifestations including growth deficiency, mental retardation, post-natal growth retardation, developmental delay and speech problems are observed. Furthermore, patients present with common signs of body and craniofacial dysmophisms, in addition to behavioral difficulties (1,3,5,6). Facial dysmorphisms, which are more remarkable in early years, include microcephaly, malformed or low set ears, arched eyebrows, depressed nasal bridge, epicanthus, strabismus, hypermetropia and/or myopia, serrated teeth, short neck and retrognathia. In addition, vertebral abnormalities are frequently observed (7–11).
It has additionally been reported that children with this chromosomal disorder present with behavioral difficulties, including aggressiveness and attention deficit disorder, and problems associated with cardiovascular and central nervous system (5,9,12). Furthermore, genito-urinary anomalies, in particular cryptorcidism and hypospadias, are observed in boys (6).
In contrast to 8p deletion syndrome, partial trisomies of the terminal 16qter are rare (1). A total of nine cases of partial distal chromosome 16 trisomy have been reported but only a few were studied with high resolution molecular techniques. Only one patient presented a pure partial trisomy 16q24.1q24.3, whereas all the others corresponded to unbalanced translocations where 16q24 was rearranged with other chromosome regions (7,8,10,11,13–15). For two of the patients, there was no detailed phenotypic information (10,15). A number of clinical characteristic features were common in all patients (low birth weight, growth retardation, intellectual disability, muscular hypotonia, small palpebral fissures, long philtrum, low set/dysplastic ears and osteochondroma), so it is difficult to characterize with precision the 16q24 trisomy phenotype or to establish a genotype-phenotype correlation (11) (Table I).
Table I.
Clinical characteristics associated with 8p23 and 16q24 regions in the literature.
| Clinical characteristics | 8p23.1 →pter | 8p23.2 →pter | 16q24.1 →qter | 16q24.1 →qter | Patient 1 | Patient 2 | Other studies | Total |
|---|---|---|---|---|---|---|---|---|
| Prematurity | − | + | ||||||
| Post-natal growth retardation | + | − | − | + | − | − | 7–12 | 7/15 |
| Low birth weight | − | − | − | + | − | − | 7,8,11 | 3/3 |
| Developmental delay | + | − | − | − | + | + | 5,9,12 | 15/20 |
| Mental retardation | + | + | − | + | − | − | 5,6,9,10 | 14/20 |
| Behavioral/neurodevelopmental (hyperactivity, aggressiveness, no self- confidence, attention deficit disorder, anxious) | + | − | − | − | + | + | 5,9,12 | 25/58 |
| Dysmorphic craniofacial features | ||||||||
| Microcephaly | + | + | − | − | + | − | 5,6,9,12 | 13/21 |
| Hypertelorism | + | + | ||||||
| Epicanthus | − | − | − | + | + | + | 7,12,14 | 4/5 |
| Broad forehead | − | + | ||||||
| Arched eyebrows | + | + | − | − | − | + | 6 | 1/1 |
| Diffuse depigmentation of retina | − | + | ||||||
| Alternating esotropia | − | + | ||||||
| Long philtrum | − | − | − | + | − | + | 10,11,14 | 3/5 |
| Thin face | − | + | ||||||
| Thin lips | + | − | − | + | − | + | 9,14 | 5/9 |
| Small mouth | − | + | ||||||
| Retrognathia | + | − | − | − | − | + | 9 | 8/8 |
| Depressed nasal bridge | + | + | − | − | − | + | 6,9 | 2/9 |
| Dysplastics/low set ears | + | − | − | + | − | + | 5,7,8,9,11,12 | 12/23 |
| Major malformations | ||||||||
| Clinodactyly | + | − | ||||||
| Laryngeal stridor/laryngomalacia | + | + | − | − | − | + | 6 | 1/1 |
| Cardiovascular system problems | + | + | − | − | + | − | 6;16 | 3/3 |
| Abdominal distension | − | + | ||||||
| Necrotizing enterocolitis | − | + | ||||||
| Genito-urinary anomalies | + | + | − | − | + | − | 5,6,9 | 9/28 |
| Central nervous system | ||||||||
| Speech problems | + | − | − | − | + | − | 5,9,12 | 5/20 |
| Dystonic posturing | − | + | ||||||
| Myelination delay | − | + |
In the present study, two cases of liveborn unrelated children with an unbalanced 8;16 translocation resulting in partial monosomy of chromosome 8 and partial trisomy of chromosome 16 were reported. The effect on the phenotype of monosomy 8 seems to be more prominent than that of trisomy 16. However, this phenotype may result from the rearranged architecture of the region, the structure and function of the genes and regions involved, and their interactions.
Patients and methods
Ethical approval
The present study was approved by the Ethics Committee of the P. & A. Kyriakou Children's Hospital (Athens, Greece) and was performed with respect to the ethical standards of the Declaration of Helsinki, as revised in 2008. Written, informed consent was obtained from the patient's families.
Patient 1
Patient 1 was a 10- and a half-year-old boy, and the second child of healthy, unrelated parents. The first child of the family is a 16-year-old healthy boy. Patient 1 was referred for developmental assessment for speech and language delay. The patient was born following an uncomplicated full term pregnancy with birth weight 3.350 kg, height 51 cm and head circumference (HC) 35 cm. The perinatal history was non-significant. At the age of 8 months the patient was diagnosed with a urinary tract infection and an X-ray investigation revealed urinary reflux (V degree), and a kidney dimercaptosuccinic acid scan revealed 20% decreased left kidney function. The developmental milestones of the patient were slightly delayed as he sat independently at the age of 9 months and walked unaided at the age of 18 months.
On developmental examination at 3 years old the patient was a sociable child, with mild dysmorphic facial and body features including microcephaly, hypertelorism, epicanthus, and clinodactyly. The patient demonstrated good ability for symbolic play and his comprehension ability was limited to one concept per sentence. His speech was limited to 3–4 simple words. His overall developmental level was equivalent to 18 months. According to the Bailey's Scales of Infant Development 2nd edition (16), his mental score was 51 and motor score was 95. Heart auscultation revealed a mild systolic murmur. On neurological examination, the patient was revealed to be slightly hypertonic with borderline microcephaly (HC=48 cm; 3%).
Echocardiography revealed a small ventricular septal defect without hemodynamic alterations. Metabolic screening revealed a mild elevation of glutamate in blood amino acids and small proteinuria involving lysine, arginin and cystin. His bone age was increased (equivalent to 6-year-old boy). Thyroid function, brain magnetic resonance imaging scans, visual and audiological examinations, urine amino acids and blood lactic acid levels were healthy.
The patient attended mainstream kindergarten and received early intervention services twice a week based on a Portage Scheme. His development was followed up at regular intervals in the Developmental Unit and was monitored according to his needs.
At the age of 3 years and 9 months, his cognitive and language skills were equivalent to the level of a 20-month-old, with severe behavioral difficulties characterized by frequent temper tantrums. At the age of 4 years, 3 months, the cognitive abilities of the patient increased to the level of a 30-month-old while his language skills remained at a 26-month level. His behavior had improved but he remained a difficult child, presenting with hyperactivity, aggressiveness and impulsiveness.
He was additionally observed at the age of 5 years and 2 months. He had made significant developmental progress and his cognitive skills were equivalent to a 4 year and 6 month level, with language skills equivalent to a 2 year and 9 month level. According to Griffiths Scales (17), his performance developmental subquotient (DQ) was 87 and his language DQ was 51. His weight was 19 kg (50th centile), his height was 107 cm (20th centile) and his HC was 49.5 cm (3rd centile). The dysmorphia of his facial features remained mild and passed unnoticed. He was well integrated in mainstream kindergarten and his parents were planning to place him in a mainstream school with extra educational help.
The patient was re-evaluated at the age of 7 years. He was well integrated into the 1st grade of mainstream primary school with special educational provision. His behavior had significantly improved and he was sociable and co-operative. His cognitive abilities were increased, with a developmental level of 5 years and 8 months with a general DQ of 86. His weight was 24 kg (25th centile), his height was 119 cm (25th centile) and his HC was 49.5 cm (below 3rd centile). Dysmorphia of his body and facial features remained mild. On neurological examination, he was revealed to be slightly hypertonic. His thyroid functions and detailed endocrinological examination (GH, IGF1, prolactin, LH, FSH, 17-OH prog., cortisol, insulin) proved normal. Previous echo-triplex results additionally proved normal.
The patient was last observed at the age of 10 years and 6 months. He was attending the 3rd grade of the same mainstream primary school with special educational support. He remained sociable with severe attention deficit disorder, impulsivity and lack of self-confidence. His cognitive deficits were more evident in reading and mathematics. His developmental level was equivalent to that of a healthy 6-year-old, with mild phonological and morphological language problems. His general DQ was 78. The dysmorphia of his body and facial features (microcephaly, hypertelorism, epicanthus and clinodactyly) was more evident. On neurological examination, he remained slightly hypertonic with brisk reflexes but without focal neurological signs. His weight was 34 kg (25th centile), his height was 140 cm (25th centile) and his HC was 49.5 cm (<3rd centile).
Patient 2
Patient 2 was a girl was born to non-consanguineous healthy parents at 36 weeks of gestational age, following a normal pregnancy and an uncomplicated delivery. Prenatal karyotype was performed due to advanced maternal age, and it was normal. The family history was unremarkable and there was no previous history of infertility or spontaneous abortion prior to this pregnancy. The birth weight was 2,400 g (25th centile), height 48 cm (75th-90th centile), and HC 30.5 cm (2nd-10th centile). Apgar scores were 9 and 10 at 1 and 5 min, respectively.
Two days following birth, the patient presented with abdominal distension and bloody stools. An X-ray revealed the presence of air outside the intestines in the abdominal cavity. Necrotizing enterocolitis with perforation was diagnosed and surgical removal of the caecum was performed, and the ileocecal valve was perforated. However, three months following surgery, she presented with intestinal obstruction caused by narrowing of the previously diseased bowel, requiring further surgical intervention. In addition, the neonatal period was complicated by laryngeal stridor due to laryngomalacia. Some dysmorphic features and dystonic posturing were noticed in early infancy. The patient acquired head control at the age of 6 months, trunk control at the age of 9 months, and autonomous deambulation at the age of 12 months. The patient started to speak at two years of age, but then stopped any further development of verbal language and developed a preference for gestural communication. Verbal comprehension was good.
Extensive studies for metabolic diseases (including blood and urine amino acids, urine organic acids, blood lactate, pyruvate and ammonia) gave normal results. Electroencephalogram, audiometric examination, cardiological evaluation including echocardiogram, X-rays of the thorax and renal ultrasound returned normal results. Ophthalmologic assessment (at 3 months of age) revealed diffuse depigmentation of the retina. Brain magnetic resonance (at 6 months of age) revealed myelination delay. At 7 months of age, the patient's height was 62 cm (10th centile), weight was 5.035 g (<3rd centile) and HC was 39.5 cm (<2nd centile). Morphological evaluation evidenced a thin face, broad forehead, low-set and posteriorly rotated ears, bilateral pits above the tragus, arched eyebrows, hypertelorism, epicanthus inversus, depressed nasal bridge, long philtrum, thin lips, small mouth with down-turned corners, and retrognathia. Neurological examination revealed developmental delay, with gross motor milestones limited to uncompleted head control. Dystonic axial posturing and fluctuating muscular tone of the four limbs was present. Alternating esotropia was additionally observed.
Cytogenetic and fluorescence in situ hybridization (FISH) analyses
Chromosome analysis was performed from 2–2.5 ml cultured blood lymphocytes using Giemsa banding and high resolution banding techniques obtained following cell culture synchronization and thymidine incorporation. FISH studies were performed using a set of probes specific for 8p (TelVysion 8p SpectrumGreen D8S504) and 16q (TelVysion 16q SpectrumOrange 16qTEL013) subtelomeres according to the manufacturer's protocol (Vysis; Abbott Molecular, Des Plaines, Illinois, USA) (18). The slides were washed and counterstained with 4′,6-diamidino-2-phenylindole, and cells were examined under a Zeiss Axioplan II, Imager.M1/Imager.Z1 fluorescence microscope equipped with a triple-bandpass filter (Zeiss GmbH, Jena, Germany). Digital images were captured and stored with Isis software version 3.4.0 (MetaSystems, Altlussheim, Germany).
Array comparative genomic hybridization (aCGH), polymerase chain reaction (PCR) and microsatellite analysis
High molecular weight genomic DNA was extracted from the patient's blood lymphocytes using aQiamp DNA Blood Midi kit (Qiagen, Inc., Valencia, CA, USA). aCGH analysis was performed with DNA from cultured amniocytes in order to characterize the extent of the deletion in Patient 1 and to justify the clinical findings in Patient 2. Molecular karyotyping was performed via oligonucleotide aCGH platforms using an 100 kb resolution array kit 44K (Agilent Technologies, Inc., Santa Clara, CA, USA). Gene dosage for 9 sequence tagged sites (STSs) from chromosome 8 was performed by PCR using the LightCycler FastStart DNA Master SYBR Green 1 Kit (Roche Diagnostics, Monza Italy), according to the manufacturer's instructions, on a Roche LightCycler 1.5 instrument (Roche Diagnostics). Primer sequences for the telomeric STSs amplified, including the genesceroid-lipofuscinosis, neuronal 8 (CLN8), CUB and Sushi multiple domains 1 (CSMD1), microcephalin 1 primary autosomal recessive 1 (MCPH1) and GATA binding protein 4 (GATA4), are listed in Table II. Altogether, the analyzed region covered ~11 Mb of DNA of the telomeric 8p region. PCR was performed using the following program: 95°C for 10 min, followed by 40 cycles of 95°C for 10 sec, 55°C for 10 sec, and 72°C for 25 sec. Copy-number/genome of each STS was evaluated by a relative quantification method using the software RelQuant (Roche Diagnostics). A 156 bp fragment of the human beta-globin gene (HBB) was used as reference DNA for normalization and amplified in separate capillaries simultaneously to the STS targets. Primer sequences for HBB were as follows: forward 5′-CAGCTCACTCAGTGTGGCAAAG-3′ and reverse 5′-AGGTTCTTTGAGTCCTTTGGGG-3. Relative standard curves were produced using 5 control DNA samples to correct for differences in efficiency of amplification between STS target and reference DNA. For each locus the test was replicated three times.
Table II.
Genotypic information of Patient 1 at the chromosome 8 STS markers obtained by quantitative polymerase chain reaction and gene dosage assay.
| STS name | Gene | Position (bp) | Deletion | Primer | Sequence (‘5-3’) | size |
|---|---|---|---|---|---|---|
| STS-N21307 | LOC286161 | 427685–427914 | Yes | F | CAGGTTGGCAAGTGAAATAC | 230 |
| R | GCAGTAGTGGCATGAAGC | |||||
| SHGC-149177 | DLGAP2 | 952948–953243 | Yes | F | GCCTCCTGGGATAAAAATCCTTT | 296 |
| R | GGTTTGCTCTCCTGATTTAGGGT | |||||
| SHGC-149177 | CLN8 | 1728163–1728478 | Yes | F | AAGAGCAAGAGGAGCAGGAAAAC | 316 |
| R | GTGAAACATGTGAATCATCAGCC | |||||
| SHGC-105022 | CSMD1 | 4126904–4127196 | Yesa | F | TTTTATTTTGGATCAGGCAACCT | 293 |
| R | TGTGCTTTGAACCACACTCCTAA | |||||
| RH119760 | CSMD1 | 4950952–4951296 | Yes | F | TATCCAGTCTCTGCATTTGATGG | 345 |
| R | AGAATCCCAAAGGAGTTACCGAA | |||||
| A004X20 | MCPH1 | 6302850–6303049 | Yes | F | TAAGTTTTCCTTCTCTTCTGTAG | 216 |
| R | AAGGACATGATGATGATT | |||||
| SHGC-77726 | MCPH1 | 6478893–6479173 | Yesa | F | GAAGTAAACTGCAACAGTTCGCC | 281 |
| R | TCTTCTTTCCGCTGTAGGGC | |||||
| RH120376 | TDH | 11224233–11224519 | No | F | AAAATCCACGCTTTGACCTAACA | 287 |
| R | TGGTAAGGGAATGAGTGTGTTCA | |||||
| RH11694 | GATA4 | 11617203–11617417 | No | F | TGCACATTGCTGTTTCTGCC | 234 |
| R | GTTTGTGGGTTAGGGAGGGT |
One in three replications provided conflicting results. STS, sequence tagged site.
Bioinformatic analyses
Sequence features of 8p and 16q regions were analysed in the University of California Santa Cruz (UCSC) Genome Browser (19) using data from the International Standards for Cytogenomic Arrays Consortium (ISCA; www.iscaconsortium.org/) database (20,21) and the corresponding data track for UCSC genes. The Basic Local Alignment Search Tool (BLAST) algorithm (http://blast.ncbi.nlm.nih.gov/Blast.cgi) was used for analysis (22). The results were used to study the nucleotide sequence similarity between the breakpoint regions.
Results
Patient 1
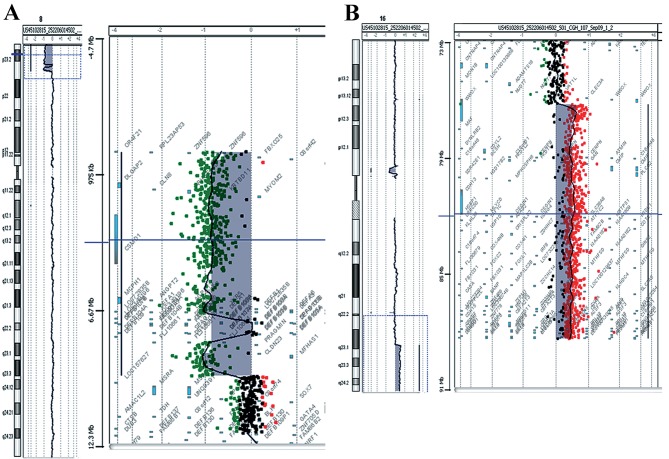
The conventional karyotype of Patient 1 revealed ‘additional’ material in the short arm of chromosome 8 (46, XY, 8p+). aCGH analysis revealed the chromosomal origin of the additional material and the exact position of the breakpoints, namely a deletion of 8p and a duplication of 16q. The 8p deletion was a 4,8 Mb deletion of the distal short arm of chromosome 8 with the proximal breakpoints between 4,814,649 bp (last deleted oligo) and 4,833,351 bp (first normal oligo), with the last oligonucleotide present in the array at 8p position 161,472 kb being deleted (Fig. 1A). The deleted region of the CSMD1 gene began at the first intron. The 16q duplication was a 5.6 Mb duplication of the long arm of chromosome 16 with the proximal breakpoint between 84,468,454 bp (normal) and 84,511,640 bp (duplicated), and the last oligonucleotide present in the array at 16q position 88,690,571 bp being duplicated (Fig. 1B). The size of the breakpoint intervals were 18,702 bp for 8p and 43,186 bp for 16q. The analysis revealed an unbalanced translocation, and the aCGH karyotype was 46,XY,der(8)t(8;16)(p23.2;q23.3)dn.arr[hg18]8p23.3p23.2(151,472–4814649)x1,16q23.3q24.3 (84,511,640–88,690,571)x3.
Figure 1.
Array comparative genomic hybridization results for Patient 1. (A) De novo 4,8 Mb deletion in the short arm of chromosome 8 located at the 8p23.1 to 8pter. (B) De novo 5,6 Mb duplication in the long arm of chromosome 16 located at the 16q24.1 to 16qter.
A list of Online Mendelian Inheritance in Man (OMIM; https://www.omim.org/) genes deleted and duplicated is presented in Table III. FISH analysis was performed to confirm the aCGH data. Three signals were detected; one on chromosome 8 and two on chromosome 16 of the 16q subtelomeric probe. FISH analysis performed in the parents revealed a normal result, indicating a de novo rearrangement.
Table III.
List of OMIM genes deleted and duplicated in both patients.
| Duplication | ||||
|---|---|---|---|---|
| Patient 1 | Patient 2 | |||
| Gene | OMIM | Gene | OMIM | |
| FBX025 | 609098 | FBX025 | 609098 | |
| DLGAP2 | 605438 | DLGAP2 | 605438 | |
| CLN8 | 607837 | CLN8 | 607837 | |
| ARHGEF10 | 608136 | ARHGEF10 | 608136 | |
| MYOM2 | 603509 | MYOM2 | 603509 | |
| CSMD1 | 608397 | CSMD1 | 608397 | |
| MCPH1 | 607117 | |||
| ANGPT2 | 601922 | |||
| AGPAT5 | 614796 | |||
| DEFB1 | 602056 | |||
| DEFA6 | 600471 | |||
| DEFA4 | 601157 | |||
| DEFA1 | 125220 | |||
| DEFA3 | 604522 | |||
| DEFA5 | 600472 | |||
| DEFB103B | 606611 | |||
| SPAG11B | 606560 | |||
| FAM90A7P | 613044 | |||
| FAM90A10P | 613047 | |||
| DEFB4A | 602215 | |||
| CLDN23 | 609203 | |||
| MFHAS1 | 605352 | |||
| ERI1 | 608739 | |||
| PPP1R3B | 610541 | |||
| TNKS | 603303 | |||
| Duplication | ||||
| Patient 1 | Patient 2 | |||
| Gene | OMIM | Gene | OMIM | |
| ATP2C2 | 613082 | WWOX | 605131 | |
| COTL1 | 606748 | MAF | 177075 | |
| USP10 | 609818 | MAFTRR | 616264 | |
| CRISPLD2 | 612434 | DYNLRB2 | 607168 | |
| ZDHHC7 | 614604 | CENPN | 611509 | |
| KIAA0513 | 611675 | ATMIN | 614693 | |
| FAM92B | 617274 | GCSH | 238330 | |
| GSE1 | 616886 | PKD1L2 | 607894 | |
| GINS2 | 610609 | BCO1 | 605748 | |
| EMC8 | 604886 | GAN | 605379 | |
| COX4I1 | 123864 | CMIP | 610112 | |
| IRF8 | 601565 | PLCG2 | 600220 | |
| LINC01082 | 614978 | SDR42E1 | 616164 | |
| LINC01081 | 614977 | HSD17B2 | 109685 | |
| FENDRR | 614975 | MPHOSPH6 | 605500 | |
| FOXF1 | 601089 | CDH13 | 601364 | |
| MTHFSD | 616820 | HSBP1 | 604553 | |
| FOXC2 | 602402 | MLYCD | 606761 | |
| FOXL1 | 603252 | OSGIN1 | 607975 | |
| FBXO31 | 609102 | SLC38A8 | 615585 | |
| MAP1LC3B | 609604 | MBTPS1 | 603355 | |
| JPH3 | 605268 | DNAAF1 | 613190 | |
| SLC7A5 | 600182 | TAF1C | 604905 | |
| CA5A | 114761 | KCNG4 | 607603 | |
| BANP | 611564 | WFDC1 | 605322 | |
| ZNF469 | 612078 | ATP2C2 | 613082 | |
| ZFPM1 | 601950 | COTL1 | 606748 | |
| IL17C | 604628 | USP10 | 609818 | |
| CYBA | 608508 | CRISPLD2 | 612434 | |
| MVD | 603236 | ZDHHC7 | 614604 | |
| SNAI3 | 612741 | KIAA0513 | 611675 | |
| RNF166 | 617178 | FAM92B | 617274 | |
| CTU2 | 617057 | GSE1 | 616886 | |
| PIEZO1 | 611184 | GINS2 | 610609 | |
| CDT1 | 605525 | EMC8 | 604886 | |
| APRT | 102600 | COX4I1 | 123864 | |
| GALNS | 612222 | IRF8 | 601565 | |
| TRAPPC2L | 610970 | LINC01082 | 614978 | |
| CBFA2T3 | 603870 | LINC01081 | 614977 | |
| ACSF3 | 614245 | FENDRR | 614975 | |
| CDH15 | 114019 | FOXF1 | 601089 | |
| ANKRD11 | 611192 | MTHFSD | 616820 | |
| SPG7 | 602783 | FOXC2 | 602402 | |
| RPL13 | 113703 | FOXL1 | 603252 | |
| CPNE7 | 605689 | FBXO31 | 609102 | |
| DPEP1 | 179780 | MAP1LC3B | 609604 | |
| CHMP1A | 164010 | JPH3 | 605268 | |
| SPATA33 | 615409 | SLC7A5 | 600182 | |
| CDK10 | 603464 | CA5A | 114761 | |
| ZNF276 | 608460 | BANP | 611564 | |
| FANCA | 607139 | ZNF469 | 612078 | |
| SPIRE2 | 609217 | ZFPM1 | 601950 | |
| TCF25 | 612326 | IL17C | 604628 | |
| MC1R | 155555 | CYBA | 608508 | |
| TUBB3 | 602661 | MVD | 603236 | |
| AFG3L1P | 603020 | SNAI3 | 612741 | |
| GAS8 | 605178 | RNF166 | 617178 | |
| GAS8-AS1 | 605179 | CTU2 | 617057 | |
| URAHP | 615805 | PIEZO1 | 611184 | |
| PRDM7 | 609759 | CDT1 | 605525 | |
| APRT | 102600 | |||
| GALNS | 612222 | |||
| TRAPPC2L | 610970 | |||
| CBFA2T3 | 603870 | |||
| ACSF3 | 614245 | |||
| CDH15 | 114019 | |||
| ANKRD11 | 611192 | |||
| SPG7 | 602783 | |||
| RPL13 | 113703 | |||
| CPNE7 | 605689 | |||
| DPEP1 | 179780 | |||
| CHMP1A | 164010 | |||
| SPATA33 | 615409 | |||
| CDK10 | 603464 | |||
| ZNF276 | 608460 | |||
| FANCA | 607139 | |||
| SPIRE2 | 609217 | |||
| TCF25 | 612326 | |||
| MC1R | 155555 | |||
| TUBB3 | 602661 | |||
| AFG3L1P | 603020 | |||
| GAS8 | 605178 | |||
| GAS8-AS1 | 605179 | |||
| URAHP | 615805 | |||
| PRDM7 | 609759 | |||
OMIM, Online Mendelian Inheritance in Man.
Patient 2
Prenatal diagnosis due to elevated maternal age revealed a normal karyotype of 46, XX. During the neonatal period and due to dysmorphic features, hypotonia and clinical complications, aCGH analysis was performed. The analysis revealed a deletion of 9.5 Mb of the distal short arm of chromosome 8 with the proximal breakpoints between 95,48,146 bp (last deleted oligo) and 95,62,020 bp (first normal oligo), with the last oligonucleotide present in the array at 8p position 151,472 kb being deleted (Fig. 2A). The deleted region of the tankyrase (TNKS) gene began at the fifth intron. A duplication of 11.7 Mb of the long arm of chromosome 16 with the proximal breakpoint between 76,961,103 bp (duplicated) and 76,938,723 bp (normal) was observed and the last oligonucleotide present in the array at 16q position 88,690,571 bp was duplicated (Fig. 2B). The size of the breakpoint intervals were 13,874 bp for 8p and 22,380 bp for 16q. The analysis revealed an unbalanced translocation and the aCGH karyotype was 46,XX,der(8)t(8;16)(p23.1;q23.1).arr[hg18]8p23.3p23.1 (151,472–9548146)x1,16q23.1q24.3(76,961,103–88,690,571)x3.
Figure 2.
Array comparative genomic hybridization results for Patient 2. (A) De novo 9,5 Mb deletion in the short arm of chromosome 8 located at the 8p23 to 8pter. (B) De novo 11,7 Mb duplication in the long arm of chromosome 16 located at the 16q23.1 to 16qter.
A list of OMIM genes deleted and duplicated is presented in Table III. Microsatellite analysis of the trio revealed that deletion and duplication occurred on maternally-derived chromosomes (Table IV).
Table IV.
Results from microsatellite analysis on Patient 2.
| Sample | 253-10 Proband | 254-10 father | 255-10 mother | Origin |
|---|---|---|---|---|
| D8S201 | 259.5 | 255.5/259.5 | 259.5/267.2 | Uninformative |
| D8S504 | 200.7 | 200.7/202.8 | 198.1/202.7 | Maternal |
| D8S264 | 138.2 | 138.1/138.1 | 126.4/126.4 | Maternal |
| D8S1781 | 259.4 | 259.4/263.2 | 251.1/263.1 | Maternal |
| D8S351 | 119.2 | 119/119 | 105/105 | Maternal |
| D8S1706 | 228/234.2 | 228/234.2 | 228/234.2 | Uninformative |
| D16S3023 | 79.5/83.5 | 83.5/83.5 | 79.5/83.6 | Uninformative |
| D16S413 | 128/132 | 130/132 | 128/132 | Uninformative |
| STS1 (chr16) | 210.8*/214.9 | 214.9/214.9 | 210.9/210.9 | Maternal |
| STS2 (chr16) | 125.2/125.2 | 125.2/125.2 | 125.2/125.2 | Uninformative |
| STS3 (chr16) | 296.5/296.5 | 294.6/296.5 | 296.6 | Uninformative |
| STS4 (chr16) | 345.32/352.4/357.8 | 352.4/359.7 | 345.4/357.38 | Maternal |
Polymorphic sequence tagged site markers were selected in the deleted and duplicated regions of chromosomes 8 and 16, respectively.
Bioinformatic analyses
A possible cause of rearrangements, duplications and deletions is the occurrence of recombination events. To search for a possible breakpoint for recombination, the BLAST algorithm was used to find sequence similarity in the breakpoint regions of the two patients. The breakpoint regions were revealed to contain similar sequences residing in Alu elements of Patient 1 and in L1 elements of Patient 2.
The rearranged regions were viewed in parallel with ISCA consortium data in the UCSC Genome Browser. The 8p region contained multiple pathogenic copy number variations (74 deletions and 31 duplications) described in the ISCA database, while rearrangements in 16q were less frequent (containing 19 deletions and 16 duplications). Manual computations of the ISCA data revealed that 66% of patients with 8p23.3-p23.1 rearrangements (deletions or duplications), and 62% of patients with 16q23.1-q24.3 rearrangements had developmental delay in their pathogenic phenotype (Fig. 3).
Figure 3.
(A) Sequence features of the 8p and 16q translocated regions. The features are presented in parallel tracks. I) Chromosome ideogram representing the translocated regions in the red square. II) Chromosome positions of deleted (in red) and duplicated (in blue) regions are shown in each patient and respective breakpoints and gene positions are highlighted. III) Pathogenic copy number variants in these regions, as published by the ISCA Consortium. Blue lines represent duplications and red lines deletions. (B) Sequence features of the respective 8p and 16q breakpoints for both patients. Presented in parallel tracks: the chromosome scale and chromosome region annotation, genes present in this chromosome region and repetitive genetic elements as annotated by the RepeatMasker tool (www.repeatmasker.org). Red color repeats denote sequences with high similarity, as revealed by BLAST alignment of the respective breakpoints for each patient. P1, patient 1; P2, patient 2.
Discussion
To the best of our knowledge, this is the first report of a rearrangement involving an 8p deletion and 16q duplication. The two patients presented in this report had subtle facial feature dysmorphia, dysmorphic body features, borderline intelligence and marginal follow up progress, low birth weight and vertebral anomalies, and one presented with cardiovascular abnormalities. The majority of the clinical characteristics of the two patients were associated with those of 8p or 16q chromosome imbalances, but it is difficult to estimate if the clinical phenotype and developmental delay were due to the rearrangement or whether they were the result of 8p monosomy and 16q trisomy separately. It has previously been recognized that deletions in the distal region of chromosome 8p are associated with growth and mental impairment, minor facial dysmorphisms, microcephaly, congenital heart defects and behavioral problems (23). According to all references, 16q trisomy is a rare abnormality due to high rates of mortality and lethality in the prenatal and neonatal period (11,24). Partial 16q trisomy is most often the result of balanced or unbalanced rearrangements, and therefore it is difficult to understand if the commonly observed phenotypic characteristics (dysmorphic facial features, developmental delay, intellectual disability, central nervous system malformations and congenital heart defects) are due to 16q or whether they are the result of changes in genome architecture (24).
More than 2/3 of patients with 8p syndrome have congenital heart defects, suggesting that 8p23.1 maybe critical for heart development (5,25). One of the candidate genes for heart disease is GATA4 because haploinsufficiency and mutations have been documented in patients and families with atrial septal defects and other cardiac defects associated with 8p23.1 deletion (4,26–29). Chen et al (30) studied a four-generation Chinese atrial septal defect family and suggested that a mutation in the GATA4 gene (c.A899C, p.K300T) may contribute to this congenital heart disease. However, the GATA4 gene was not deleted in either patient in the present study. The fact that Patient 1 has heart problems suggested either that other genes were responsible for these problems, or that the rearrangement resulted in a structural alteration affecting the function of genes associated with the heart. The CSMD1 gene (8p23.2) was deleted in Patient 2 but only partially deleted in Patient 1. According to the literature, CSMD1 loss of function is correlated with head and neck squamous cell carcinoma (31,32), and liver (33,34), lung, breast and skin cancers (31). Deletion of this gene has been reported in a case of craniofacial and body dysmorphisms and mental retardation (35). In Patient 2, two of the deleted genes were TNKS and MCPH1. These genes are involved in meiosis and mitosis mechanisms. The TNKS gene, located at 8p23.1, is involved in sister chromatid cohesion and deletions result in anaphase arrest (36). Páez et al (4) identified deletions of TNKS gene in patients with mental retardation and behavioral problems. TNKS protein positively regulates the Wnt/β-catenin signaling pathway (37). This pathway is critical for healthy embryonic development and cellular differentiation (38). Furthermore, TNKS is a candidate gene for Cornelia de Lange Syndrome (CdLS) (3,36), a syndrome characterized by distinctive facial features including well-defined curved and confluent eyebrows, long eyelashes, anteverted nares, micrognathia and downturned corners of the mouth with a thin upper lip. Patient 2 resembled the CdLS facial phenotype, and she is expected to have psychomotor retardation, language acquisition difficulties and behavioral disorders in the autistic spectrum, typical aspects of CdLS. The MCPH1 gene, additionally located in 8p23.1, is involved in preventing cells from prematurely entering mitosis, and truncated mutations have been associated with premature chromosome condensation and were observed in patients with microcephaly, growth impairment and mental retardation (36,39). Another gene located in 8p23.1 is RP1 like 1 (RP1L1). Its expression is restricted to the postnatal retina, potentially being involved in retinal development (40). The RP1L1 gene may not be haploinsufficient in Patient 2, who was diagnosed with diffuse depigmentation of the retina. This gene maybe under the control of translocated regulatory elements, being in the proximity of the breakpoint, and may have resulted in this retinal disorder.
In total, >30 cases with distal 8p deletion have been described in the literature, and 9 with 16q24 duplication, but only a few have been characterized with high resolution molecular techniques (1,11). The 8p region is more often reported to be involved in rearrangements than 16q. Giglio et al (41) demonstrated that the olfactory receptor (OR) gene clusters are the substrate for the formation of intrachromosomal rearrangements involving chromosome 8p. Different rearrangements, most of them recurring, are associated with the distal 8p region. Among them there are inv dup(8p), del(8p22) and small marker chromosomes der(8)(p23-pter) (41). Furthermore, seven individuals with balanced and unbalanced translocations between 4p16 and 8p23 demonstrated that the breakpoints fell within the 4p and 8p OR-gene clusters (42).
BLAST alignment in the 8p and 16q regions revealed high similarity regions with several Alu elements in Patient 1 and two similar long interspersed nuclear element 1 elements in Patient 2. Retroelements are known to facilitate recombination events (43). Consequently, a potential mechanism of their appearance maybe unequal cross over between repetitive DNA regions with high sequence similarity (44). The deleted and duplicated regions are regions often correlated with developmental delay, according to the ISCA Consortium (20).
Novel diagnostic methods with great potential have facilitated the study and interpretation of the consequences of chromosome aberrations and revealed that the pathogenicity may be due to complex molecular mechanisms (45,46). A number of the genes identified as deleted or duplicated in these cases may have resulted in developmental delay, but developmental delay may also be the result of rearrangements and changes of important parts of gene structure functional elements, truncated or fusion genes. A multidisciplinary effort aiming to study all cases with 8p;16q rearrangements with combined and accurate methods and tools (cytogenetic, molecular cytogenetic, NGS mapping) along with their clinical phenotypes may elucidate the involvement of the rearrangement, genes involved, participation of control elements and/or interactions with polymorphic regions, and potentially a clear phenotype-genotype correlation.
References
- 1.Reddy KS. A paternally inherited terminal deletion, del(8)(p23.1)pat, detected prenatally in an amniotic fluid sample: A review of deletion 8p23.1 cases. Prenat Diagn. 1999;19:868–872. doi: 10.1002/(SICI)1097-0223(199909)19:9<868::AID-PD641>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 2.Fagan KA, Morris RB. Del(4)(q33-qter): Another case report of a child with mild dysmorphism. J Med Genet. 1989;26:776–778. doi: 10.1136/jmg.26.12.776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ballarati L, Cereda A, Caselli R, Selicorni A, Recalcati MP, Maitz S, Finelli P, Larizza L, Giardino D. Genotype-phenotype correlations in a new case of 8p23.1 deletion and review of the literature. Eur J Med Genet. 2011;54:55–59. doi: 10.1016/j.ejmg.2010.10.003. [DOI] [PubMed] [Google Scholar]
- 4.Páez MT, Yamamoto T, Hayashi K, Yasuda T, Harada N, Matsumoto N, Kurosawa K, Furutani Y, Asakawa S, Shimizu N, Matsuoka R. Two patients with atypical interstitial deletions of 8p23.1: Mapping of phenotypical traits. Am J Med Genet A. 2008;146A:1–1165. doi: 10.1002/ajmg.a.32205. [DOI] [PubMed] [Google Scholar]
- 5.Devriendt K, Matthijs G, Van Dael R, Gewillig M, Eyskens B, Hjalgrim H, Dolmer B, McGaughran J, Bröndum-Nielsen K, Marynen P, et al. Delineation of the critical deletion region for congenital heart defects, on chromosome 8p23.1. Am J Hum Genet. 1999;64:1119–1126. doi: 10.1086/302330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shin WJ, Kim SD, Kim KH. The general anesthesia experience of deletion 8p syndrome patient-A case report. Korean J Anesthesiol. 2011;61:332–335. doi: 10.4097/kjae.2011.61.4.332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brisset S, Joly G, Ozilou C, Lapierre JM, Gosset P, LeLorc'h M, Raoul O, Turleau C, Vekemans M, Romana SP. Molecular characterization of partial trisomy 16q24.1-qter: Clinical report and review of the literature. Am J Med Genet. 2002;113:339–345. doi: 10.1002/ajmg.b.10740. [DOI] [PubMed] [Google Scholar]
- 8.Ferrero GB, Belligni E, Sorasio L, Delmonaco AG, Oggero R, Faravelli F, Pierluigi M, Silengo M. Phenotype resembling donnai-barrow syndrome in a patient with 9qter;16qter unbalanced translocation. Am J Med Genet A. 2006;140:892–894. doi: 10.1002/ajmg.a.31188. [DOI] [PubMed] [Google Scholar]
- 9.Hutchinson R, Wilson M, Voullaire L. Distal 8p deletion (8p23. 1-8pter): A common deletion? J Med Genet. 1992;29:407–411. doi: 10.1136/jmg.29.6.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zahn S, Ehrbrecht A, Bosse K, Kalscheuer V, Propping P, Schwanitz G, Albrecht B, Engels H. Further delineation of the phenotype maps for partial trisomy 16q24 and jacobsen syndrome by a subtle familial translocation t(11;16)(q24.2;q24.1) Am J Med Genet A. 2005;139:19–24. doi: 10.1002/ajmg.a.30995. [DOI] [PubMed] [Google Scholar]
- 11.Zhou Y, Yao Q, Cui YX, Yao B, Fan K, Xia XY, Hu YA, Li XJ. Clinical and cytogenetic characterization of a boy with a de novo pure partial trisomy 16q24.1q24.3 and complex chromosome rearrangement. Am J Med Genet A. 2013;161A:1–900. doi: 10.1002/ajmg.a.35782. [DOI] [PubMed] [Google Scholar]
- 12.Burnside RD, Pappas JG, Sacharow S, Applegate C, Hamosh A, Gadi IK, Jaswaney V, Keitges E, Phillips KK, Potluri VR, et al. Three cases of isolated terminal deletion of chromosome 8p without heart defects presenting with a mild phenotype. Am J Med Genet A. 2013;161A:1–828. doi: 10.1002/ajmg.a.35699. [DOI] [PubMed] [Google Scholar]
- 13.Baker E, Hinton L, Callen DF, Altree M, Dobbie A, Eyre HJ, Sutherland GR, Thompson E, Thompson P, Woollatt E, Haan E. Study of 250 children with idiopathic mental retardation reveals nine cryptic and diverse subtelomeric chromosome anomalies. Am J Med Genet. 2002;107:285–293. doi: 10.1002/ajmg.10159. [DOI] [PubMed] [Google Scholar]
- 14.Giardino D, Finelli P, Gottardi G, Clerici D, Mosca F, Briscioli V, Larizza L. Cryptic subtelomeric translocation t(2;16)(q37;q24) segregating in a family with unexplained stillbirths and a dysmorphic, slightly retarded child. Eur J Hum Genet. 2001;9:881–886. doi: 10.1038/sj.ejhg.5200730. [DOI] [PubMed] [Google Scholar]
- 15.Maher ER, Willatt L, Cuthbert G, Chapman C, Hodgson SV. Three cases of 16q duplication. J Med Genet. 1991;28:801–802. doi: 10.1136/jmg.28.11.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bayley N. Scales of Infant Development. 2nd edition. The Psychological Corporation; San Antonio, TX: 1993. [Google Scholar]
- 17.Griffiths R. The Abilities of Young Children. Bucks: Association for Research in Infant and Child Development. The test Agency; Oxford: 1984. A comprehensive system of measurement for the first eight years of life; pp. 101–172. [Google Scholar]
- 18.Manolakos E, Peitsidis P, Eleftheriades M, Dedoulis E, Ziegler M, Orru S, Liehr T, Petersen MB. Prenatal detection of full monosomy 21 in a fetus with increased nuchal translucency: Molecular cytogenetic analysis and review of the literature. J Obstet Gynaecol Res. 2010;36:435–440. doi: 10.1111/j.1447-0756.2009.01140.x. [DOI] [PubMed] [Google Scholar]
- 19.Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kaminsky EB, Kaul V, Paschall J, Church DM, Bunke B, Kunig D, Moreno-De-Luca D, Moreno-De-Luca A, Mulle JG, Warren ST, et al. An evidence-based approach to establish the functional and clinical significance of copy number variants in intellectual and developmental disabilities. Genet Med. 2011;13:777–784. doi: 10.1097/GIM.0b013e31822c79f9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miller DT, Adam MP, Aradhya S, Biesecker LG, Brothman AR, Carter NP, Church DM, Crolla JA, Eichler EE, Epstein CJ, et al. Consensus statement: Chromosomal microarray is a first-tier clinical diagnostic test for individuals with developmental disabilities or congenital anomalies. Am J Hum Genet. 2010;86:749–764. doi: 10.1016/j.ajhg.2010.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 23.Digilio MC, Marino B, Guccione P, Giannotti A, Mingarelli R, Dallapiccola B. Deletion 8p syndrome. Am J Med Genet. 1998;75:534–536. doi: 10.1002/(SICI)1096-8628(19980217)75:5<534::AID-AJMG15>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 24.Lonardo F, Perone L, Maioli M, Ciavarella M, Ciccone R, Monica MD, Lombardi C, Forino L, Cantalupo G, Masella L, Scarano F. Clinical, cytogenetic and molecular-cytogenetic characterization of a patient with a de novo tandem proximal-intermediate duplication of 16q and review of the literature. Am J Med Genet A. 2011;155A:1–777. doi: 10.1002/ajmg.a.33852. [DOI] [PubMed] [Google Scholar]
- 25.Johnson MC, Hing A, Wood MK, Watson MS. Chromosome abnormalities in congenital heart disease. Am J Med Genet. 1997;70:292–298. doi: 10.1002/(SICI)1096-8628(19970613)70:3<292::AID-AJMG15>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 26.Mei M, Yang L, Zhan G, Wang H, Ma D, Zhou W, Huang G. Analysis of genomic copy number variations in two unrelated neonates with 8p deletion and duplication associated with congenital heart disease. Zhonghua Er Ke Za Zhi. 2014;52:460–463. (In Chinese) [PubMed] [Google Scholar]
- 27.Pehlivan T, Pober BR, Brueckner M, Garrett S, Slaugh R, Van Rheeden R, Wilson DB, Watson MS, Hing AV. Gata4 haploinsufficiency in patients with interstitial deletion of chromosome region 8p23.1 and congenital heart disease. Am J Med Genet. 1999;83:201–206. doi: 10.1002/(SICI)1096-8628(19990319)83:3<201::AID-AJMG11>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 28.Wat MJ, Shchelochkov OA, Holder AM, Breman AM, Dagli A, Bacino C, Scaglia F, Zori RT, Cheung SW, Scott DA, Kang SH. Chromosome 8p23.1 deletions as a cause of complex congenital heart defects and diaphragmatic hernia. Am J Med Genet A. 2009;149A:1–1677. doi: 10.1002/ajmg.a.32896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang YQ, Wang J, Liu XY, Chen XZ, Zhang W, Wang XZ, Liu X, Fang WY. Novel GATA4 mutations in patients with congenital ventricular septal defects. Med Sci Monit. 2012;18:CR344–CR350. doi: 10.12659/MSM.882877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen J, Qi B, Zhao J, Liu W, Duan R, Zhang M. A novel mutation of GATA4 (K300T) associated with familial atrial septal defect. Gene. 2016;575:473–477. doi: 10.1016/j.gene.2015.09.021. [DOI] [PubMed] [Google Scholar]
- 31.Ma C, Quesnelle KM, Sparano A, Rao S, Park MS, Cohen MA, Wang Y, Samanta M, Kumar MS, Aziz MU, et al. Characterization CSMD1 in a large set of primary lung, head and neck, breast and skin cancer tissues. Cancer Biol Ther. 2009;8:907–916. doi: 10.4161/cbt.8.10.8132. [DOI] [PubMed] [Google Scholar]
- 32.Toomes C, Jackson A, Maguire K, Wood J, Gollin S, Ishwad C, Paterson I, Prime S, Parkinson K, Bell S, et al. The presence of multiple regions of homozygous deletion at the CSMD1 locus in oral squamous cell carcinoma question the role of CSMD1 in head and neck carcinogenesis. Genes Chromosomes Cancer. 2003;37:132–140. doi: 10.1002/gcc.10191. [DOI] [PubMed] [Google Scholar]
- 33.Midorikawa Y, Yamamoto S, Tsuji S, Kamimura N, Ishikawa S, Igarashi H, Makuuchi M, Kokudo N, Sugimura H, Aburatani H. Allelic imbalances and homozygous deletion on 8p23.2 for stepwise progression of hepatocarcinogenesis. Hepatology. 2009;49:513–522. doi: 10.1002/hep.22698. [DOI] [PubMed] [Google Scholar]
- 34.Zhu Q, Gong L, Liu X, Wang J, Ren P, Zhang W, Yao L, Han X, Zhu S, Lan M, et al. Loss of heterozygosity at D8S262: An early genetic event of hepatocarcinogenesis. Diagn Pathol. 2015;10:70. doi: 10.1186/s13000-015-0308-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Naseer MI, Chaudhary AG, Rasool M, Kalamegam G, Ashgan FT, Assidi M, Ahmed F, Ansari SA, Zaidi SK, Jan MM, Al-Qahtani MH. Copy number variations in Saudi family with intellectual disability and epilepsy. BMC Genomics. 2016;17(Suppl 9):S757. doi: 10.1186/s12864-016-3091-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Baynam G, Goldblatt J, Walpole I. Deletion of 8p23.1 with features of Cornelia de Lange syndrome and congenital diaphragmatic hernia and a review of deletions of 8p23.1 to 8pter? A further locus for Cornelia de Lange syndrome. Am J Med Genet A. 2008;146A:1–1570. doi: 10.1002/ajmg.a.32095. [DOI] [PubMed] [Google Scholar]
- 37.Roy S, Liu F, Arav-Boger R. Human Cytomegalovirus Inhibits the PARsylation Activity of Tankyrase-A potential strategy for suppression of the Wnt pathway. Viruses. 2015;8:pii: E8. doi: 10.3390/v8010008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Clevers H, Nusse R. Wnt/β-catenin signaling and disease. Cell. 2012;149:1192–1205. doi: 10.1016/j.cell.2012.05.012. [DOI] [PubMed] [Google Scholar]
- 39.Trimborn M, Bell SM, Felix C, Rashid Y, Jafri H, Griffiths PD, Neumann LM, Krebs A, Reis A, Sperling K, et al. Mutations in microcephalin cause aberrant regulation of chromosome condensation. Am J Hum Genet. 2004;75:261–266. doi: 10.1086/422855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Conte I, Lestingi M, den Hollander A, Alfano G, Ziviello C, Pugliese M, Circolo D, Caccioppoli C, Ciccodicola A, Banfi S. Identification and characterisation of the retinitis pigmentosa 1-like1 gene (RP1L1): A novel candidate for retinal degenerations. Eur J Hum Genet. 2003;11:155–162. doi: 10.1038/sj.ejhg.5200942. [DOI] [PubMed] [Google Scholar]
- 41.Giglio S, Calvari V, Gregato G, Gimelli G, Camanini S, Giorda R, Ragusa A, Guerneri S, Selicorni A, Stumm M, et al. Heterozygous submicroscopic inversions involving olfactory receptor-gene clusters mediate the recurrent t(4;8)(p16;p23) translocation. Am J Hum Genet. 2002;71:276–285. doi: 10.1086/341610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Giglio S, Broman KW, Matsumoto N, Calvari V, Gimelli G, Neumann T, Ohashi H, Voullaire L, Larizza D, Giorda R, et al. Olfactory receptor-gene clusters, genomic-inversion polymorphisms, and common chromosome rearrangements. Am J Hum Genet. 2001;68:874–883. doi: 10.1086/319506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Konkel MK, Batzer MA. A mobile threat to genome stability: The impact of non-LTR retrotransposons upon the human genome. Semin Cancer Biol. 2010;20:211–221. doi: 10.1016/j.semcancer.2010.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Robberecht C, Voet T, Esteki M Zamani, Nowakowska BA, Vermeesch JR. Nonallelic homologous recombination between retrotransposable elements is a driver of de novo unbalanced translocations. Genome Res. 2013;23:411–418. doi: 10.1101/gr.145631.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Newman S, Hermetz KE, Weckselblatt B, Rudd MK. Next-generation sequencing of duplication CNVS reveals that most are tandem and some create fusion genes at breakpoints. Am J Hum Genet. 2015;96:208–220. doi: 10.1016/j.ajhg.2014.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Utami KH, Hillmer AM, Aksoy I, Chew EG, Teo AS, Zhang Z, Lee CW, Chen PJ, Seng CC, Ariyaratne PN, et al. Detection of chromosomal breakpoints in patients with developmental delay and speech disorders. PLoS One. 2014;9:e90852. doi: 10.1371/journal.pone.0090852. [DOI] [PMC free article] [PubMed] [Google Scholar]