Abstract
The development of an efficient delivery system is critical for the successful treatment of cardiovascular diseases using non-viral gene therapies. Cytoplasmic and nuclear membrane barriers reduce delivery efficiency by impeding the transfection of foreign genes. Thus, a gene delivery system capable of transporting exogenous genes may improve gene therapy. The present study used a novel strategy involving ultrasound-targeted microbubbles and peptide nucleic acid (PNA)-binding nuclear localization signals (NLS). Ultrasound-targeted microbubble destruction (UTMD) and PNA-binding NLS were used to improve the cytoplasmic and nuclear importation of the plasmid, respectively. Experiments were performed using antibody-targeted microbubbles (AT-MCB) that specifically recognize the SV40T antigen receptor expressed on the membranes of 293T cells, resulting in the localization of ultrasound microbubbles to 293T cell membranes. Furthermore, PNA containing NLS was inserted into the enhanced green fluorescent protein (EGFP)-N3 plasmid DNA (NLS-PNA-DNA), which increased nuclear localization. The nuclear import and gene expression efficiency of the AT-MCB with PNA-binding NLS were compared with AT-MCB alone or a PNA-binding NLS. The effect of the AT-MCB containing PNA-binding NLS on transfection was investigated. The ultrasound and AT-MCB delivery significantly enhanced the cytoplasmic intake of exogenous genes and maintained high cell viability. The nuclear import and gene expression of combined microbubble- and PNA-transfected cells were significantly greater compared with cells that were transfected with AT-MCB or DNA with only PNA-binding NLS. The quantity of EGFP-N3 plasmids in the nuclei was increased by >5.0-fold compared with control microbubbles (CMCB) and NLS-free plasmids. The gene expression was ~1.7-fold greater compared with NLS-free plasmids and 1.3-fold greater compared with control microbubbles. In conclusion, UTMD combined with AT-MCB and a PNA-binding NLS plasmid significantly improved transfection efficiency by increasing cytoplasmic and nuclear DNA import. This method is a promising strategy for the noninvasive and effective delivery of target genes or drugs for the treatment of cardiovascular diseases.
Keywords: ultrasound, microbubbles, peptide nucleic acid, nuclear localization signals, gene transfection
Introduction
The safe and efficient delivery of therapeutic genes across the plasma membrane of the cell is crucial for effective gene therapy. Although viral vectors may be used for gene transfection, their application is limited due to potential side effects. By contrast, ultrasound techniques may have great potential for non-viral gene delivery in vitro and in vivo in the absence of severe adverse effects. However, due to cytoplasmic and nuclear membrane barriers, transfection efficiency is relatively low (1). Numerous methods have been assessed to address this issue and increase transfection efficiency.
Controversy surrounds the development of an efficient method to deliver DNA or drugs into cells. One of these methods is ultrasound-targeted microbubble destruction (UTMD) (2–4). Compared with other direct DNA delivery methods, including microinjection and electroporation, UTMD may be simpler to implement (5,6). Sonication (ultrasound), which may target gene delivery to a specific area, alters the transient permeability of plasma membranes to facilitate intake (5). In the present study, UTMD technology was used to facilitate the entry of plasmid (p)DNA into the cytoplasm without causing severe cell damage.
Due to the nuclear membrane barrier, delivery of genes to the nucleus is challenging. Certain studies have demonstrated that pDNA microinjection results in <3% nuclear import and low expression (7–9). It is therefore important to overcome this obstacle to increase nuclear import during gene transfer. A previous study used numerous nuclear localization signals (NLS) to promote nuclear gene import; however, efficiency remained unsatisfactory (10).
The present study examined gene transfection by UTMD using 293T cells with a microbubble contrast agent. A specific NLS was bound by peptide nucleic acid (PNA) to pDNA. The antibody-targeted microbubbles (AT-MCB) recognized and bound to the antigen on the cell membrane. Subsequently, the DNA with NLS was guided to bring the pDNA into the cell, and this may have improved nuclear intake and transfection efficiency. AT-MCB may improve cellular uptake of pDNA, and PNA-binding NLS may promote nuclear import of pDNA. Therefore, combining these two methods may significantly enhance transfection efficiency.
Materials and methods
Antibody-targeted microbubble preparation
Electrostatic adsorption has been reported to be an efficient method of constructing AT-MCB (11). Normal saline (5 ml) was poured into a small bottle with SonoVue freeze-dried powder (25 mg; Bracco Suisse S.A., Geneva, Switzerland) and shaken well for 10 sec to form a microbubble suspension. The diameters of the microbubbles were 0.8–10 µm, with the majority being 2–5 µm. A monoclonal anti-SV40Tag antibody (ab82118; Abcam, Cambridge, MA, USA) conjugated to fluorescein isothiocyanate was dissolved 1:50 in PBS. The microbubble suspension was mixed with the diluted antibody at a ratio of 2:1, and the pH of the reaction liquid was adjusted to pH 4.0–5.0 prior to incubation at 4°C for 2 h, during which the mixture was shaken several times. The clear liquid at the bottom of the container (containing the unbound antibodies) was subsequently removed and washed with a buffer. Following 10–15 min, the liquid separated into layers, and the bottom clear liquid (containing the unbound antibodies) was again removed. Finally, PBS was added to suspend the floating foam to target the microbubble suspension for an anti-SV40Tag antibody.
Targeted microbubble identification
The prepared targeted microbubble suspension was observed under a fluorescence microscope to determine the combination of antibodies and microbubbles, as well as the status and distribution of the targeted microbubbles (Fig. 1).
Figure 1.
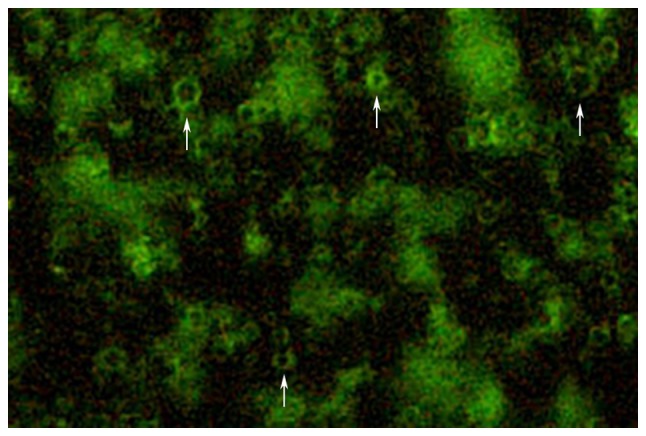
Antibody binding to microbubbles. Arrows indicate the fluorescein isothiocyanate-conjugated antibody bound to microbubbles in the microbubble suspension (magnification, ×100).
Enhanced green fluorescent protein (EGFP)-N3 plasmid construction
To hybridize EGFP-N3 and PNA, EGFP-N3 was added to the sample at a proportion of EGFP-N3: biotin-PNA=3:100 in a graduated 200-µl Eppendorf tube to cause the plasmid EGFP-N3 to completely hybridize with the biotin PNA, which produced a sepharose gel. Subsequently, a 1-µl sample was taken from the Eppendorf tube (60 V at constant voltage for 6 h). The band was observed under a blue light transilluminator. A specific NLS sequence (PKKKRKV) was designed to embed into biotin PNA. The redundant PKKKRKV-PNA was removed by liquid chromatography to measure the optical density (OD) using an ultraviolet spectrophotometer. The purified sample was 450 µl at 8.2 µg/ml, with an OD260/280 of 1.72. The biotinylated PNA/DNA compound was mixed with a streptavidin SV40T antigen NLS and incubated for 20 min at 4°C; subsequently, centrifugal washing at 500 × g for 30 sec was performed twice to remove the unbound NLS and construct the NLS-PNA/EGFP-N3 karyotheca-targeted gene (Fig. 2).
Figure 2.
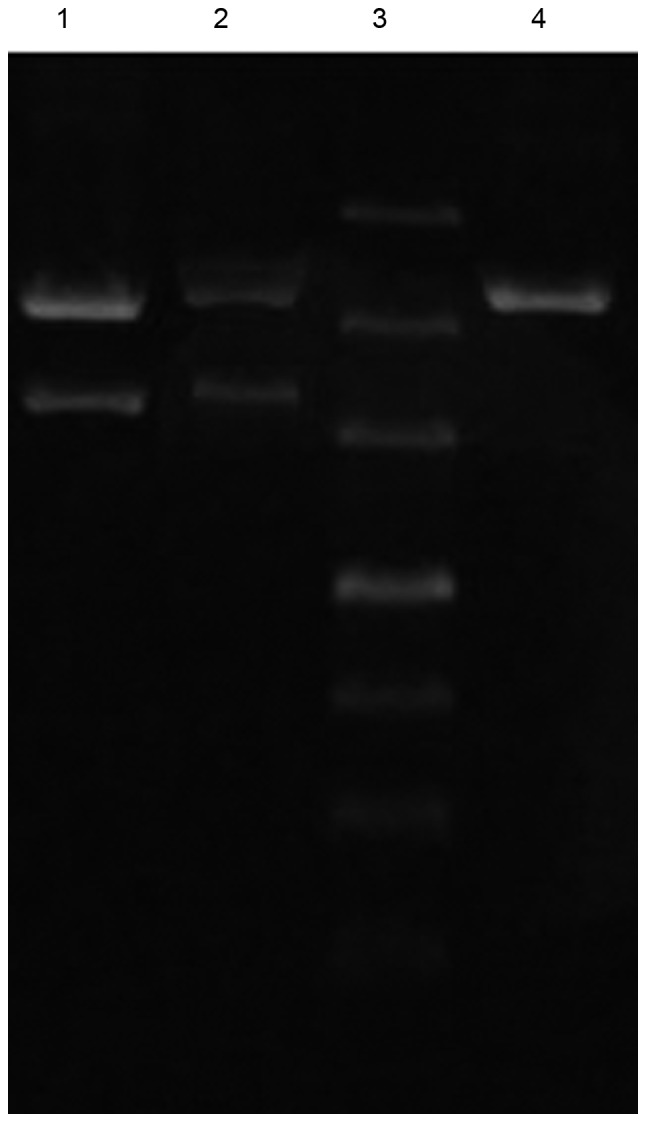
Enhanced green fluorescent protein-N3 plasmid construction. Lane 1, plasmid DNA; lane 2, plasmid DNA + PNA; lane 3, marker; lane 4, purified plasmid DNA + PNA. PNA, peptide nucleic acid.
Cell culture
The 293T cells were obtained from the China Center for Type Culture Collection (Wuhan, China), and cultured in Dulbecco's modified Eagle's medium (DMEM; ScienCell Research Laboratories, Carlsbad, CA, USA) containing 10% fetal bovine serum (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA). The cells were cultured in 10-cm culture dishes at 37°C with 5% CO2. Adherent cells were removed by trypsinization and seeded at a density of 2×105 cells per well (in 2 ml DMEM) in 6-well plates (Corning Incorporated, Corning, NY, USA) 24 h prior to transfection.
UTMD-mediated transfection of EGFP-N3 plasmid
The present study was divided into three groups, and each group had three subgroups. Group 1: control, CMCB + DNA, AT-MCB + DNA; Group 2: control, CMCB + DNA, CMCB + NLS-PNA-DNA; Group 3: AT-MCB + DNA, CMCB + NLS-PNA-DNA, AT-MCB + NLS-PNA-DNA. The control group received no treatment and the negative group was CMCB + DNA, which were treated with ordinary microbubbles (CMCB) and pDNA. 293T cells were transfected with CMCB and NLS-PNA-DNA, AT-MCB and pDNA, AT-MCB and NLS-PNA-DNA in group CMCB + NLS-PNA-DNA, AT-MCB + DNA, AT-MCB + NLS-PNA-DNA respectively.
Transfection was performed when the cells reached 65–75% confluence. The culture medium was replaced with 2 ml Opti-mimimal essential medium (Gibco; Thermo Fisher Scientific, Inc.) per well. Plasmid (10 µg) was added to each well along with microbubbles prior to transfection. Ultrasound was subsequently used to irradiate the cell, plasmid and microbubble suspension (UGT2007; Ultrasonic Research Institute of Chongqing Medical University, Chongqing, China). The parameters were set to different acoustic intensities and exposure times, and the transducer was sterilized prior to immersion in the cell suspension.
It has been previously demonstrated that gene transfection efficiency in vitro may be affected by alterations in ultrasound parameters, including duration and intensity (12,13). Transfections were performed using the optimal ultrasound exposure parameters, which were determined by our previous study: 1×107 microbubbles/ml and 1.5 W/cm2 for 45 sec with a 20% duty cycle (14). The various parameters were assessed by cell viability and pDNA uptake.
Cell viability
The 293T cells were seeded in 96-well plates (5×103 cells/well in 100 µl DMEM. The viability of the cells was measured using a Cell Counting kit-8 (CCK-8; Dojindo Laboratories, Kumamoto, Japan). Following incubation at 37°C for 24 h, 10 µl CCK-8 was added per well and cells were incubated for 2 h at 37°C. The absorbance was measured at a wavelength of 450 nm using an automatic microplate reader (Lambda 1050; PerkinElmer, Inc., Waltham, MA, USA). The percentage cell viability was calculated as follows: Cell viability (%) = (ODsample / ODcontrol) × 100.
Cellular uptake of pDNA
EGFP-N3 was used to transfect the 293T cells in the presence or absence of UTMD. Following 48 h of incubation at 37°C in the incubator, the 293T cells were observed under an inverted fluorescence microscope to determine the presence of the EGFP-N3 plasmid (15). The transfected cells were allowed to incubate at 37°C for a further 6 h after which they were removed by trypsinization and washed in PBS. The percentage of cells that had endocytosed EGFP-N3 was determined using flow cytometry (Beckman Coulter, Inc., Brea, CA, USA) and calculated as follows: EGFP-N3 uptake (%) = (EGFP-N3-positive cell population / total cell population) × 100 using FlowJo version 7.6.1 (FlowJo LLC, Ashland, OR, USA).
Nuclear uptake of pDNA
The transfected cells in different groups were fixed with 4% paraformaldehyde for 15 min and rinsed with PBS three times. The nuclear stain DAPI (Beyotime Biotechnology Inc., Nantong, China) was added for 20 min to determine the cellular localization of fluorescence plasmids under an inverted fluorescence microscope (Olympus Corporation, Tokyo, Japan). ImageJ software version 1.46 (National Institutes of Health, Bethesda, MD, USA) was used to quantify the fluorescence intensity (FL) of the cell (FLcell) and the nucleus (FLnucleus). Nuclear uptake (%) = (FLnucleus / FLcell) × 100%.
Reverse transcription-polymerase chain reaction (RT-PCR)
To evaluate the relative mRNA expression levels of EGFP-N3, total mRNA was extracted from 293T cells using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.). A total of 2 µg RNA of each sample was reverse-transcribed into cDNA using an RT-for-PCR kit (Takara Bio, Inc., Otsu, Japan). RT-PCR was performed using a Light Cycler 480 instrument with software version 1.5 (Roche Diagnostics GmbH, Mannheim, Germany). The amplification was performed using a Light Cycler 480 SYBR Green Master Mix (Roche Diagnostics GmbH). The program was performed as follows: 10 sec complete denaturation at 95°C and followed by 30 cycles of exponential amplification. Each cycle consisted of a 30 sec denaturation step at 95°C, a 20 sec annealing step at 60°C, and a 20 sec incubation at 72°C for extension. The primers were as follows: Forward, 5′-TTTATGGTGAGCAAGGGCGAG-3′ and reverse, 5′-TTTTGGTGCAGATGAACTTCAG-3′ for EGFP-N3; and forward, 5′-TCAAGAAGGTGGTGAAGCAGG-3′ and reverse, 5′-TCAAAGGTGGAGGAGTGGGT-3′ for GAPDH. The results were normalized to GAPDH expression (16). Each experiment was replicated three times.
Western blot analysis
EGFP-N3 protein expression levels were determined by western blot analysis. Total proteins were extracted from the 293T cells with RIPA lysis buffer product. The lysate was incubated on ice for 30 min and then centrifuged at 16,000 × g at 4°C for 30 min in a centrifuge (HI650; Hunan Xiangyi Laboratory Instrument Development Co., Ltd., Hunan, China). The supernatant containing proteins was collected and 20 µg protein from each sample were subjected to 10% sodium dodecyl sulfate-polyacrylamide gel and electrophoresis. Following electrophoresis, the proteins were transferred to polyvinylidene fluoride (PVDF) membranes. The membranes were incubated in TBST solution containing 5% non-fat milk to block the nonspecific reaction. The membranes were incubated with an anti-EGFP antibody (ab6556; 1:1,000; Abcam) and anti-GAPDH antibody (ab9485; 1:1,000; Abcam) at 4°C overnight. Then, the membranes were washed with TBST and incubated with horseradish peroxidase (HRP)-conjugated secondary antibodies (ab6721; 1:10,000; Abcam) for 1 h at room temperature. Following which, the membranes were washed with TBST and incubated in developing solution (NCI5079; Thermo Fisher Scientific, Inc.). Finally, the membranes were exposed to X-rays to detect the expression bands. The density of the respective bands was quantitated by BandScan software version 5.1 (Glyko; BioMarin Pharmaceutical, Inc., Novato, CA, USA). The result of protein expression were normalized to the protein expression of GAPDH.
Statistical analysis
Statistical analyses were performed using SPSS software version 18.0 (SPSS, Inc., Chicago, IL, USA). Data are expressed as the mean ± standard deviation. Groups were compared using Student-Newman-Keuls q-test or independent-samples t-test. P<0.05 was considered to indicate a statistically significant difference.
Results
Cytoplasmic uptake of pDNA is increased by AT-MCB
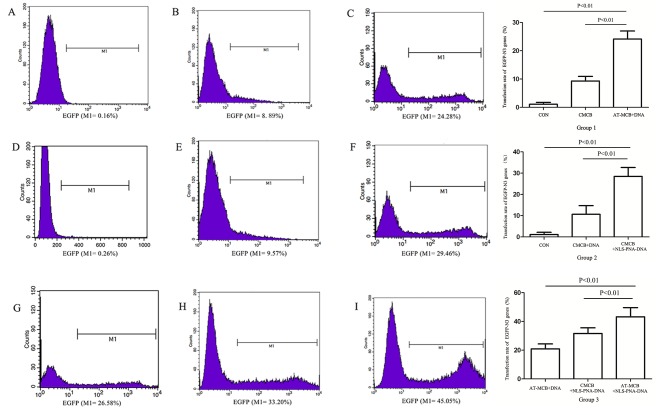
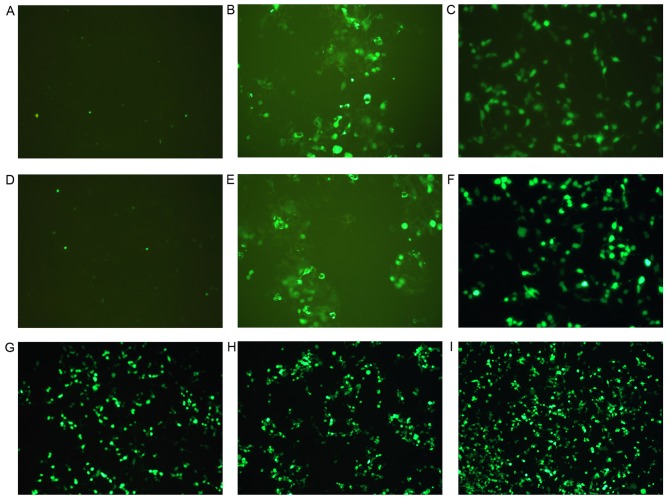
Transfections were performed using the optimal ultrasound exposure parameters, which were determined by our previous study (14): 1×107 microbubbles/ml and 1.5 W/cm2 for 45 sec with a 20% duty cycle. After 24 h, flow cytometry was used to analyze the cytoplasmic uptake of pDNA. The percentage of fluorescent 293T cells following exposure to optimal ultrasound parameters in the presence of AT-MCB was 45%, which was greater compared with ordinary microbubbles (CMCB; 9%; Fig. 3). The cell viability was >85% under the optimal exposure conditions. These results suggested that the optimal parameters of ultrasound exposure combined with AT-MCB may be an effective and safe method of gene delivery. The differences between all three groups and the blank control group were statistically significant (P<0.01), as were the differences between the three groups and the negative control group. Cells were observed under a fluorescence microscope, and those in the AT-MCB + NLS-PNA-DNA group exhibited the greatest fluorescence. AT-MCB + DNA and CMCB + NLS-PNA-DNA had the greatest quantity of fluorescence-labeled cells in each of the subgroups (Fig. 4).
Figure 3.
Flow cytometric analysis and a graph of the results following gene transfection in the three groups (group 1, 2 and 3). Histograms reveal the expression of EGFP by cells from: (A) Group 1, control; (B) group 1, CMCB + DNA; (C) group 1, AT-MCB + DNA; (D) group 2, control; (E) group 2, CMCB + DNA; (F) group 2, CMCB + NLS-PNA-DNA; (G) group 3, AT-MCB + DNA; (H) group 3, CMCB + NLS-PNA-DNA; and (I) group 3, AT-MCB + NLS-PNA-DNA. EGFP, enhanced green fluorescent protein; CMCB, ordinary microbubbles; AT-MCB, antibody targeted microbubbles; NLS, nuclear localization signal; PNA, peptide nucleic acid.
Figure 4.
Fluorescence of cells following gene transfection in the three groups (group 1, 2 and 3). Images reveal the expression of enhanced green fluorescent protein by cells from: (A) Group 1, control; (B) group 1, CMCB + DNA; (C) group 1, AT-MCB + DNA; (D) group 2, control; (E) group 2, CMCB + DNA; (F) group 2, CMCB + NLS-PNA-DNA; (G) group 3, AT-MCB + DNA; (H) group 3, CMCB + NLS-PNA-DNA; and (I) group 3, AT-MCB + NLS-PNA-DNA. EGFP, enhanced green fluorescent protein; CMCB, ordinary microbubbles; AT-MCB, antibody targeted microbubbles; NLS, nuclear localization signal; PNA, peptide nucleic acid (magnification, ×60).
Cellular and nuclear uptake of pDNA is increased by AT-MCB combined with PNA-binding NLS
The mRNA expression levels of EGFP-N3 were greater in the AT-MCB + NLS-PNA-DNA group compared with the AT-MCB + DNA and CMCB + NLS-PNA-DNA groups (P<0.01; Fig. 5).
Figure 5.
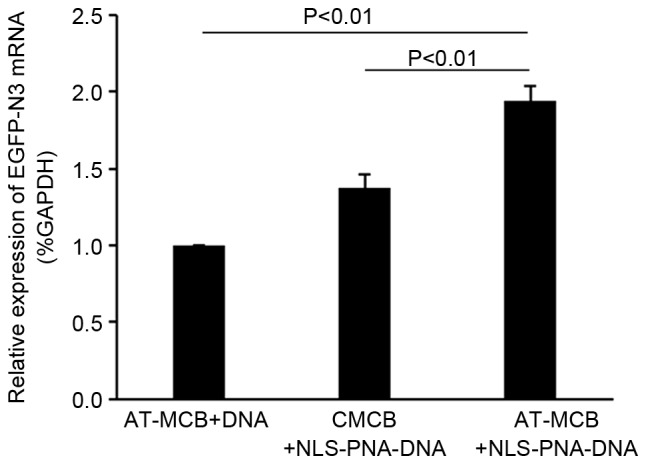
mRNA expression levels of EGFP-N3, as assessed using reverse transcription-polymerase chain reaction. Data are expressed as the mean ± standard deviation. EGFP, enhanced green fluorescent protein; CMCB, ordinary microbubbles; AT-MCB, antibody targeted microbubbles; NLS, nuclear localization signal; PNA, peptide nucleic acid.
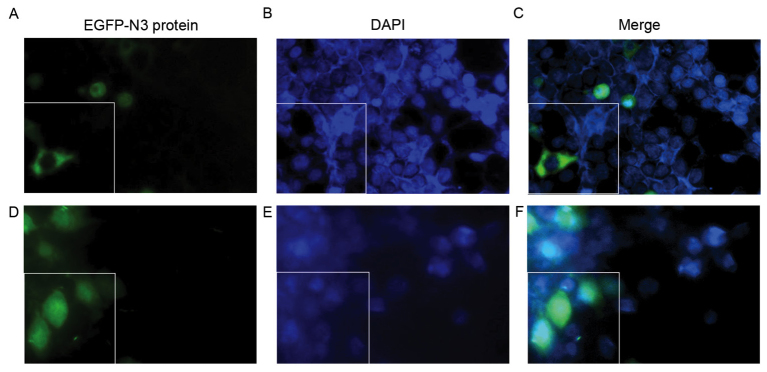
Following 48 h of transfection, western blot analysis revealed that the protein expression levels of EGFP were significantly greater in the AT-MCB + NLS-PNA-DNA group compared with the AT-MCB + DNA and CMCB + NLS-PNA-DNA groups (P<0.01; Fig. 6). Fig. 7 presents the subcellular localization of NLS-free DNA and NLS-DNA plasmids, which were both transfected with AT-MCB.
Figure 6.
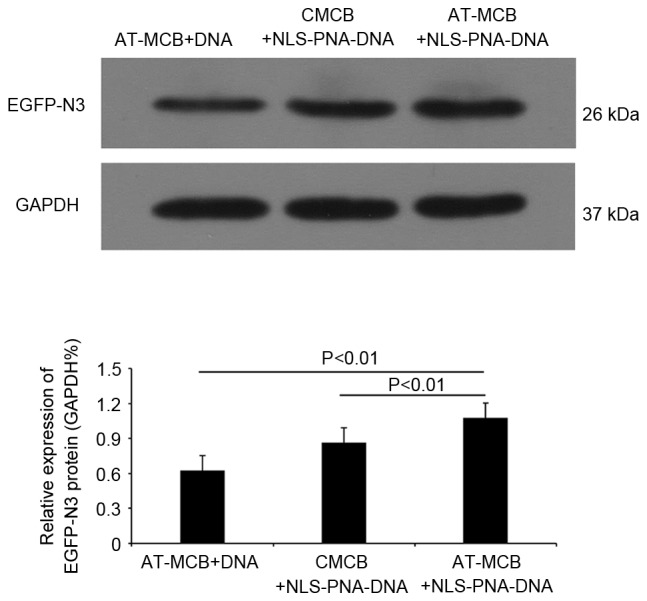
Protein expression levels of EGFP-N3, as assessed by western blot analysis. Data are expressed as the mean ± standard deviation. EGFP, enhanced green fluorescent protein; CMCB, ordinary microbubbles; AT-MCB, antibody targeted microbubbles; NLS, nuclear localization signal; PNA, peptide nucleic acid.
Figure 7.
Subcellular localization of the plasmids was observed using fluorescence microscopy. 293T cells were transfected with EGFP-N3 and were subsequently stained with DAPI, a fluorochrome that combines with DNA in the cell nucleus. (A-C) 293T cells were transfected with NLS-free EGFP-N3 plasmid DNA by UTMD. The cell nuclei contained no EGFP-N3. (D-F) 293T cells were transfected with NLS-PNA-EGFP-N3 plasmid DNA by UTMD. EGFP-N3 was present in the cell nuclei. EGFP-N3 plasmids appear green; DAPI appears blue. The boxed images present a typical single cell (magnification, ×100). EGFP, enhanced green fluorescent protein; NLS, nuclear localization signal; UTMD, ultrasound-targeted microbubble destruction; PNA, peptide nucleic acid.
Nuclear uptake of pDNA is increased by AT-MCB combined with PNA-binding NLS
Following 48 h of transfection, the fluorescence of the cells following gene transfection in the three groups were observed. As shown in Fig. 7, the subcellular localization of NLS-free DNA and NLS-DNA plasmids were presented under the fluorescence microscope, which were both transfected with AT-MCB. The fluorescence intensity in the nuclei of the NLS-PNA-DNA group was markedly greater compared with the NLS-free DNA group (>5.0-fold).
Discussion
Transfection efficiency is a critical issue in gene therapy. Poor transfection efficiency remains a major obstacle to non-viral gene therapy, and is due to genes being unable to cross the cytoplasmic and nuclear membranes (17). In the present study UTMD was combined with PNA-binding NLS to promote transportation through cytoplasmic and nuclear membranes. This strategy significantly increased the entrance of pDNA into the cytoplasm and nucleus, with the expression of the transfected gene at least 4.5-fold greater compared with the control group (CMCB). UTMD is considered to be an important method of gene delivery. Numerous studies have suggested that microbubbles, which serve as cavitation nuclei, facilitate cavitation during ultrasound exposure (5,18,19). To achieve good transfection efficiency, it is important for the pDNA to translocate from the cytoplasm to the nucleus. The cavitation of microbubbles leads to the transient formation of nanopores on the cell membrane that release large amounts of energy. These reversible pores are formed by sonoporation and may be observed by scanning electron microscopy (20). Therefore, exogenous pDNA or other molecules may enter cells through these nanopores, without causing irreversible cellular damage. Various types of microbubbles and ultrasound parameters have been investigated to improve transfection efficiency (21). In the present study, commercially available SonoVue lipid microbubbles were used and the optimized ultrasound exposure parameters were as follows: Microbubble concentration, 1×107/ml; acoustic intensity, 1.5 W/cm2; and exposure time, 45 sec. Ultrasound alone only slightly increased gene delivery efficiency, with a transfection rate of <5%, despite cell viability of >80%, suggesting that UTMD-mediated gene delivery is safe but inefficient.
To increase the uptake of exogenous genes into the cytoplasm, AT-MCB were used, which specifically recognized an antigen present on the cell membrane (22). Thus, microbubbles were localized to the cell membrane in greater numbers compared with CMCB. Following UTMD, more nanopores were formed for the entry of exogenous pDNA. However, the high cytoplasmic uptake of exogenous genes did not result in high expression, as the nuclear membrane prevents translocation of these genes from the cytoplasm to the nucleus.
NLS are special signal peptides that contain arginine, lysine and other basic amino acids, including the SV40 large T antigen, which was the first NLS demonstrated to stimulate gene translocation into the nuclei of non-dividing cells (23,24). NLS guide their sequence to the nucleus, and may bind to nucleic acid molecules to promote nuclear uptake. Under the guidance of NLS, the combined DNA and nuclear membrane receptor of NLS may form a complex called the nuclear pore targeting complex (25–27). PNA is a polynucleic acid analog that has the deoxyribose-phosphate backbone replaced with a peptide backbone (28). PNA may hybridize with cDNA in a double-stranded helix, resulting in a greater affinity compared with DNA-DNA. This hybrid is one of the most successful known DNA or RNA analogs that, due to the neutral charge of the PNA backbone, does not introduce a repulsive force to drive the hybridized strands apart. PNA has demonstrated the potential to invade duplex DNA and hybridize to plasmids to form a stable PNA-DNAPNA triplex hybrid. Using this strategy, NLS were connected to PNA (29).
In the present study, a specific NLS was designed and inserted into pDNA by combining PNA to promote nuclear import (30). Combining the NLS with genes and subsequently to an ultrasound-mediated AT-MCB increased the nuclear membrane permeability, which further improved nuclear gene transduction. These results indicated that PNA-binding NLS significantly promoted the entry of the plasmid from the cytoplasm to the nucleus. In addition, the protein expression levels of EGFP-N3 were ~1.5-fold greater in cells transfected with PNA-NLS compared with those transfected with normal pDNA. This result was consistent with increased cytoplasmic and nuclear import.
In conclusion, the present study demonstrated the efficiency of exogenous pDNA in delivering genes to cells. The findings of the present study indicate that UTMD combined with AT-MCB and a PNA-binding NLS plasmid may improve the import of exogenous genes by enhancing cytoplasmic and nuclear intake, thus providing an experimental basis for gene therapy for in vivo experiments and clinical application.
Acknowledgements
The present study was supported by grants from the National Natural Science Foundation of China (grant nos. 81201109 and 81471674).
References
- 1.Jafari M, Soltani M, Naahidi S, Karunaratne DN, Chen P. Nonviral approach for targeted nucleic acid delivery. Curr Med Chem. 2012;19:197–208. doi: 10.2174/092986712803414141. [DOI] [PubMed] [Google Scholar]
- 2.Dijkmans PA, Juffermans LJ, Musters RJ, van Wamel A, ten Cate FJ, van Gilst W, Visser CA, de Jong N, Kamp O. Microbubbles and ultrasound: From diagnosis to therapy. Eur J Echocardiogr. 2004;5:245–256. doi: 10.1016/j.euje.2004.02.001. [DOI] [PubMed] [Google Scholar]
- 3.Bouakaz A, Versluis M, de Jong N. High-speed optical observations of contrast agent destruction. Ultrasound Med Biol. 2005;31:391–399. doi: 10.1016/j.ultrasmedbio.2004.12.004. [DOI] [PubMed] [Google Scholar]
- 4.De Jong N, Emmer M, van wamel A, Versluis M. Ultrasonic characterization of ultrasound contrast agents. Med Biol Eng Comput. 2009;47:861–873. doi: 10.1007/s11517-009-0497-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bekeredjian R, Chen S, Frenkel PA, Grayburn PA, Shohet RV. Ultrasound-targeted microbubble destruction can repeatedly direct highly specific plasmid expression to the heart. Circulation. 2003;108:1022–1026. doi: 10.1161/01.CIR.0000084535.35435.AE. [DOI] [PubMed] [Google Scholar]
- 6.Juffermans LJ, van Dijk A, Jongenelen CA, Drukarch B, Reijerkerk A, de Vries HE, Kamp O, Musters RJ. Ultrasound and microbubble-induced intra- and intercellular bioeffects in primary endothelial cells. Ultrasound Med Biol. 2009;35:1917–1927. doi: 10.1016/j.ultrasmedbio.2009.06.1091. [DOI] [PubMed] [Google Scholar]
- 7.Miller AM, Munkonge FM, Alton EW, Dean DA. Identification of protein cofactors necessary for sequence-specific plasmid DNA nuclear import. Mol Ther. 2009;17:1897–1903. doi: 10.1038/mt.2009.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zabner J, Fasbender AJ, Moninger T, Poellinger KA, Welsh MJ. Cellular and molecular barriers to gene transfer by a cationic lipid. J Biol Chem. 1995;270:18997–19007. doi: 10.1074/jbc.270.32.18997. [DOI] [PubMed] [Google Scholar]
- 9.Young JL, Dean DA. Nonviral gene transfer strategies for the vasculature. Microcirculation. 2002;9:35–49. doi: 10.1038/sj.mn.7800120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ma H, Zhu J, Maronski M, Kotzbauer PT, Lee VM, Dichter MA, Diamond SL. Non-classical nuclear localization signal peptides for high efficiency lipofection of primary neurons and neuronal cell lines. Neuroscience. 2002;112:1–5. doi: 10.1016/S0306-4522(02)00044-1. [DOI] [PubMed] [Google Scholar]
- 11.Klibanov AL. Targeted delivery of gas-filled microspheres, contrast agents for ultrasound imaging. Adv Drug Deliv Rev. 1999;37:139–157. doi: 10.1016/S0169-409X(98)00104-5. [DOI] [PubMed] [Google Scholar]
- 12.Suzuki R, Takizawa T, Negishi Y, Hagisawa K, Tanaka K, Sawamura K, Utoguchi N, Nishioka T, Maruyama K. Gene delivery by combination of novel liposomal bubbles with perfluoropropane and ultrasound. J Control Release. 2007;117:130–136. doi: 10.1016/j.jconrel.2006.09.008. [DOI] [PubMed] [Google Scholar]
- 13.Suzuki R, Takizawa T, Negishi Y, Utoguchi N, Sawamura K, Tanaka K, Namai E, Oda Y, Matsumura Y, Maruyama K. Tumor specific ultrasound enhanced gene transfer in vivo with novel liposomal bubbles. J Control Release. 2008;125:137–144. doi: 10.1016/j.jconrel.2007.08.025. [DOI] [PubMed] [Google Scholar]
- 14.Deng Q, Chen JL, Zhou Q, Hu B, Chen Q, Huang J, Guo RQ. Ultrasound microbubbles combined with the NFκB binding motif increase transfection efficiency by enhancing the cytoplasmic and nuclear import of plasmid DNA. Mol Med Rep. 2013;8:1439–1445. doi: 10.3892/mmr.2013.1672. [DOI] [PubMed] [Google Scholar]
- 15.Zhang L, Liu Y, Xiang G, Lv Q, Huang G, Yang Y, Zhang Y, Song Y, Zhou H, Xie M. Ultrasound-triggered microbubble destruction in combination with cationic lipid microbubbles enhances gene delivery. J Huazhong Univ Sci Technolog Med Sci. 2011;31:39–45. doi: 10.1007/s11596-011-0147-3. [DOI] [PubMed] [Google Scholar]
- 16.Yuan Y, Yan L, Wu QQ, Zhou H, Jin YG, Bian ZY, Deng W, Yang Z, Shen DF, Zeng XF, et al. Mnk1 (mitogen-activated protein kinase-interacting kinase 1) deficiency aggravates cardiac remodeling in mice. Hypertension. 2016;68:1393–1399. doi: 10.1161/HYPERTENSIONAHA.116.07906. [DOI] [PubMed] [Google Scholar]
- 17.Pérez-Martínez FC, Guerra J, Posadas I, Ceña V. Barriers to non-viral vector-mediated gene delivery in the nervous system. Pharm Res. 2011;28:1843–1858. doi: 10.1007/s11095-010-0364-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mehier-Humbert S, Bettinger T, Yan F, Guy RH. Plasma membrane poration induced by ultrasound exposure: Implication for drug delivery. J Control Release. 2005;104:213–222. doi: 10.1016/j.jconrel.2005.01.007. [DOI] [PubMed] [Google Scholar]
- 19.Wan C, Li F, Li H. Gene therapy for ocular diseases meditated by ultrasound and microbubbles (Review) Mol Med Rep. 2015;12:4803–4814. doi: 10.3892/mmr.2015.4054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Newman CM, Bettinger T. Gene therapy progress and prospects: Ultrasound for gene transfer. Gene Ther. 2007;14:465–475. doi: 10.1038/sj.gt.3302925. [DOI] [PubMed] [Google Scholar]
- 21.Duvshani-Eshet M, Machluf M. Therapeutic ultrasound optimization for gene delivery: A key factor achieving nuclear DNA localization. J Control Release. 2005;108:513–528. doi: 10.1016/j.jconrel.2005.08.025. [DOI] [PubMed] [Google Scholar]
- 22.Tsutsui JM, Xie F, Porter RT. The use of microbubbles to target drug delivery. Cardiovasc Ultrasound. 2004;2:23. doi: 10.1186/1476-7120-2-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kimmelman J. Protection at the cutting edge: The case for central review of human gene transfer research. CMAJ. 2003;169:781–782. [PMC free article] [PubMed] [Google Scholar]
- 24.Benimetskaya L, Guzzo-Pernell N, Liu ST, Lai JC, Miller P, Stein CA. Protamine-fragment peptides fused to an SV40 nuclear localization signal delivery oligonucleotides that produce antisense effects in prostate and bladder carcinoma cells. Bioconjug Chem. 2002;13:177–178. doi: 10.1021/bc010068j. [DOI] [PubMed] [Google Scholar]
- 25.Imamoto N. Diversity in nucleocytoplasmic transport pathways. Cell Struct Funct. 2000;25:207–216. doi: 10.1247/csf.25.207. [DOI] [PubMed] [Google Scholar]
- 26.Nakanishi M, Akuta T, Nagoshi E, Eguchi A, Mizuguchi H, Senda T. Nuclear targeting of DNA. Eur J Phar Sci. 2001;13:17–24. doi: 10.1016/S0928-0987(00)00203-7. [DOI] [PubMed] [Google Scholar]
- 27.Lange A, Mills RE, Lange CJ, Stewart M, Devine SE, Corbett AH. Classical nuclear localization signals: Definition, function, and interaction with importin alpha. J Biol Chem. 2007;282:5101–5105. doi: 10.1074/jbc.R600026200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nielsen PE, Egholm M, Berg RH, Buchardt O. Sequence-selective recognition of DNA by strand displacement with athymine-substituted polyamide. Science. 1991;254:1497–1500. doi: 10.1126/science.1962210. [DOI] [PubMed] [Google Scholar]
- 29.Zelphati O, Liang X, Nguyen C, Barlow S, Sheng S, Shao Z, Felgner PL. PNA-dependent gene chemistry: Stable coupling of peptides and oligonucleotides to plasmid DNA. Biotechniques. 2000;28:304–310. doi: 10.2144/00282rr01. [DOI] [PubMed] [Google Scholar]
- 30.Munkonge FM, Amin V, Hyde SC, Green AM, Pringle IA, Gill DR, Smith JW, Hooley RP, Xenariou S, Ward MA, et al. Identification and functional characterization of cytoplasmic determinants of plasmid DNA nuclear import. J Biol Chem. 2009;284:26978–26987. doi: 10.1074/jbc.M109.034850. [DOI] [PMC free article] [PubMed] [Google Scholar]