Abstract
The canonical Wnt/β-catenin signaling is important in the differentiation of human mesenchymal stem cells into osteoblasts. Accumulating evidence suggests that the expression of β-catenin is, in part, regulated by specific microRNAs (miRNAs). The aim of the present study was to investigate the putative roles of miRNAs in osteoblast differentiation. Polymerase chain reaction (PCR) arrays were used to identify miRNAs that were differentially expressed between differentiated and non-differentiated periodontal ligament stem cells (PDLSCs), and reverse transcription-quantitative PCR (RT-qPCR) was used for validation. Since miR-214 was revealed to be significantly downregulated during PDLSC differentiation, its function was further investigated via silencing and overexpression. In addition, osteogenic differentiation of PDLSCs was evaluated at 10 and 21 days following induction, using Alizarin red staining and RT-qPCR analysis for mRNA expression levels of the osteogenic differentiation markers alkaline phosphatase (ALP), osteocalcin and bone sialoprotein. Furthermore, the potential target genes of miR-214 were investigated using a dual-luciferase reporter assay, RT-qPCR and western blot analysis, whereas a TOPflash/FOPflash reporter plasmid system followed by a luciferase assay was used to examine the effects of miR-214 on Wnt/β-catenin signaling. The present results demonstrated that miR-214 was significantly downregulated during the osteoblastic differentiation of PDLSCs. Notably, its overexpression inhibited PDLSC differentiation, whereas its knockdown promoted PDLSC differentiation, as revealed by alterations in mRNA expression of osteoblast-specific genes and ALP. In addition, miR-214 was demonstrated to directly interact with the 3′-untranslated region of the β-catenin gene CTNNB1, and suppressed Wnt/β-catenin signaling through the inhibition of β-catenin. The results of the present study suggested that miR-214 may participate in the regulation of the Wnt/β-catenin signaling pathway, and may have potential as a candidate target for the development of preventive or therapeutic agents for the treatment of patients with osteogenic disorders.
Keywords: Wnt/β-catenin pathway, miR-214, periodontal ligament stem cell, differentiation, β-catenin
Introduction
Periodontitis is a chronic inflammatory disease which results in the loss of periodontal tissue, and is clinically manifested with the loss of periodontal attachment, the formation of periodontal pockets, alveolar bone osteopenia and ultimately teeth exfoliation (1). Periodontal ligament stem cells (PDLSCs), a newly recognized subpopulation of mesenchymal stem cells (MSCs), have been suggested as potential seed cells for periodontal tissue regeneration, for the formation of cementum-periodontal ligament complexes and adjacent bone tissues (2–4). However, effective strategies for the regulation of their osteogenic differentiation potency have yet to be developed. Therefore, the elucidation of the molecular mechanisms underlying the osteogenic differentiation of PDLSCs is critical prior to their use in tooth regeneration and osteogenic tissue engineering applications.
Numerous regulatory pathways, including Wnt, transforming growth factor-β and bone morphogenetic protein-mediated signaling, are implicated in the progression of osteogenesis (5–7). In particular, the canonical Wnt/β-catenin signaling pathway has been identified as critical for the differentiation of human MSCs into osteoblasts. Wnt/β-catenin signaling has been demonstrated to be important in MSC fate determination and differentiation, including osteogenic (8), chondrogenic (9), adipogenic (10) and myogenic differentiation (11). However, the molecular mechanisms underlying the role of Wnt/β-catenin signaling in PDLSC differentiation, as well as its potential therapeutic applications have yet to be explored.
Accumulating evidence suggests that osteoblastic induction and differentiation are also regulated by post-transcriptional mechanisms, partly through temporarily-expressed microRNAs (miRNAs) (12). miRNAs are a class of endogenous small noncoding RNA molecules, with a length of 18–22 nucleotides. Mature miRNAs bind to the 3′-untranslated region (UTR) of target mRNAs and repress their translation or induce their degradation (13). Mizuno et al (12) reported that miR-125b inhibited the osteoblastic differentiation of stem cells through the downregulation of cell proliferation. miR-27 has been reported to regulate adipogenesis, myeloblast differentiation and skeletal muscle development (14,15). However, to the best of our knowledge, the putative roles of miRNAs in PDLSC differentiation have only been investigated at a preliminary level (16), and the molecular mechanisms underlying the involvement of miRNAs in PDLSC differentiation have yet to be elucidated.
The aim of the present study was to investigate the differential expression of miRNAs during the osteogenic differentiation of PDLSCs and explore their biological actions. The present results demonstrated that inhibition of miR-214 resulted in the cellular accumulation of β-catenin and the activation of Wnt signaling, by targeting β-catenin gene expression, and thus potentiated stem cell differentiation.
Materials and methods
PDLSC isolation and cultivation
Human premolars were obtained from 5 men aged 18–24 years (mean age, 20.2) who underwent molar extraction at the Department of Stomatology of the First Affiliated Hospital of Jinan University (Guangzhou, China) from July 2015 to May 2016. Written informed consent was obtained prior to study inclusion and the present study was approved by the Ethics Committee of the First Affiliated Hospital of Jinan University. Tissue from the periodontal ligament was isolated as previously described (17). Briefly, following extraction, the periodontal ligament was gently scraped from the middle portion of the root surface, minced into 1 mm3 cubes and placed into 6-well culture dishes. The explants were cultured in Minimum Essential Medium Eagle-α modification (αMEM; Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA), supplemented with 10% fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc.), 0.292 mg/ml glutamine, 100 U/ml penicillin and 100 µg/ml streptomycin. The cultures were incubated at 37°C in a 5% CO2 humidified atmosphere for 10–14 days. Stro-1-positive MSCs were isolated using immunomagnetic Dynabeads (cat. no. 110.41 and 110.42; Invitrogen; Thermo Fisher Scientific, Inc.), according to the manufacturer's instructions. Following washing, bead-bound cells were isolated after a sequential digestion of the extracellular matrix with pronase (0.15%, w/v) for 2 h and collagenase (0.2%, w/v) overnight at 37°C. The cells were centrifuged with 200 × g for 5 min at 4°C and subsequently suspended in αMEM with 10% FBS, and then seeded in 6-well culture dishes at a density of ~1×105 cells/well. Cells were cultured as monolayers until they reached confluence (10–12 days) at 37°C in a humidified atmosphere containing 5% CO2, with a change of culture medium every 3–4 days.
PDLSC differentiation
Differentiation was induced 3 days post confluence. The culture media was exchanged for osteogenic medium, which contained 10% FBS and DAG (100 nM dexamethasone, 50 µM ascorbic acid, and 5 mM β-glycerophosphate) in α-MEM. The medium was changed every 2–3 days during the course of incubation (0–21 days). Cells cultured in basal media (αMEM with 10% FBS) served as a control. These undifferentiated and differentiated cells were used in subsequent experiments.
RNA extraction
Total RNA and miRNA were isolated from the undifferentiated and differentiated PDLSCs with TRizol (Invitrogen; Thermo Fisher Scientific, Inc.) or an miRNeasy Mini kit (Qiagen, Inc., Valencia, CA, USA), respectively, according to the manufacturers' instructions. Potential genomic DNA contamination was removed from the samples by digestion with RNase-free DNase (Qiagen, Inc.) for 15 min at room temperature. The RNA concentration was measured using a NanoDrop 1000 Spectrophotometer (NanoDrop; Thermo Fisher Scientific, Inc., Pittsburgh, PA, USA), and the quality of miRNA and mRNA was evaluated using an Agilent 2100 Bioanalyzer system (Agilent Technologies, Inc., Santa Clara, CA, USA).
Polymerase chain reaction (PCR) arrays
The expression levels of 84 miRNAs related to osteoblastic differentiation in PDLSCs were profiled using miScript miRNA PCR Array System (cat. no. 331221; Qiagen, Inc.). Briefly, genomic DNA was eliminated from the RNA samples (1 µg) isolated from undifferentiated and differentiated PDLSCs, using an miRNeasy Mini kit (cat. no. 217004; Qiagen, Inc.), at 42°C for 5 min, and then transferred on ice for ~1 min for the removal of any residual DNA contamination. The purified RNA sample was used for cDNA synthesis using the reverse transcription mixture provided in the miScript II RT kit (cat. no. 218160; Qiagen, Inc.). The mixture was incubated at 42°C for 15 min and then at 95°C for 5 min. cDNA was subsequently used for quantitative PCR (qPCR) with the RT2 SYBR-Green qPCR Mastermix (cat. no. 218073; Qiagen, Inc.), according to the manufacturer's protocol. Data analysis was performed using the manufacturer's integrated web-based software package for the PCR Array System, using the 2−∆∆Cq method (18). Hierarchical clustering analysis and heatmap visualization were performed using CIMminer (http://discover.nci.nih.gov/cimminer).
Reverse transcription qPCR (RT-qPCR)
For detection of miRNAs and mRNAs, cDNA synthesis was performed from total RNA (0.2–0.5 µg) using the PrimeScript reverse transcription reagent kit (cat. no. DRR037A; Takara Biotechnology Co., Ltd., Dalian, China) according to the manufacturer's instructions. Quantitative analysis of the change in expression levels was performed using SYBR Premix Ex Taq™ (DRR420A; Takara Biotechnology Co., Ltd.) using an ABI 7300 system (Applied Biosystems; Thermo Fisher Scientific, Inc.). PCR cycling conditions were as follows: Initial incubation at 95°C for 30 sec, followed by 40 cycles of denaturation at 95°C for 5 sec and annealing at 60°C for 31 sec. Quantitative normalization was performed on the expression of RNU6 small nucleolar RNA or GAPDH, for miRNA and mRNA, respectively. The sequences of the primers are listed in Table I. Relative expression levels between samples were calculated using the 2−ΔΔCq method (18). Experiments were conducted in triplicate.
Table I.
Primer sequences use for polymerase chain reactions.
| Gene | Forward primer (5′-3′) | Reverse primer (5′-3′) |
|---|---|---|
| miR-214 | AGCCGACAGCAGGCACAGACA | AGCCGACAGCAGGCACAGACA |
| Let-7a | GTCGTATCCAGTGCAGGGTCCGAG | GTATTCGCACTGGATACGACAACT |
| miR-100 | GCGGCAACCCGTAGATCCGAA | GTGCAGGGTCCGAGGT |
| miR-125a-5p | TCCCTGAGACCCTTTAACCTGTGA | ACAGGTGAGGTTCTTGGGAGCC |
| miR-98 | ACACTCCCUAUACAACUUAC | GGGAAAGUAGUGAGGCCTCAGA |
| miR-142-5p | GGCCCATAAAGTAGAAAGC | TTTGGCACTAGCACATT |
| miR-128a | ATAGAATTCTTGAATACTGTGA | CCGGGATCCTAAGCAATAGCTTTCA |
| AGTACACTGC | CAAATT | |
| miR-184 | CTGGTAGGTGGACGGAGAACTG | TCAACTGGTGTCGTGGAGTCGGC |
| miR-19a | GTCGTATCCAGTGCAGGGTCCGAG | CTGGAGTGTGCAAATCTATGC |
| GTATTCGCACTGGATACGACTCAGTTT | ||
| miR-27b | CCGGCCTTCACAGTGGCTA | CGGGTCGGTGGCAGAACTT |
| U6 | CGCTTCACGAATTTGCGTGTCAT | GCTTCGGCAGCACATATACTAAAAT |
| ALP | CCACGTCTTCACATTTGGTG | AGACTGCGCCTGGTAGTTGT |
| BSP | AAAGTGAGAACGGGGAACCT | GATGCAAAGCCAGAATGGAT |
| OCN | GAGGGCAATAAGGTAGTGAA | CATAGATGCGTTTGTAGGC |
| GAPDH | GAAGATGGTGATGGGATTTC | GAAGGTGAAGGTCGGAGT |
miR, micro RNA; ALP, alkaline phosphatase; BSP, bone sialoprotein; OCN, osteocalcin.
Transfection
The miR-214 mimic (sense, 5′-aca gca gg cac aga cag gca gu-3′ and antisense, 5′-ugc cug ucu gug ccu gcu guu u-3′), mimic negative control (NC; sense, 5′-uuc ucc gaa cgu guc acg utt-3′ and antisense, 5′-acg uga cac guu cgg aga att-3′), miR-214 inhibitor (5′-ugc cug ucu gug ccu gcu guu u-3′) and inhibitor negative control (5′-cag uac uuu ugu gua gua caa-3′) were purchased from Shanghai GenePharma Co., Ltd. (Shanghai, China). PDLSCs (1×106 cells/well) were plated in a six-well plate 1 day prior to transfection. PDLSCs were then transfected with miR-214 mimic, mimic NC, miR-214 inhibitor or inhibitor NC, at a final concentration of 25 nM, using siLentFect Lipid Reagent (Bio-Rad Laboratories, Inc., Hercules, CA, USA). Subsequently, cells were cultured in serum-free Dulbecco's modified Eagle's medium: Nutrient Mixture F-12 (Shanghai GeneChem Co., Ltd., Shanghai, China). Following 4 h of incubation at 37°C, the medium was replaced with DMEM/F12 supplemented with 10% FBS. The cells were harvested for further experiments at 48 h after transfection.
Alizarin red staining
Cells were seeded into 24-well plates at a density of ~1×105 cells/well. When the cells had reached ~80% confluence, the culture medium was replaced with standard osteogenic differentiation induction medium as aforementioned and cultured for a further 3 weeks. The induction medium was replaced every 3 days. Following 3 weeks of culture, cells were stained with Alizarin red (pH 4.1) and analyzed under an inverted microscope (TE2000U; Nikon Corporation, Tokyo, Japan). The staining was quantified as previously described (19).
β-catenin/transcription factor (TCF) transcription reporter assay
TOPflash and FOPflash reporters (EMD Millipore, Billerica, MA, USA) contain wild type (WT) and mutated TCF-4 consensus binding sites, respectively, and are used to evaluate β-catenin-dependent signaling events that drive the expression of TCF. These reporters have been described previously (20). Briefly, 1×105 cells/well were seeded into a 24-well plate followed by transfection with TOPflash or FOPflash constructs, along with the miR-214 mimic, the miR-214 inhibitor or the corresponding NC miRNAs. All transfections were performed using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol. At 24 h following transfection, the β-catenin luciferase assay was performed using a Dual-Luciferase Assay System kit (Promega Corporation, Madison, WI, USA).
miRNA target prediction
To identify potential targets of miR-214, two public available algorithms, TargetScan (http://www.targetscan.org/) and PicTar (http://pictar.bio.nyu.edu/) were used. The interaction between miR-214 and the 3′-UTR of the putative target was predicted by RNAhybrid (http://bibiserv.techfak.uni-bielefeld.de/rnahybrid/).
Luciferase assay
The 3′-UTR of the β-catenin mRNA CTNNB1 (accession no. NM_001098209.1), with wild-type or mutant (Mut) binding sites for miR-214, was amplified from human cDNA by PCR, and subcloned into the pGL3 vector MCS cloning site (Promega Corporation) to generate the pGL3-WT-CTNNB1-3′-UTR or pGL3-Mut-CTNNB1-3′-UTR plasmids, respectively. CTNNB1 3′-UTR forward primer, 5′-TCTAGAATACAATGACTTTTTAGCTG-3′; CTNNB1 3′-UTR reverse primer, 5′-TCTAGATTAGCCAAG-3′. PDLSCs were seeded in duplicate in 24-well plates and co-transfected with 1–2 µg/ml pGL3-WT-CTNNB1-3′-UTR or pGL3-Mut-CTNNB1-3′-UTR plasmids and 25 nM miR-214 mimic or miR-214 inhibitor using Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) in accordance with the manufacturer's instructions. Corresponding NCs (Shanghai GenePharma Co., Ltd.) were transfected using the same procedure. Following incubation for 48 h, luciferase activity was analyzed using the Dual-Luciferase Reporter Assay System (Promega Corporation). Renilla luciferase was used for normalization.
Western blot analysis
Cells were lysed on ice using radioimmunoprecipitation assay lysis buffer (Beyotime Institute of Biotechnology, Haimen, China) and protein concentrations were determined using a bicinchoninic acid assay. Proteins (40 µg) were separated by 10% SDS-PAGE and transferred onto a polyvinylidene difluoride membrane (EMD Millipore) and incubated with 5% fat-free skim milk in TBS + 0.05% Tween 20, for 1 h at room temperature. Membranes were subsequently incubated overnight at 4°C with the following antibodies: Anti-CTNNB1 (rabbit monoclonal; 1:1,000; ab16051), anti-β-catenin (rabbit monoclonal; 1:1,000; ab32572), anti-TCF4 (rabbit polyclonal; 1:1,000; ab185736) or anti-β-actin (rabbit polyclonal; 1:1,000; ab8227) (all from Abcam, Cambridge, MA, USA). Secondary antibody incubations were performed with horseradish peroxidase-conjugated mouse anti-rabbit antibody (1:10,000; ab99702; Abcam) for 1 h at room temperature. Enhanced chemiluminescence substrate was used to visualize signals (EMD Millipore). β-actin was used as an endogenous protein for normalization. Relative band intensities were determined by densitometry using Quantity One 4.6.2 software (Bio-Rad Laboratories, Inc.).
Statistical analysis
Statistical analyses were performed using SPSS 13.0 (SPSS, Inc., Chicago, IL, USA). Data were expressed as the mean ± standard deviation of 3 independent experiments. Comparison of two groups was achieved using an unpaired two-tailed t-test. Differences between multiple groups were evaluated by one-way analysis of variance followed by Tukey's multiple comparison test. P<0.05 were considered statistically significant.
Results
miRNA profiling during PDLSC differentiation
Hierarchical clustering of PCR array results demonstrated systematic variations in the expression of various miRNAs between differentiated and non-differentiated PDLSCs (Fig. 1A). To validate the miRNA PCR array analysis findings, 5′upregulated and 5 downregulated miRNAs with the largest fold-change in expression were selected and their expression levels were assessed during PDLSC differentiation by RT-qPCR. The results revealed that let-7a, miR-100, miR-125b-5p, miR-98 and miR-142-5p were overexpressed during PDSLC differentiation, whereas the expression of miR-128a, miR-214, miR-184, miR-19a and miR-27b was suppressed (P<0.01; Fig. 1B); these results were in accordance with the expression patterns detected by the PCR array. These findings indicated that the expression of this set of miRNAs is differentially regulated during PDLSC differentiation. Among them, miR-214 was demonstrated to undergo the greatest change in expression during differentiation. Therefore, miR-214 was further investigated. The expression levels of miR-214 decrease in a time-dependent manner following the induction of osteoblastic differentiation of PDLSCs, compared with undifferentiated cells (Fig. 1C). These results suggested that miR-214 may be involved in the regulation of PDLSC osteoblastic differentiation.
Figure 1.
miRNA expression in differentiated and non-differentiated PDLSCs. (A) Heat map of miRNA profiles represents the differentially expressed miRNAs between differentiated and non-differentiated PDLSCs. Green indicates low expression levels; red indicates high expression levels. (B) RT-qPCR was performed to determine the expression of miRNAs in differentiated and undifferentiated PDLSCs. (C) RT-qPCR was used to assess the expression levels of miR-214 at the indicated time points in differentiated and non-differentiated PDLSCs. miR-214 expression was normalized to U6 small nuclear RNA. **P<0.01 vs. non-differentiated cells. miRNA, microRNA; PDLSC, periodontal ligament stem cell; RT-qPCR, reverse transcription quantitative polymerase chain reaction.
miR-214 inhibition promotes osteoblastic differentiation of PDLSCs
To investigate the roles of miR-214 in the osteoblastic differentiation of PDLSCs, cells were transfected with a miR-214 mimic or a miR-214 inhibitor and stained with Alizarin Red S. The results demonstrated that miR-214 knockdown significantly promoted osteogenesis in PDLSCs compared with control; conversely, miR-214 overexpression inhibited PDLSC osteoblastic differentiation compared with control (Fig. 2A). In addition, RT-qPCR analysis was performed in order to examine the mRNA expression levels of the osteogenic marker gene ALP and the mineralization markers OCN and BSP. The results demonstrated that miR-214 silencing significantly upregulated ALP, OCN and BSP mRNA expression compared with control, while miR-214 overexpression produced the opposite effects (Fig. 2B-D). Since the expression of these markers is indicative of osteoblastic differentiation status, these results suggested that miR-214 may be important in osteogenesis.
Figure 2.
miR-214 inhibition promotes the osteoblastic differentiation of PDLSCs. PDLSCs were transfected with a miR-214 mimic, a miR-214 inhibitor or NC miRNA. (A) Following 21 days of osteoblastic differentiation induction, Alizarin red staining demonstrated that PDLSCs transfected with a miR-214 inhibitor formed more mineralized nodules compared with control. Reverse transcription-quantitative polymerase chain reaction was used to assess the mRNA expression levels of the osteogenic differentiation markers (B) ALP, (C) osteocalcin and (D) BSP on days 10 and 21 post-induction. Data are expressed as the mean ± standard deviation of 3 independent experiments. **P<0.01, with comparisons indicated by lines. miR, microRNA; PDLSC, periodontal ligament stem cell; NC, negative control; ALP, alkaline phosphatase; BSP, bone sialoprotein.
miR-214 inhibits β-catenin gene expression
Bioinformatics analysis was used to predict potential target genes for miR-214, and results identified a putative miR-214 binding site in the 3′UTR of the β-catenin gene CTNNB1 (Fig. 3A). A luciferase reporter assay was used to investigate the putative miR-214-dependent post-transcriptional regulation of CTNNB1. The present results demonstrated that miR-214 knockdown significantly increased the luciferase activity in cells co-transfected with the plasmid containing wild-type CTNNB1 3′-UTR, whereas it produced no effect on mutant CTNNB1 3′-UTR-transfected cells (Fig. 3B).
Figure 3.
miR-214 directly binds to and inhibits the expression of the β-catenin CTNNB1 gene. (A) Schematic representation of the firefly luciferase reporter constructs for CTNNB1, indicating the putative interaction sites between miR-214 and the 3′-UTR of CTNNB1. (B) PDLSCs were co-transfected with luciferase reporter constructs containing wt or mut CTNNB1 3′-UTR and miR-214 mimic, miR-214 inhibitor or NC miRNA and luciferase activity was detected (n=3). (C) Western blot analysis was used to assess the protein expression levels of β-catenin in PDLSCs following transfection with miR-214 mimic, miR-214 inhibitor or NC miRNA (n=3). Data are expressed as the mean ± standard deviation of 3 independent experiments. **P<0.01, with comparisons indicated by lines. miR, microRNA; UTR, untranslated region; PDLSC, periodontal ligament stem cell; wt, wild-type; mt, mutant; NC, negative control.
To further investigate whether miR-214 may regulate the expression of β-catenin, the protein expression levels of β-catenin in cells transfected with a miR-214 mimic or a miR-214 inhibitor were evaluated by western blotting. The results revealed that miR-214 overexpression significantly downregulated the protein expression levels of β-catenin compared with control, while miR-214 inhibition significantly increased β-catenin protein expression compared with control (Fig. 3C).
Relationship between miR-214 expression and activity of the β-catenin/Wnt pathway
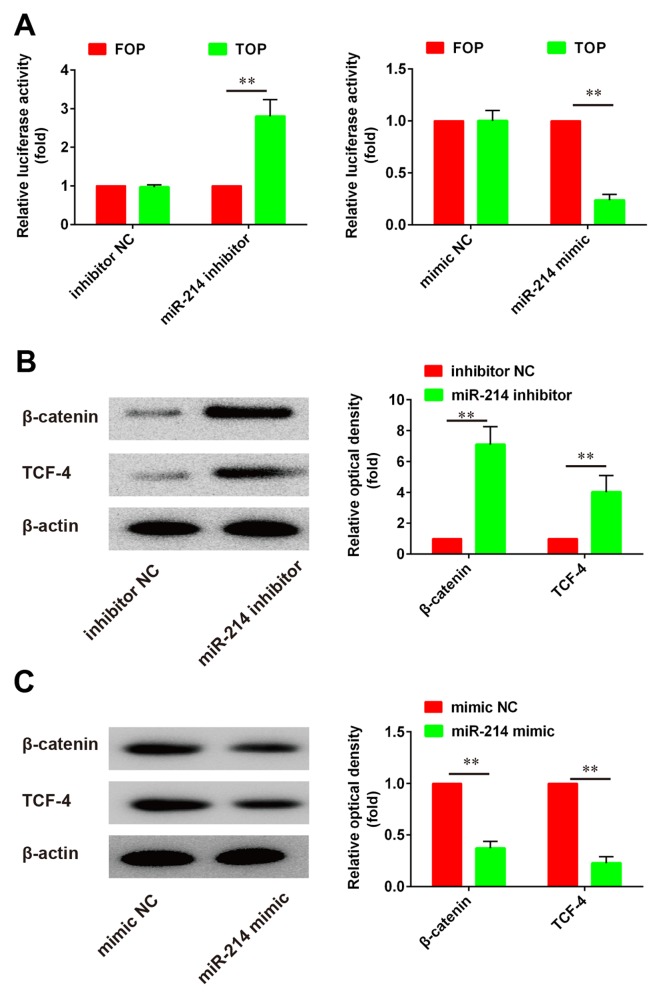
The Wnt/β-catenin signaling pathway has been reported to serve critical roles in the regulation of cell growth and differentiation. To evaluate the effects of miR-214 on Wnt/β-catenin signaling in PDLSCs, a luciferase assay based on a TOPflash/FOPflash reporter plasmid system was used to evaluate β-catenin-dependent transcriptional activity. The present results demonstrated that Wnt/β-catenin signaling was activated following miR-214 knockdown, whereas miR-214 overexpression produced the opposite effects, as evidenced by the alterations in luciferase activity (Fig. 4A). Western blot analysis was used to investigate the protein expression levels of β-catenin and TCF-4. The present results revealed that β-catenin and TCF-4 protein expression was significantly upregulated following miR-214 inhibition compared with control (Fig. 4B). By contrast, following miR-214 overexpression using a miR-214 mimic, β-catenin and TCF-4 protein expression was significantly suppressed compared with control (Fig. 4C). These findings suggested that miR-214 inhibition may promote the osteoblastic differentiation of PDLSCs, through the regulation of the Wnt/β-catenin signaling pathway.
Figure 4.
miR-214 modulates the activity of the Wnt/β-catenin signaling pathway. (A) A luciferase assay based on a TOPflash/FOPflash reporter plasmid system was used to evaluate β-catenin-dependent transcriptional activity. The TOPflash reporter construct contained wild-type TCF4 consensus binding sites, whereas the FOPflash construct contained mutated TCF4 binding sites that cannot be activated by β-catenin, and therefore is used as a negative control. Transfection with the miR-214 inhibitor resulted in increased luciferase activity, indicating increased transcriptional activity of the β-catenin/TCF4 complex. By contrast, transfection with the miR-214 mimic resulted in decreased β-catenin-dependent transcriptional activity. (B) Western blot analysis of β-catenin and TCF-4 protein expression in PDLSCs transfected with a miR-214 inhibitor. (C) Western blot analysis of β-catenin and TCF-4 protein expression in PDLSCs transfected with a miR-214 mimic. Data are expressed as the mean ± standard deviation of 3 independent experiments. **P<0.01, with comparisons indicated by lines. miR, microRNA; TCF, T cell factor; PDLSC, periodontal ligament stem cell; NC, negative control.
Discussion
In the present study a number of miRNAs were identified that were differentially expressed between differentiated and non-differentiated PDLSCs. Among them, miR-214 was further investigated by silencing and overexpression. The results suggested that miR-214 may be implicated in the mechanisms underlying osteoblastic differentiation of PDLSCs, as its overexpression inhibited osteogenesis, whereas its knockdown had the opposite effect. Furthermore, the β-catenin gene CTNNB1 was identified as a direct target of miR-214, and miR-214 was demonstrated to promote osteoblastic differentiation by modulating Wnt/β-catenin signaling.
miRNAs are small non-coding RNA molecules, with a length of ~22 nucleotides, that bind to target mRNAs and prevent their translation or promote their degradation (21). Through the post-transcriptional regulation of gene expression, miRNAs have been demonstrated to modulate numerous biological processes, including tumorigenesis, cellular proliferation and differentiation. miR-214 has previously been suggested to be implicated in osteogenic differentiation. Shi et al (22) reported that miR-214 suppressed osteogenic differentiation of C2C12 myoblasts by targeting the transcription factor Osterix, whereas Yang et al (23) demonstrated that miR-214 attenuated osteogenesis by inhibiting the fibroblast growth factor (FGF) receptor 1/FGF signaling pathway. These findings suggested that targeting miR-214 may hold potential as a novel therapeutic strategy for the treatment of patients with postmenopausal osteoporosis. However, the exact roles of miR-214 in the regulation of PDLSC differentiation, as well as the molecular mechanisms underlying its actions have yet to be elucidated.
The Wnt/β-catenin signaling pathway has been implicated in the mechanisms guiding cell fate decision in MSCs (8), and miRNAs have been reported to be involved in the regulation of Wnt/β-catenin signaling (24). Su et al (25) demonstrated that miR-200a regulated the epithelial-mesenchymal transition of cancer cells, via regulating the activity of the Wnt/β-catenin signaling pathway. In addition, Chen et al (26) reported that miR-709 inhibited adipocyte differentiation, by targeting glycogen synthase kinase 3β and subsequently activating Wnt/β-catenin signaling. Similarly, miR-27 has been demonstrated to promote odontoblastic differentiation in MDPC-23 cells by targeting adenomatous polyposis coli and activating Wnt/β-catenin signaling, through the accumulation of β-catenin (27). In the present study, the roles of Wnt/β-catenin signaling during osteoblastic differentiation were investigated in PDLSCs. The present results suggested that miR-214 may regulate the expression of β-catenin through the direct interaction with the 3′-UTR of CTNNB1. Furthermore, a TOPflash/FOPflash reporter luciferase assay was used to investigate the Wnt/β-catenin-dependent transcriptional activity of the TCF4 gene, and results suggested a significant association between miR-214 expression and Wnt/β-catenin signaling activity.
In conclusion, the present results suggested that miR-214 may regulate the Wnt/β-catenin signaling pathway by targeting CTNNB1, thus participating in the mechanisms underlying PDLSC differentiation. The present findings provide valuable insight into the molecular mechanisms underlying the complex processes of PDLSC osteoblastic differentiation.
References
- 1.Armitage GC. Periodontal diagnoses and classification of periodontal diseases. Periodontol. 2000;34:9–21. doi: 10.1046/j.0906-6713.2002.003421.x. [DOI] [PubMed] [Google Scholar]
- 2.Hiraga T, Ninomiya T, Hosoya A, Takahashi M, Nakamura H. Formation of bone-like mineralized matrix by periodontal ligament cells in vivo: A morphological study in rats. J Bone Miner Metab. 2009;27:149–157. doi: 10.1007/s00774-009-0039-9. [DOI] [PubMed] [Google Scholar]
- 3.Huang GT, Gronthos S, Shi S. Mesenchymal stem cells derived from dental tissues vs. those from other sources: Their biology and role in regenerative medicine. J Dent Res. 2009;88:792–806. doi: 10.1177/0022034509340867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kato T, Hattori K, Deguchi T, Katsube Y, Matsumoto T, Ohgushi H, Numabe Y. Osteogenic potential of rat stromal cells derived from periodontal ligament. J Tissue Eng Regen Med. 2011;5:798–805. doi: 10.1002/term.379. [DOI] [PubMed] [Google Scholar]
- 5.Lamplot JD, Qin J, Nan G, Wang J, Liu X, Yin L, Tomal J, Li R, Shui W, Zhang H, et al. BMP9 signaling in stem cell differentiation and osteogenesis. Am J Stem Cells. 2013;2:1–21. [PMC free article] [PubMed] [Google Scholar]
- 6.Coleman DT, Gray AL, Stephens CA, Scott ML, Cardelli JA. Repurposed drug screen identifies cardiac glycosides as inhibitors of TGF-β-induced cancer-associated fibroblast differentiation. Oncotarget. 2016;7:32200–32209. doi: 10.18632/oncotarget.8609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ishitani T, Kishida S, Hyodo-Miura J, Ueno N, Yasuda J, Waterman M, Shibuya H, Moon RT, Ninomiya-Tsuji J, Matsumoto K. The TAK1-NLK mitogen-activated protein kinase cascade functions in the Wnt-5a/Ca(2+) pathway to antagonize Wnt/beta-catenin signaling. Mol Cell Biol. 2003;23:131–139. doi: 10.1128/MCB.23.1.131-139.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boland GM, Perkins G, Hall DJ, Tuan RS. Wnt 3a promotes proliferation and suppresses osteogenic differentiation of adult human mesenchymal stem cells. J Cell Biochem. 2004;93:1210–1230. doi: 10.1002/jcb.20284. [DOI] [PubMed] [Google Scholar]
- 9.Hartmann C, Tabin CJ. Dual roles of Wnt signaling during chondrogenesis in the chicken limb. Development. 2000;127:3141–3159. doi: 10.1242/dev.127.14.3141. [DOI] [PubMed] [Google Scholar]
- 10.Ross SE, Hemati N, Longo KA, Bennett CN, Lucas PC, Erickson RL, MacDougald OA. Inhibition of adipogenesis by Wnt signaling. Science. 2000;289:950–953. doi: 10.1126/science.289.5481.950. [DOI] [PubMed] [Google Scholar]
- 11.Shang Y, Zhang C, Wang S, Xiong F, Zhao C, Peng F, Feng S, Yu M, Li M, Zhang Y. Activated beta-catenin induces myogenesis and inhibits adipogenesis in BM-derived mesenchymal stromal cells. Cytotherapy. 2007;9:667–681. doi: 10.1080/14653240701508437. [DOI] [PubMed] [Google Scholar]
- 12.Mizuno Y, Yagi K, Tokuzawa Y, Kanesaki-Yatsuka Y, Suda T, Katagiri T, Fukuda T, Maruyama M, Okuda A, Amemiya T, et al. miR-125b inhibits osteoblastic differentiation by down-regulation of cell proliferation. Biochem Biophys Res Commun. 2008;368:267–272. doi: 10.1016/j.bbrc.2008.01.073. [DOI] [PubMed] [Google Scholar]
- 13.Liang H, Li X, Wang L, Yu S, Xu Z, Gu Y, Pan Z, Li T, Hu M, Cui H, et al. MicroRNAs contribute to promyelocyte apoptosis in As2O3-treated APL cells. Cell Physiol Biochem. 2013;32:1818–1829. doi: 10.1159/000356615. [DOI] [PubMed] [Google Scholar]
- 14.Feng J, Iwama A, Satake M, Kohu K. MicroRNA-27 enhances differentiation of myeloblasts into granulocytes by post-transcriptionally downregulating Runx1. Br J Haematol. 2009;145:412–423. doi: 10.1111/j.1365-2141.2009.07632.x. [DOI] [PubMed] [Google Scholar]
- 15.McDaneld TG, Smith TP, Doumit ME, Miles JR, Coutinho LL, Sonstegard TS, Matukumalli LK, Nonneman DJ, Wiedmann RT. MicroRNA transcriptome profiles during swine skeletal muscle development. BMC Genomics. 2009;10:77. doi: 10.1186/1471-2164-10-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou Q, Zhao ZN, Cheng JT, Zhang B, Xu J, Huang F, Zhao RN, Chen YJ. Ibandronate promotes osteogenic differentiation of periodontal ligament stem cells by regulating the expression of microRNAs. Biochem Biophys Res Commun. 2011;404:127–132. doi: 10.1016/j.bbrc.2010.11.079. [DOI] [PubMed] [Google Scholar]
- 17.Seo BM, Miura M, Gronthos S, Bartold PM, Batouli S, Brahim J, Young M, Robey PG, Wang CY, Shi S. Investigation of multipotent postnatal stem cells from human periodontal ligament. Lancet. 2004;364:149–155. doi: 10.1016/S0140-6736(04)16627-0. [DOI] [PubMed] [Google Scholar]
- 18.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 19.Li B, Qu C, Chen C, Liu Y, Akiyama K, Yang R, Chen F, Zhao Y, Shi S. Basic fibroblast growth factor inhibits osteogenic differentiation of stem cells from human exfoliated deciduous teeth through ERK signaling. Oral Dis. 2012;18:285–292. doi: 10.1111/j.1601-0825.2011.01878.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Han L, Yang Y, Yue X, Huang K, Liu X, Pu P, Jiang H, Yan W, Jiang T, Kang C. Inactivation of PI3K/AKT signaling inhibits glioma cell growth through modulation of β-catenin-mediated transcription. Brain Res. 2010;1366:9–17. doi: 10.1016/j.brainres.2010.09.097. [DOI] [PubMed] [Google Scholar]
- 21.Jones-Rhoades MW, Bartel DP. Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Mol Cell. 2004;14:787–799. doi: 10.1016/j.molcel.2004.05.027. [DOI] [PubMed] [Google Scholar]
- 22.Shi K, Lu J, Zhao Y, Wang L, Li J, Qi B, Li H, Ma C. MicroRNA-214 suppresses osteogenic differentiation of C2C12 myoblast cells by targeting Osterix. Bone. 2013;55:487–494. doi: 10.1016/j.bone.2013.04.002. [DOI] [PubMed] [Google Scholar]
- 23.Yang L, Ge D, Cao X, Ge Y, Chen H, Wang W, Zhang H. MiR-214 Attenuates osteogenic differentiation of mesenchymal stem cells via targeting FGFR1. Cell Physiol Biochem. 2016;38:809–820. doi: 10.1159/000443036. [DOI] [PubMed] [Google Scholar]
- 24.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 25.Su J, Zhang A, Shi Z, Ma F, Pu P, Wang T, Zhang J, Kang C, Zhang Q. MicroRNA-200a suppresses the Wnt/β-catenin signaling pathway by interacting with β-catenin. Int J Oncol. 2012;40:1162–1170. doi: 10.3892/ijo.2011.1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen H, Mo D, Li M, Zhang Y, Chen L, Zhang X, Li M, Zhou X, Chen Y. miR-709 inhibits 3T3-L1 cell differentiation by targeting GSK3β of Wnt/β-catenin signaling. Cell Signal. 2014;26:2583–2589. doi: 10.1016/j.cellsig.2014.07.017. [DOI] [PubMed] [Google Scholar]
- 27.Wang T, Xu Z. miR-27 promotes osteoblast differentiation by modulating Wnt signaling. Biochem Biophys Res Commun. 2010;402:186–189. doi: 10.1016/j.bbrc.2010.08.031. [DOI] [PubMed] [Google Scholar]