Abstract
Glioblastoma multiforme (GBM) is defined by the World Health Organization as the most aggressive form of grade IV glioma, characterized by unrestrained cellular proliferation. microRNAs (miRs) serve important roles in the pathogenesis of GBM. However, the function of miR-194-5p in GBM remains unknown. In the present study, the miR-194-5p levels in GBM tissues and cells were evaluated using the reverse transcription-quantitative polymerase chain reaction. Cellular proliferation was tested by MTT analysis. Cellular apoptosis was analyzed by fluorescence-activated cell sorting. The protein level of insulin-like growth factor 1 receptor, the target gene of miR-194-5p, was evaluated by western blotting. The interaction between miR-194-5p and the target gene was confirmed by the dual-luciferase reporter assay. It was demonstrated that miR-194-5p inhibited cell growth and promoted apoptosis. In conclusion, the results of the present study indicated the tumor suppressive role of miR-194-5p.
Keywords: micro RNA-194-5p, cell growth, apoptosis, glioblastoma multiforme
Introduction
Glioblastoma multiforme (GBM) develops from the lineage of star-shaped glial cells (1). GBM is a highly prevalent cancer, accounting for 12–15% of all brain tumors and ~70% of all diagnosed gliomas (2,3). GBM is defined by the World Health Organization as the most aggressive form of grade IV glioma, characterized by unrestrained cellular proliferation, strong resistance to apoptosis, diffuse infiltration, vigorous angiogenesis and widespread genomic instability (4). The annual incidence of malignant glioma is >5/100,000 population (5). Currently, the clinical outcome of glioma has improved (6–8); however, the median survival rate of patients with GBM remains at only 12–15 months (9), thus, novel therapeutic targets are required.
Increasing evidence has confirmed that microRNAs (miRNAs/miRs) serve essential roles in the pathogenesis of various tumors. miRNAs are a class of small RNAs, ~18–22 nucleotides in length (10–12). miRNAs suppress protein expression through the inhibition of translation or the induction of mRNA degradation, by binding to the 3′ untranslated region (3′UTR) of target mRNAs (13). miRNAs are involved in a diverse range of biological processes, including cellular proliferation, migration, apoptosis and differentiation (14–18).
The insulin-like growth factor (IGF) signaling axis is critical to the growth, development and maintenance of a number of tissue types (19,20). A previous study demonstrated that the IGF 1 receptor (IGF1R) is an independent prognostic factor associated with decreased survival time in patients with GBM (21). The present study investigated the role of miR-194-5p in GBM and identified IGF1R as a target of miR-194-5p. IGF1R may warrant further investigation as a potential novel therapeutic target.
Materials and methods
Patients and tissue samples
A total of 10 GBM tissues and 10 corresponding adjacent normal tissues were collected from the Department of Neurosurgery, The People's Hospital of Leshan City (Leshan, China). The use of human tissues in the present study was evaluated and approved by the Ethic Committee of The People's Hospital of Leshan City. All patients enrolled in the present study provided written informed consent, and all specimens were handled and made anonymous according to the ethical and legal standards of China. All GBM tissues were confirmed and evaluated by a pathologist, and were frozen at −80°C immediately following surgery.
Cell culture
The normal human astrocytes (NHA; cat. no. 43923), and human glioblastoma cell lines U373 and U118, were purchased from the Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China). All cells were cultured in Dulbecco's modified Eagle's medium (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) with 10% fetal bovine serum (Gibco; Thermo Fisher Scientific, Inc.) at 37°C with 5% CO2. The U373 cell line is known to be contaminated with the U251 astrocytoma cell line and the U118 cell line is known to be contaminated with the U138 astrocytoma cell line (22).
Analysis of miR-194-5p in tissues and cell lines
Total RNA was extracted from cells using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol. The level of miR-194-5p was evaluated using a TaqMan MiRNA Assay (Applied Biosystems; Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol. U6 small nuclear RNA was used as an internal loading control. Reverse transcription-quantitative polymerase chain reaction was performed on an ABI 7900HT instrument (Applied Biosystems; Thermo Fisher Scientific, Inc.). The primers were synthesized and tested by Sangon Biotech Co., Ltd. (Shanghai, China). The primer sequences were as follows: U6 forward, GTG GAC CGC ACA AGC TCG CT, and reverse, TTG TTG AAC GGC ACT GTG TAT AGC A; miR-194-5p forward, AGT GTG ACG TTG ACA TCC GT, and reverse, GCA GCT CAG TAA CAG TCC GC. Relative miR-194-5p expression was calculated according to the comparative Cq method (23) and normalized to the expression of U6.
Overexpression and inhibition of miR-194-5p in vitro
miR-194-5p was overexpressed and inhibited in vitro by transfection with miR-194-5p mimics and an miR-194-5p antisense oligonucleotide (ASO), respectively. The miR-194-5p mimics and miR-194-5p ASO were purchased from Sangon Biotech Co., Ltd. The miRNA sequences were as follows: miR-194-5p mimic, 5′-AAGGCAGGGCCCCCGCUCCCC-3′; miR-194-5p ASO, 5′-GGAGCGGGGGCCCUGCCUUUU-3; scrambled miRNA control, 5′-CAGUACUUUUGUGUAGUACAA-3. The cells were seeded (1×106 cells/well) and cultured overnight. Transfection was performed for 30 min using Lipofectamine 2000 (Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol.
Cellular proliferation assay
After 24, 48 and 72 h of transfection, cellular proliferation was assessed by the MTT assay. Cells were seeded into 96-well plates at a density of 5×105 cells/well. MTT reagent was added into the medium at a final concentration of 0.1 mg/ml. A total of 100 µl DMSO was added. Optical density was measured by a microplate reader at 570 nm.
Cellular apoptosis analysis
A cell suspension (5×105 cells/ml) was prepared in annexin V-fluorescein isothiocyanate (FITC; Abcam, Shanghai, China) binding buffer and Annexin V-FITC was added and incubated for 15 min at room temperature. Subsequently, propidium iodide (PI; Abcam, Shanghai, China) was added, samples were analyzed by flow cytometry using an LSR II flow cytometer (BD Biosciences, Franklin Lakes, NJ, USA) at an excitation wavelength of 488 nm (argon-ion laser or solid state laser), and an emission wavelength of 530 nm for FITC and 590 nm for PI (24). Data analysis was performed using the BD FACSDiva software version 8.0.1 (BD Biosciences).
Prediction of the possible targets of miR-149-5p
Targetscan software (www.targetscan.org) was used to predict the potential targets of miR-149-5p.
Dual-luciferase reporter assays
Cells were seeded at 1×105 cells/well and were serum starved for 6 h prior to transfection. The wild-type (WT) 3′UTR of IGF1R and mutated controls were cloned and inserted into the dual-luciferase-reporter plasmid (500 ng; Shengong Biotechnology Company) and the pGL3-control (100 ng; Promega Corporation, Madison, WI, USA). miR-194-5p mimics and controls were subsequently transfected into U373 cells containing wild-type or mutant 3′UTR plasmids with Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.). Cells were harvested and luciferase activities were analyzed after 24 h using the Dual-Luciferase Reporter assay system (Promega Corporation). The mutant IGF1R 3′UTRs were generated using a Site-Directed Mutagenesis kit (Promega Corporation). The primers used to perform mutagenesis were as follows: IGF1R forward, 5′-ctgtccctgatgctgaaggcaggcagaatgacttc-3′, and reverse, 5′-gaagtcattctgctctgccttcagctcagggacag-3′. The kit was used according to the manufacturer's protocol.
Western blot analysis
Whole-cell lysates were prepared as previously described (25). The cell lysates were centrifuged at 4°C, 12,000 × g for 10 min, extracted and the proteins were quantified using a bicinchoninic acid protein quantification kit (cat. no. ab102536; Abcam, Cambridge, UK). Proteins (30 µg) were separated via 4–10% SDS-PAGE. Membranes were electrotransferred to nitrocellulose membranes (EMD Millipore, Billerica, MA, USA). The membranes were blocked with 5% non-fat milk overnight at 4°C and incubated with the anti-IGF1R antibody (cat. no. 3027; 1:1,000; Cell Signaling Technology, Inc., Danvers, MA, USA) overnight at 4°C. β-actin (cat. no. 4967; 1:1,000; Cell Signaling Technology, Inc.) was used as the endogenous control, incubated overnight at 4°C. Then the membranes were incubated with an anti-rabbit horseradish peroxidase-conjugated secondary antibody (cat. no. 7074; 1:1,000; Cell Signaling Technology, Inc.) for 2 h at room temperature. The bands were visualized using enhanced chemiluminescence (GE Healthcare Life Sciences, Little Chalfont, UK).
Statistical analysis
Data are presented as the mean ± standard deviation of 3 independent experiments. Statistical analysis was performed using SPSS software (version 10.0; SPSS, Inc., Chicago, IL, USA). The difference between two groups was analyzed using a two-tailed Student's t-test. One-way analysis of variance was used to analyze the difference between multiple groups, followed by the Student-Newman-Keuls method. P<0.05 was considered to indicate a statistically significant difference.
Results
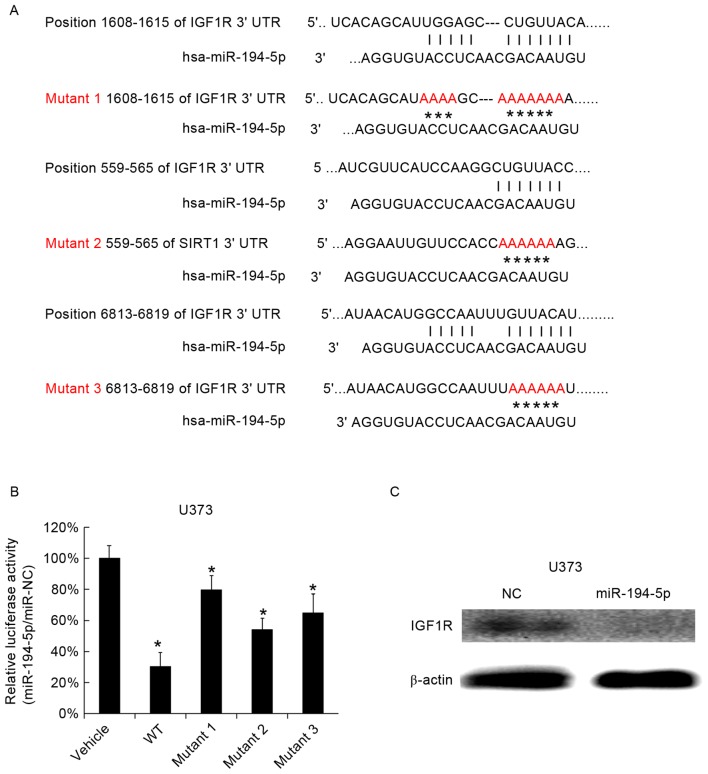
miR-194-5p targets IGF1R
IGF1R is an independent prognostic factor associated with decreased survival of patients with GBM (21). Therefore, miR-194-5p was selected for investigation, as it was identified to target IGF1R. A total of three miR-194-5p binding sites were identified in the 3′UTR of the IGF1R gene and three mutants were subsequently generated (Fig. 1A). The three mutated versions of the IGF1R 3′UTRs were cloned into luciferase reporter plasmids. miR-194-5p mimics and the mutant 3′UTRs were cotransfected into U373 cells, and it was observed that the miR-194-5p mimics reduced the luciferase activity of the WT 3′UTR, while the mutated 3′UTRs partially reduced the inhibitory function of miR-194-5p, scrambled miRNA was used as a vehicle (Fig. 1B). miR-194-5p mimics were subsequently transfected into U373 cells and the IGF-1R protein level was observed to be reduced 48 h following transfection (Fig. 1C). The results indicated that miR-194-5p may serve a role in GBM via IGF1R, thus miR-194-5p was selected for further study.
Figure 1.
IGF1R is targeted by miR-194-5p in U373 cells. (A) The three miR-194-5p binding sites in IGF1R and the mutated sites are listed. (B) miR-194-5p mimics were transfected into U373 cells containing WT or mutant 3′UTR plasmids and a dual-luciferase reporter assay was performed. (C) Western blotting of IGF1R was performed following transfection with miR-194-5p mimics. The data are presented as the mean ± standard deviation. Experiments were repeated at least three times. *P<0.05 vs. the vehicle. IGF1R, insulin-like growth factor 1 receptor; miR, microRNA; WT, wild-type; 3′UTR, 3′ untranslated region; NC, negative control.
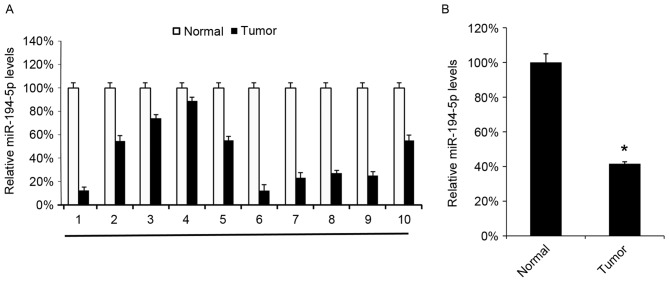
Decreased miR-194-5p in GBM tissues
miR-194-5p levels were evaluated in 10 GBM tissues and compared with the miR-194-5p levels in the 10 corresponding adjacent normal tissues. All the GBM tissues were observed to express decreased miR-194-5p levels compared with the corresponding adjacent normal tissues (Fig. 2A). The mean levels of the 10 GBM tissues also exhibited a decreased level of miR-194-5p compared with the normal tissues (Fig. 2B).
Figure 2.
Expression of miR-194-5p in GBM tissues. (A) Levels of miR-194-5p were analyzed by the reverse transcription-quantitative polymerase chain reaction in 10 GBM tissues and the corresponding adjacent healthy tissues. (B) The mean values of miR-194-5p in the 10 tissues were calculated. The data are presented as the mean ± standard deviation. Experiments were repeated at least three times. *P<0.05 vs. the normal tissues. miR, microRNA; GBM, glioblastoma multiforme.
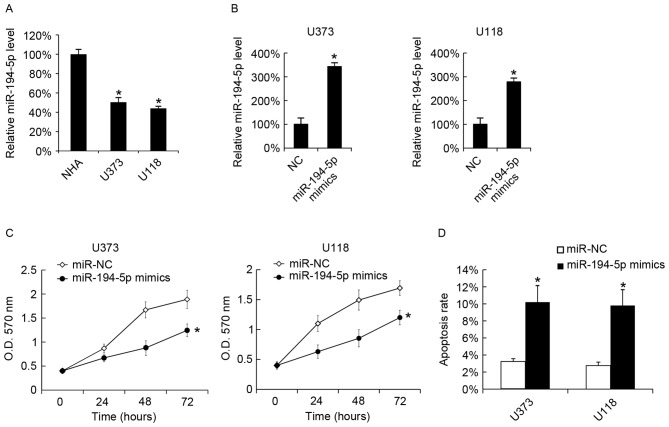
Effect of miR-194-5p mimics on GBM cell growth and apoptosis
miR-194-5p was overexpressed in GBM cells to test the function of miR-194-5p. The miR-194-5p levels in U373, U118 and NHA cells were analyzed by RT-qPCR, and it was observed that U373 and U118 cells expressed decreased levels of miR-194-5p compared with NHA cells (Fig. 3A). miR-194-5p was successfully overexpressed in U373 and U118 cells by miR-194-5p mimic transfection (Fig. 3B). MTT analysis revealed that the miR-194-5p mimics inhibited the growth of U373 and U118 cells (Fig. 3C). The cellular apoptotic rate was analyzed 48 h following miR-194-5p mimic transfection, and it was observed that miR-194-5p upregulated the apoptotic rate of U373 and U118 cells (Fig. 3D).
Figure 3.
Transfection with miR-194-5p mimics inhibits U373 and U118 cellular proliferation, and promotes cellular apoptosis. (A) miR-194-5p levels in NHA, U373 and U118 cells were analyzed by RT-qPCR. miR-194-5p mimics were transfected into (B) U373 and (C) U118 cells, and the miR-194-5p levels were evaluated by RT-qPCR 48 h following transfection. (C) After 24, 48 and 72 h of transfection, cellular proliferation was analyzed using the MTT assay. (D) A total of 48 h following miR-194-5p mimic transfection, cellular apoptosis was measured. The data are presented as the mean ± standard deviation. Experiments were repeated at least three times. *P<0.05 vs. the miR-NC. miR, microRNA; NHA, normal human astrocyte; RT-qPCR, reverse transcription-quantitative polymerase chain reaction; NC, negative control; O.D., optical density.
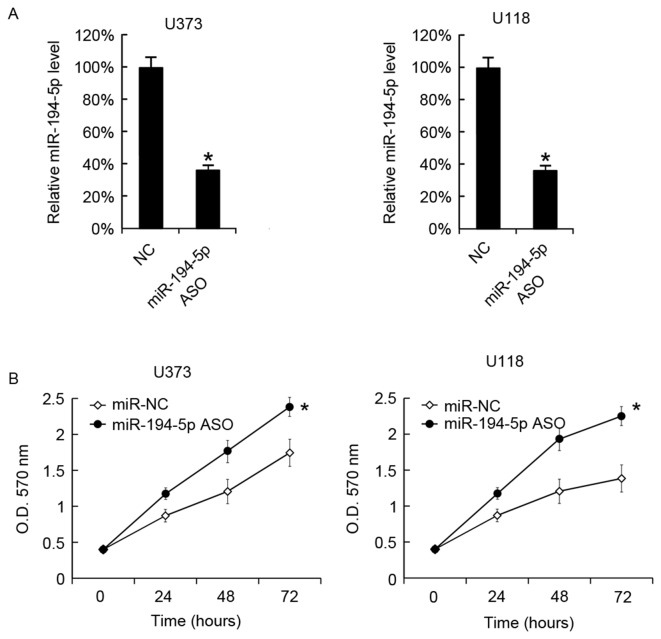
Effect of miR-194-5p ASO on cell growth
miR-194-5p was successfully downregulated in U373 and U118 cells by miR-194-5p ASO transfection. After 48 h, the miR-194-5p levels in cells were analyzed by RT-qPCR (Fig. 4A). Cellular proliferation in the two cell lines was analyzed using the MTT assay, and it was observed that miR-194-5p ASO promoted the cellular proliferation of U373 and U118 cells (Fig. 4B).
Figure 4.
Inhibition of miR-194-5p increases U373 and U118 cell growth. (A) miR-194-5p ASO was transfected into U373 and U118 cells, and the miR-194-5p levels were evaluated using the reverse transcription-quantitative polymerase chain reaction 48 h following transfection. (B) After 24, 48 and 72 h of transfection, cellular proliferation was analyzed using the MTT assay. The data are presented as the mean ± standard deviation. Experiments were repeated at least three times. *P<0.05 vs. the miR-NC. miR, microRNA; ASO, antisense oligonucleotide; NC, negative control; O.D., optical density.
Discussion
miR-194-5p inhibited cellular proliferation, invasion and migration in non-small cell lung cancer by targeting forkhead box A1 (26). In the present study, miR-194-5p was demonstrated to inhibit cellular proliferation and apoptosis in GBM by targeting IGF1R.
GBM immunotherapy studies observed that macrophages accumulate during GBM progression and may be targeted by inhibition of colony-stimulating factor-1 receptor (CSF-1R) (27,28). In clinical trials, several approaches to inhibit CSF-1R are currently being used, including antibodies and small molecules (29–31). Quail et al (32) studied acquired resistance to CSF-1R inhibition immunotherapy in glioma in 2016 and demonstrated that the phosphatidylinositol 3-kinase pathway activity was elevated in recurrent GBM, driven by macrophage-derived IGF1 and IGF1R. IGF1R inhibition with CSF-1R inhibition in recurrent tumors significantly prolonged overall survival in mice. In the present study, miR-194-5p was observed to target IGF1R in GBM cell lines. Therefore, it was hypothesized that miR-194-5p may be useful in GBM therapy. It was hypothesized that overexpression of miR-194-5p may inhibit tumor cell growth and promote cell apoptosis, and that inhibition of IGF1R may promote the effect of the macrophage-targeted immunotherapy. The role of miR-194-5p in macrophage-targeted immunotherapy requires further investigation.
In conclusion, the results of the present study demonstrated that miR-194-5p inhibited cellular proliferation and promoted cell apoptosis. The target gene of miR-194-5p is IGF1R. The present study demonstrated that miR-194-5p inhibited GBM cell growth and promoted apoptosis via IGF1R.
References
- 1.Parsons DW, Jones S, Zhang X, Lin JC, Leary RJ, Angenendt P, Mankoo P, Carter H, Siu IM, Gallia GL, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ohgaki H, Kleihues P. Genetic pathways to primary and secondary glioblastoma. Am J Pathol. 2007;170:1445–1453. doi: 10.2353/ajpath.2007.070011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ohgaki H, Kleihues P. Population-based studies on incidence, survival rates, and genetic alterations in astrocytic and oligodendroglial gliomas. J Neuropathol Exp Neurol. 2005;64:479–489. doi: 10.1093/jnen/64.6.479. [DOI] [PubMed] [Google Scholar]
- 4.Ramakrishna R, Pisapia D. Recent molecular advances in our understanding of glioma. Cureus. 2015;7:e287. doi: 10.7759/cureus.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fisher JL, Schwartzbaum JA, Wrensch M, Wiemels JL. Epidemiology of brain tumors. Neurol Clin. 2007;25:867–90, vii. doi: 10.1016/j.ncl.2007.07.002. [DOI] [PubMed] [Google Scholar]
- 6.Stupp R, Hegi ME, van den Bent MJ, Mason WP, Weller M, Mirimanoff RO, Cairncross JG, European Organisation for Research and Treatment of Cancer Brain Tumor and Radiotherapy Groups. National Cancer Institute of Canada Clinical Trials Group Changing paradigms-an update on the multidisciplinary management of malignant glioma. Oncologist. 2006;11:165–180. doi: 10.1634/theoncologist.11-2-165. [DOI] [PubMed] [Google Scholar]
- 7.Oermann E, Collins BT, Erickson KT, Yu X, Lei S, Suy S, Hanscom HN, Kim J, Park HU, Eldabh A, et al. CyberKnife® enhanced conventionally fractionated chemoradiation for high grade glioma in close proximity to critical structures. J Hematol Oncol. 2010;3:22. doi: 10.1186/1756-8722-3-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ottenhausen M, Krieg SM, Meyer B, Ringel F. Functional preoperative and intraoperative mapping and monitoring: Increasing safety and efficacy in glioma surgery. Neurosurg Focus. 2015;38:E3. doi: 10.3171/2014.10.FOCUS14611. [DOI] [PubMed] [Google Scholar]
- 9.Paw I, Carpenter RC, Watabe K, Debinski W, Lo HW. Mechanisms regulating glioma invasion. Cancer Lett. 2015;362:1–7. doi: 10.1016/j.canlet.2015.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xie B, Ding Q, Han H, Wu D. miRCancer: A microRNA-cancer association database constructed by text mining on literature. Bioinformatics. 2013;29:638–644. doi: 10.1093/bioinformatics/btt014. [DOI] [PubMed] [Google Scholar]
- 11.Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O'Briant KC, Allen A, et al. Circulating microRNAs as stable blood-based markers for cancer detection; Proc Natl Acad Sci USA; 2008; pp. 10513–10518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shimono Y, Zabala M, Cho RW, Lobo N, Dalerba P, Qian D, Diehn M, Liu H, Panula SP, Chiao E, et al. Downregulation of miRNA-200c links breast cancer stem cells with normal stem cells. Cell. 2009;138:592–603. doi: 10.1016/j.cell.2009.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 14.Babae N, Bourajjaj M, Liu Y, Van Beijnum JR, Cerisoli F, Scaria PV, Verheul M, Van Berkel MP, Pieters EH, Van Haastert RJ, et al. Systemic miRNA-7 delivery inhibits tumor angiogenesis and growth in murine xenograft glioblastoma. Oncotarget. 2014;5:6687–6700. doi: 10.18632/oncotarget.2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hwang H, Mendell J. microRNAs in cell proliferation, cell death, and tumorigenesis. Br J Cancer. 2006;94:776–780. doi: 10.1038/sj.bjc.6603023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Frankel LB, Wen J, Lees M, Høyer-Hansen M, Farkas T, Krogh A, Jäättelä M, Lund AH. microRNA-101 is a potent inhibitor of autophagy. EMBO J. 2011;30:4628–4641. doi: 10.1038/emboj.2011.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang H, Li Y, Lai M. The microRNA network and tumor metastasis. Oncogene. 2010;29:937–948. doi: 10.1038/onc.2009.406. [DOI] [PubMed] [Google Scholar]
- 18.Dontula R, Dinasarapu A, Chetty C, Pannuru P, Herbert E, Ozer H, Lakka SS. microRNA 203 modulates glioma cell migration via Robo1/ERK/MMP-9 signaling. Genes cancer. 2013;4:285–296. doi: 10.1177/1947601913500141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.LeRoith D, Roberts CT., Jr The insulin-like growth factor system and cancer. Cancer Lett. 2003;195:127–137. doi: 10.1016/S0304-3835(03)00159-9. [DOI] [PubMed] [Google Scholar]
- 20.Denduluri SK, Idowu O, Wang Z, Liao Z, Yan Z, Mohammed MK, Ye J, Wei Q, Wang J, Zhao L, Luu HH. Insulin-like growth factor (IGF) signaling in tumorigenesis and the development of cancer drug resistance. Genes Dis. 2015;2:13–25. doi: 10.1016/j.gendis.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maris C, D'Haene N, Trépant AL, Le Mercier M, Sauvage S, Allard J, Rorive S, Demetter P, Decaestecker C, Salmon I. IGF-IR: A new prognostic biomarker for human glioblastoma. Br J Cancer. 2015;113:729–737. doi: 10.1038/bjc.2015.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Capes-Davis A, Theodosopoulos G, Atkin I, Drexler HG, Kohara A, MacLeod RA, Masters JR, Nakamura Y, Reid YA, Reddel RR, Freshney RI. Check your cultures! A list of cross-contaminated or misidentified cell lines. Int J Cancer. 2010;127:1–8. doi: 10.1002/ijc.25242. [DOI] [PubMed] [Google Scholar]
- 23.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 24.Vermes I, Haanen C, Steffens-Nakken H, Reutellingsperger C. A novel assay for apoptosis. Flow cytometric detection of phosphatidylserine expression on early apoptotic cells using fluorescein labelled Annexin V. J Immunol Methods. 1995;184:39–51. doi: 10.1016/0022-1759(95)00072-I. [DOI] [PubMed] [Google Scholar]
- 25.Song B, Zhang C, Li G, Jin G, Liu C. miR-940 inhibited pancreatic ductal adenocarcinoma growth by targeting MyD88. Cell Physiol Biochem. 2015;35:1167–1177. doi: 10.1159/000373941. [DOI] [PubMed] [Google Scholar]
- 26.Zhu X, Li D, Yu F, Jia C, Xie J, Ma Y, Fan S, Cai H, Luo Q, Lv Z, Fan L. miR-194 inhibits the proliferation, invasion, migration, and enhances the chemosensitivity of non-small cell lung cancer cells by targeting forkhead box A1 protein. Oncotarget. 2016;7:13139–13152. doi: 10.18632/oncotarget.7545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Josephs DH, Bax HJ, Karagiannis SN. Tumour-associated macrophage polarisation and re-education with immunotherapy. Front Biosci (Elite Ed) 2015;7:293–308. doi: 10.2741/E735. [DOI] [PubMed] [Google Scholar]
- 28.Seton-Rogers S. Tumour microenvironment: Teaching old macrophages new tricks. Nat Rev Cancer. 2013;13:753. doi: 10.1038/nrc3616. [DOI] [PubMed] [Google Scholar]
- 29.Ruffell B, Coussens LM. Macrophages and therapeutic resistance in cancer. Cancer Cell. 2015;27:462–472. doi: 10.1016/j.ccell.2015.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ries CH, Cannarile MA, Hoves S, Benz J, Wartha K, Runza V, Rey-Giraud F, Pradel LP, Feuerhake F, Klaman I, et al. Targeting tumor-associated macrophages with anti-CSF-1R antibody reveals a strategy for cancer therapy. Cancer Cell. 2014;25:846–859. doi: 10.1016/j.ccr.2014.05.016. [DOI] [PubMed] [Google Scholar]
- 31.Tap WD, Wainberg ZA, Anthony SP, Ibrahim PN, Zhang C, Healey JH, Chmielowski B, Staddon AP, Cohn AL, Shapiro GI, et al. Structure-guided blockade of CSF1R kinase in tenosynovial giant-cell tumor. N Engl J Med. 2015;373:428–437. doi: 10.1056/NEJMoa1411366. [DOI] [PubMed] [Google Scholar]
- 32.Quail DF, Bowman RL, Akkari L, Quick ML, Schuhmacher AJ, Huse JT, Holland EC, Sutton JC, Joyce JA. The tumor microenvironment underlies acquired resistance to CSF-1R inhibition in gliomas. Science. 2016;352:aad3018. doi: 10.1126/science.aad3018. [DOI] [PMC free article] [PubMed] [Google Scholar]