Abstract
The present study aimed to investigate the role of microRNA (miR)-99b-5p in spinal cord injury (SCI). Reverse transcription-quantitative polymerase chain reaction demonstrated that, compared with control mice, the expression levels of miR-99b-5p were upregulated in the mouse spinal cord following SCI. Mechanistic target of rapamycin (mTOR) was predicted to be the possible target of miR-99b-5p according to TargetScan and microrna databases. Dual-luciferase reporter assay verified that miR-99b-5p was able to target mTOR. Furthermore, the results of an apoptosis analysis demonstrated that there were few apoptotic neurons in the control group, whereas SCI induced a significant increase in the number of apoptotic cells. Conversely, apoptosis was inhibited following transfection with a miR-99b-5p inhibitor. The effects of miR-99b-5p on neurite growth were also evaluated. The results of an immunofluorescence analysis indicated that neurite growth was normal in the control group, whereas SCI induced a reduction in neurite growth, which was rescued following transfection with a miR-99b-5p inhibitor. The protein expression levels of mTOR were detected in the three groups by western blotting. The results demonstrated that, compared with the control group, the protein expression levels of mTOR were significantly reduced in SCI neurons, whereas transfection with a miR-99b-5p inhibitor suppressed the SCI-induced reduction of mTOR. In conclusion, treatment with a miR-99b-5p inhibitor may attenuate SCI-induced harmful alterations in spinal cord neurons via the regulation of mTOR expression.
Keywords: microRNA-99b-5p, mechanistic target of rapamycin, spinal cord injury, apoptosis, neurite growth
Introduction
It has been reported that 250,000–500,000 people per year sustain a spinal cord injury (SCI) globally, which is commonly caused by a traumatic event that is debilitating and leads to permanent motor/sensory deficits (1). The pathophysiology of SCI can be divided into primary and secondary mechanisms of injury: the primary mechanism of injury determines a patient's neurologic grade on admission; while the secondary mechanism of injury exacerbates damage and limits restorative processes (2,3). SCI results in permanent motor and sensory deficits (4), and adversely affects patients' families and presents a societal burden (2). Neuronal apoptosis, axonal retraction and spinal nerve fiber sprouting are all abnormally altered in response to SCI (5,6). Current treatments, for instance, neurorehabilitation in the form of exercise/physical training has been demonstrated to be beneficial for the recovery of function in patients (7). Although the complex pathophysiological processes have been studied recently, a cure for SCI has yet to be discovered due to the abnormal response of cells and tissues to the injury, and treatment strategies must be able to rescue damaged neural connections and create new neural connections (8).
Mechanistic target of rapamycin (mTOR) is a key factor in intracellular signaling pathways, which serves pivotal roles in protein synthesis, and cell survival and proliferation (9–11). Central nervous system (CNS) injury has been reported to result in mTOR activity reduction (12). Furthermore, phosphatase and tensin homolog (PTEN)/mTOR has been revealed to promote axonal sprouting (13).
MicroRNAs (miRNAs) are small (18–24 nucleotides), non-coding RNA molecules, which modulate gene expression via interacting with mRNAs or gene promoters (14). In addition, miRNAs also inhibit mRNA translation in the nervous system (15), and miRNA dysregulation in the CNS is correlated with traumatic injury (16). The present study aimed to analyze potential miRNA-based interventions for SCI. TargetScan and microrna databases were used to explore miRNAs that could target mTOR, the seed sequence of miR-99b-5p was found to be complementary to the position 289–295 of mTOR 3′UTR and was subsequently used in the present study.
Materials and methods
Generation of SCI
Adult 40 male ICR mice (weight, 30 g; age, 6 weeks) were maintained in a controlled environment with controlled temperature (23±1°C), relative humidity (70±10%), artificial 12-h light-dark cycle and free access to food and water, in Tianjin Medical University. Experiments were conducted according to the ethical standards of Tianjin Medical University General Hospital, and the present study was approved by the ethics committee of Tianjin Medical University General Hospital (Tianjing, China).
Mice were randomly divided into 2 different groups including control (n=15) and SCI group (n=15). The SCI model was established as previously described (16). Mice were anesthetized with 10% choral hydrate (0.35 ml/100 g, i.p.) before surgery. Briefly, an incision was made through the skin, subcutis and muscle, and a thoracic (T)11-lumbar (L)1 laminectomy was performed. Subsequently, a contusion injury was delivered using a customized designed electromagnetic force-driven at the T11 region following the laminectomy, after which the skin was sutured impactor, as previously described (16). Mice were kept warm and allowed to recover from the anesthesia. The majority of mice in the SCI group presented flaccid paralysis in the lower extremities, whereas some others presented spastic features. And for mice in control group, the same procedure was followed but without the contusion injury. In the present study, mice that presented flaccid paralysis were used in subsequent experiments, including total mRNA extraction from L4-6 spinal cord and cell culture. As for the generation of an in vitro SCI model, 10 male ICR mice were used as follows. Following sacrifice of mice, L4-6 spinal cord tissue was extracted from the mice and digested with 0.125% trypsin and 0.02% EDTA at 37°C for 20 min, after which digestion was terminated with Dulbecco's modified Eagle's medium (DMEM, Invitrogen; Thermo Fisher Scientific, Inc. Waltham, MA, USA). Briefly, spinal cords were dissociated and cells were isolated and purified as previously described (11), grown as a mixed culture for 2 days. Then the medium was refreshed with high glucose DMEM containing 20% fetal bovine serum (Invitrogen; Thermo Fisher Scientific, Inc.). After 10 days, neurons were isolated and refreshed for further purification. Finally, neurons at a concentration of 3×104/cm2 were seeded into polylysine-coated 35 mm petri dishes. Subsequently, pre-heated neurobasal culture medium (Invitrogen; Thermo Fisher Scientific, Inc.), containing 1% B27, 0.5 mM glutamine and 1% penicillin/streptomycin, was added and neurons were cultured at 37°C in an incubator containing 5% CO2. The in vitro SCI model of neurons were performed by grinding, described previously (8). 24 h later, neurons were randomly divided into 3 different groups (control group, SCI group and SCI + miR-99b-5p inhibitor group) and collected for the following experiments.
Bioinformatic analysis
TargetScan (www.targetscan.org) and miRanda (www.microrna.org) were used for the prediction of miRNAs that could target mTOR. Reverse transcription-quantitative polymerase chain reaction (RT-qPCR) analysis. Small RNAs were extracted and purified using the mirVana miRNA Isolation kit (Ambion; Thermo Fisher Scientific, Inc., Waltham, MA, USA) according to the manufacturer's protocol. Reverse transcriptase (Takara Biotechnology Co., Ltd., Dalian, China) was used to generate cDNA from the RNA. The miRNA expression levels were analyzed in an ABI 7500 system (Applied Biosystems; Thermo Fisher Scientific, Inc.) using SYBR Green I dye (Bio-Rad Laboratories, Inc., Hercules, CA, USA). U6 was used as an internal reference for miRNA. The mRNA expression levels of mTOR were quantified using the SYBR PrimeScript RT-PCR kit II (Takara Bio, Inc., Shiga, Japan) and were normalized to GAPDH. The thermal cycling conditions were as follows: pre-denaturation at 95°C for 60 sec and 40 cycles of 95°C for 15 sec and 60°C for 30 sec.
The primer sequences were as follows: miR-99b-5p (MIMAT0000689), 5′-CACCCGUAGAACCGACCUUGCG-3′; U6, forward 5′-GCTTCGGCAGCACATATACTAAAAT-3′, reverse 5′-CGCTTCACGAATTTGCGTGTCAT-3′; mTOR, forward 5′-ACAGCCCAGGCCGCATTGTC-3′, reverse, 5′-TCCAGGGACTCCGTCAGGCG-3′; and GAPDH, forward 5′-CAATGACCCCTTCATTGACC-3′ and reverse 5′-GACAAGCTTCCCGTTCTCAG-3′. The relative expression levels were determined using the 2−ΔΔCq method (17). All tests were performed in triplicate.
Cell transfection
Cells were seeded into 24-well plates at the concentration of 1×105 cells/well. Transfection was conducted using Lipofectamine RNAiMAX (Invitrogen; Thermo Fisher Scientific, Inc.). The neurons were transfected with 30 µM miR-99b-5p mirVana miRNA inhibitor (5′-CGCAAGGUCGGUUCUACGGGUG-3′) and a control inhibitor (5′-CAGUACUUUUGUGUAGUACAA-3′) by Lipofectamine 2000 transfection reagent at 37°C for 48 h (Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol.
Apoptosis analysis
The levels of cell apoptosis in the various groups were analyzed using a fluorescein isothiocyanate (FITC) apoptosis detection kit (BD biosciences, San Jose, CA, USA). A total of 48 h post-transfection, neurons (1×105) were suspended in 300 µl 1× binding buffer followed by addition of 5 µl Annexin V-FITC and 5 µl propidium iodide (PI) and were incubated for ~15 min at room temperature in the dark. A flow cytometer (BD Biosciences) was used to assess the apoptotic rate of the various groups according to the manufacturer's protocol.
Dual luciferase reporter assay
HEK-293T cells were seeded into 96-well plates at a density of 3×104 cells/well and cultured for ~24 h at room temperature. Then a mixture of 200 ng p-mTOR-wild type (WT) or p-mTOR-mutant (Mut) and 5 pmol miR-99b-5p inhibitor was transfected into HEK-293T cells with LipoFiter™ (Hanbio Biotechnology, Co, Ltd., Hangzhou, China). A total of 48 h post-transfection, cells were collected. Firefly and Renilla luciferase activities were analyzed using the Dual Luciferase Assay system (Promega Corporation, Madison, WI, USA). The Firefly luciferase activity was used as an internal control.
Immunofluorescence
Cells (5×103) were washed with pre-heated PBS for two or three times, after which they were fixed with 4% paraformaldehyde for 15–20 min at 4°C. Following the removal of paraformaldehyde, the cells were washed three times with 0.01 M PBS (10 min/wash). Cells were then blocked with 3% bovine serum albumin (Shanghai Biyuntian Biotechnology Co., Ltd., Shanghai, China) and 0.3% Triton X-100 blocking buffer at room temperature for ~1 h. Subsequently, cells were incubated with a primary antibody of NeuN (1:100, cat. no. 24307, Cell Signaling Technology, Danvers, MA, USA) at 4°C overnight, and were then incubated with a secondary antibody of Alexa Fluor 488-labeled donkey anti-sheep antibody (1:1,000, cat. no A-11015; Invitrogen; Thermo Fisher Scientific, Inc.) for ~2 h at room temperature in the dark. Finally, the cell images were captured under a laser scanning confocal microscope (FV1000) with FLUOVIEW image system v.1.4a (both from Olympus Corporation, Tokyo, Japan), assembled in Photoshop 7.0 software (Adobe Systems, Inc., San Jose, CA, USA).
Western blotting
Neurons from the three different groups were lysed on ice with radioimmunoprecipitation assay lysis buffer (Beyotime Institute of Biotechnology, Beijing, China) supplemented with protease inhibitors. Protein concentrations were determined by BCA protein assay kit (Beyotime Institute of Biotechnology). Protein samples (15 µg) were then separated by 10% SDS-PAGE and were transferred to nitrocellulose membranes. The membranes were blocked with 5% skimmed milk at room temperature for 1 h, and were incubated with anti-mTOR (1:1,000, cat. no. 2792, Cell Signaling Technology, Inc.) or GAPDH antibodies (1:1,000, cat. no. 5174; Cell Signaling Technology, Inc.) at 4°C overnight. After washing with TBS containing 1% Tween (TBST) three times, the membranes were incubated with horseradish peroxidase-labeled secondary antibody (1: 20,000, cat. no. 7074; Cell Signaling Technology, Inc.) for ~1 h at room temperature. After washing three times with TBST, the blots were visualized using a chemiluminescent reagent (Hanbio Biotechnology Co. Ltd.). Densitometric analyses of the western blot bands were performed using Gel-Pro Analyzer software version 6.0 (Media Cybernetics, Rockville, MD, USA) using GAPDH as a control.
Statistical analysis
Data are presented as the mean ± standard deviation of more than three independent experiments. Data were analyzed using SPSS statistical analysis software version 17.0 (SPSS, Inc., Chicago, IL, USA). Baseline and post-treatment of SCI injury variables were compared by paired Student's t-test. Statistically significant differences between groups were calculated by repeated measures of two-way analysis of variance with post hoc Tukey's test for multiple comparisons. P<0.05 was considered to indicate a statistically significant difference.
Results
SCI upregulates the expression levels of miR-99b-5p in the mouse spinal cord
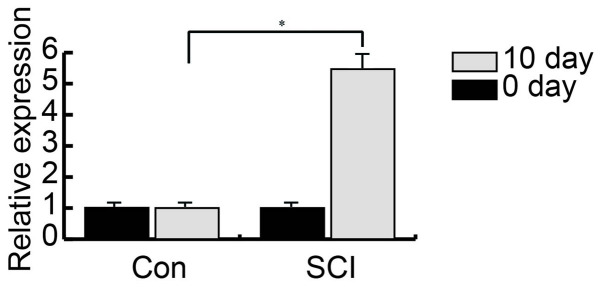
The expression levels of miR-99b-5p were detected in control and SCI mice using RT-qPCR. The results demonstrated that, compared with in the control mice, the expression levels of miR-99b-5p were significantly upregulated in SCI mice (Fig. 1).
Figure 1.
SCI upregulates the expression levels of miR-99b-5p in the mouse spinal cord. The expression levels of miR-99b-5p were significantly upregulated in SCI mice compared with in Con mice. *P<0.05. Con, control; miR, microRNA; SCI, spinal cord injury.
Possible target of miR-99b-5p
TargetScan and miRanda databases were used to predict target genes of miR-99b-5p. The score between mTOR and miR-99b-5p was 90 and was relatively higher compared with the other targets (Fig. 2A).
Figure 2.
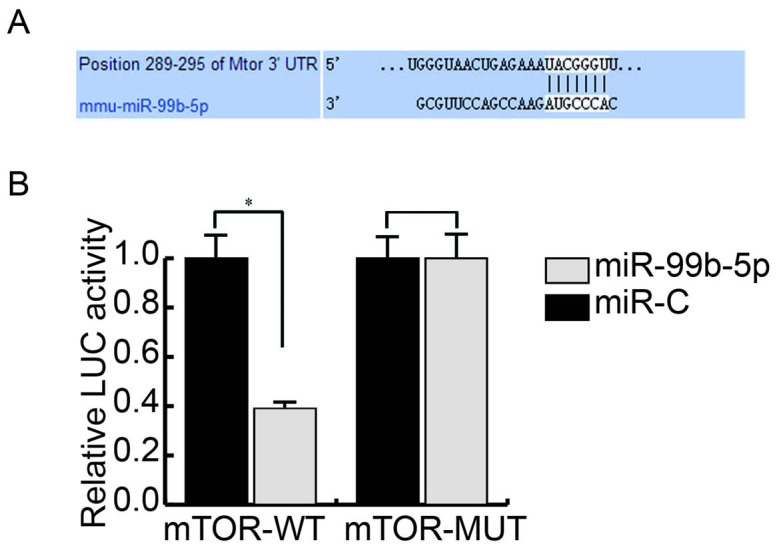
miR-99b-5p targets mTOR. (A) mTOR was predicted as a potential target gene of miR-99b-5p. (B) Interaction between mTOR and miR-99b-5p was analyzed using a LUC reporter assay. LUC activity was lower in cells co-transfected with the mTOR-WT plasmid and miR-99b-59 compared with the control group; no obvious difference was detected in fluorescence between the groups transfected with mTOR-MUT. *P<0.05. C, control; LUC, luciferase; miR, microRNA; mTOR, mechanistic target of rapamycin; MUT, mutant; UTR, untranslated region; WT, wild type.
Interaction between mTOR and miR-99b-5p
Dual-luciferase reporter assay was used to determine whether miR-99b-5p interacts with mTOR. Luciferase activity in cells co-transfected with the mTOR-WT plasmid and miR-99b-5p was lower compared with the control group; however, there was no obvious difference in fluorescence between the groups transfected with the mTOR-MUT plasmid (Fig. 2B).
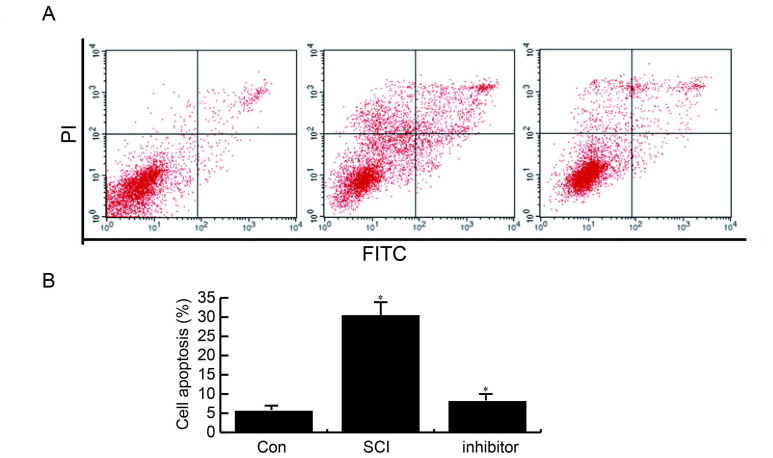
miR-99b-5p inhibitor suppresses SCI-induced neuronal apoptosis
Cell apoptosis in the three different groups was examined using an apoptosis analysis. Compared with the control group, the number of apoptotic cells was significantly increased in the SCI group; however, this enhancement was inhibited following transfection with a miR-99b-5p inhibitor (Fig. 3).
Figure 3.
miR-99b-5p inhibitor suppresses SCI-induced neuronal apoptosis. (A) Compared with the Con group, the number of apoptotic cells was increased in the SCI group; however, miR-99b-5p inhibitor transfection suppressed SCI-induced apoptosis. (B) Statistical analysis of apoptosis assay. *P<0.05 SCI group vs. control group; inhibitor group vs. SCI group. Con, control; FITC, fluorescein isothiocyanate; miR, microRNA; PI, propidium iodide; SCI, spinal cord injury.
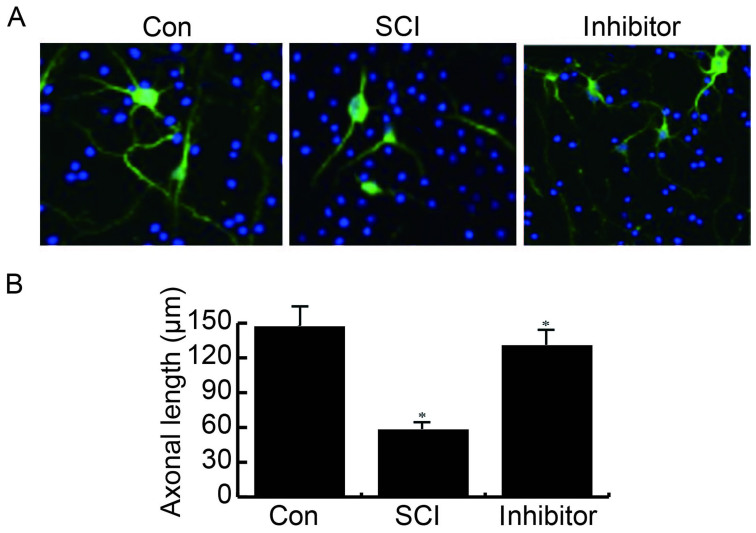
miR-99b-5p inhibitor rescues SCI-induced neurite growth inhibition
The effects of the miR-99b-5p inhibitor were determined on neurite growth. The results of an IF analysis demonstrated that in the control group, neurite growth was normal, whereas SCI markedly reduced neurite growth, which was rescued by transfection with a miR-99b-5p inhibitor (Fig. 4).
Figure 4.
miR-99b-5p inhibitor rescues SCI-induced neurite growth inhibition. (A) Neurite growth was normal in the Con group; however, it was markedly reduced by SCI and rescued by miR-99b-5p inhibitor transfection; magnification, ×200. (B) Statistical analysis of immunofluorescence. *P<0.05 SCI group vs. control group; inhibitor group vs. SCI group. Con, control; miR, microRNA; SCI, spinal cord injury.
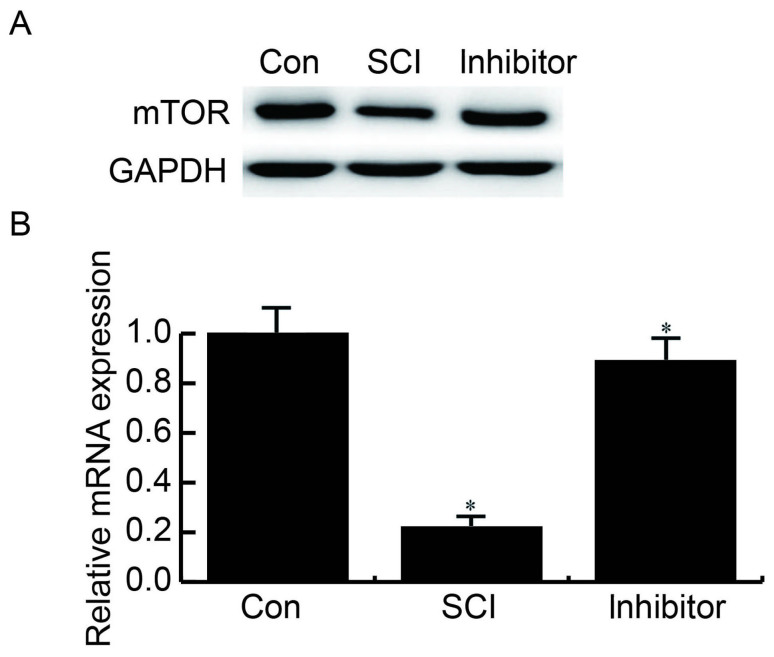
miR-99b-5p inhibitor reverses SCI-induced reduced mTOR expression
The protein and mRNA expression levels of mTOR in the three groups were assessed by western blotting and RT-qPCR, respectively. The results demonstrated that, compared with the control group, the expression levels of mTOR were reduced in SCI neurons, whereas transfection with a miR-99b-5p inhibitor reversed the SCI-induced reductions in mTOR (Fig. 5).
Figure 5.
miR-99b-5p inhibitor rescues SCI-induced reduced mTOR expression. Compared with the Con group, significantly reduced expression levels of mTOR were detected in SCI neurons, whereas transfection with a miR-99b-5p inhibitor reversed SCI-induced reduced mTOR expression. (A) Protein expression levels of mTOR; (B) mRNA expression levels of mTOR. *P<0.05 SCI group vs. control group; inhibitor group vs. SCI group. Con, control; miR, microRNA; mTOR, mechanistic target of rapamycin; SCI, spinal cord injury.
Discussion
Apoptosis and neuroplasticity have been reported to contribute to functional deficiency in patients with SCI (18,19). Despite research focusing on the pathophysiological processes of SCI, there remains no effective treatment for SCI (8).
Plasticity alterations, including axonal regeneration, are known to occur in the spinal cord following SCI (20). Furthermore, miRNA expression is altered following CNS injury (21), and miRNAs are considered novel targets for axonal regeneration (22). The present study aimed to determine whether the expression levels of miR-99b-5p were altered in the mouse spinal cord following SCI. The results of RT-qPCR demonstrated that, compared with mice in the control group, the expression levels of miR-99b-5p were significantly increased in SCI mice.
miRNAs have been revealed to inhibit mRNA translation in the nervous system (15); however, to the best of our knowledge, the SCI-associated target of miR-99b-5p is currently unknown. It has previously been reported that miR-99b-5p regulates the mTOR signaling pathway (23,24); therefore, the present study hypothesized and aimed to verify that miR-99b-5p directly targets mTOR. Initially, the present study used TargetScan and miRanda databases to predict the possible targets of miR-99b-5p. As expected, mTOR was predicted as a target gene of miR-99b-5p. A previous study demonstrated that mTOR activity is associated with CNS injury (12). However, whether miR-99b-5p could interact with mTOR remains unknown; therefore, the present study conducted a dual-luciferase reporter assay to determine whether miR-99b-5p interacted with mTOR. The results demonstrated that luciferase activity in neurons co-transfected with mTOR-WT and miR-99b-5p was lower compared with in the control group, thus suggesting that miR-99b-5p may interact with mTOR.
The effects of miR-99b-5p/mTOR in cultured injured neurons were also identified. An apoptosis analysis was used to examine the effects of miR-99b-5p on neuronal apoptosis. In the SCI group, the number of apoptotic cells was significantly increased compared with in control group, whereas transfection with a miR-99b-5p inhibitor significantly inhibited SCI-induced neuronal apoptosis.
SCI generates progressive degeneration that lasts for weeks or months, and secondary responses to SCI include neuronal/glial apoptosis and axonal retraction in nerve fibers (5,6). Therefore, the effects of a miR-99b-5p inhibitor were determined on neurite growth. The results of an IF analysis demonstrated that in the control group, neurite growth was normal. However, neurite growth in the SCI neurons was markedly reduced, whereas transfection with a miR-99b-5p inhibitor significantly rescued neurite growth. These findings were similar to the results of previous studies, which suggested that neuronal apoptosis, axonal retraction and nerve fiber sprouting were abnormally altered in patients with SCI (5,6).
CNS reorganization (25,26) is a property that may explain functional recovery following injury (27). For example, the lumbosacral spinal cord may learn numerous tasks; in a previous study, cats were reported to stand and step on a treadmill with training following complete transection injuries due to CNS reorganization (28). The corresponding molecules responsible for these alterations are currently unclear. Notably, PTEN/mTOR has been reported to promote axonal sprouting in adult retinal ganglion cells following optic nerve injury (12). In addition, mTOR activation has been demonstrated to increase axonal growth in injured peripheral nerves (12). Therefore, the present study detected the protein expression levels of mTOR in the different groups by RT-qPCR and western blotting. Compared with the control group, mTOR was significantly reduced in injured neurons, whereas the miR-99b-5p inhibitor suppressed SCI-induced reductions in mTOR.
In conclusion, the present study demonstrated that a miR-99b-5p inhibitor could attenuate SCI-induced harmful alterations in spinal cord neurons. Although more studies are required to confirm the hypothesis that the effects of miR-99a-5p on SCI may be associated with mTOR signaling, the results provided a potential miRNA-based intervention for SCI. However, whether the miR-99b-5p inhibitor may be used as a preventative intervention for SCI requires further investigation.
Acknowledgements
The present study was funded by the National Natural Science Foundation for Young Scholars of China (grant no. 81401784), the Key Project of National Natural Science Foundation of China (grant no. 81330042), the Special Program for Sino-Russian Joint Research, which was Sponsored by the Ministry of Science and Technology, China (grant no. 2014DFR31210), and the Key Program, which was Sponsored by the Tianjin Science and Technology Committee, China (grant nos. 13RCGFSY19000 and 14ZCZDSY00044).
References
- 1.W.H.O. Spinal Cord Injury Fact Sheet N. 384. 2013. http://www.who.int/mediacentre/factsheets/fs384/en/ [Nov 11;2013 ]. http://www.who.int/mediacentre/factsheets/fs384/en/
- 2.Dumont RJ, Okonkwo DO, Verma S, Hurlbert RJ, Boulos PT, Ellegala DB, Dumont AS. Acute spinal cord injury, part I: Pathophysiologic mechanisms. Clin Neuropharmacol. 2001;24:254–264. doi: 10.1097/00002826-200109000-00003. [DOI] [PubMed] [Google Scholar]
- 3.Sekhon LH, Fehlings MG. Epidemiology, demographics, and pathophysiology of acute spinal cord injury. Spine (Phila Pa 1976) 2001;26(24 Suppl):S2–S12. doi: 10.1097/00007632-200112151-00002. [DOI] [PubMed] [Google Scholar]
- 4.Devivo MJ. Epidemiology of traumatic spinal cord injury: Trends and future implications. Spinal Cord. 2012;50:365–372. doi: 10.1038/sc.2011.178. [DOI] [PubMed] [Google Scholar]
- 5.Crowe MJ, Bresnahan JC, Shuman SL, Masters JN, Beattie MS. Apoptosis and delayed degeneration after spinal cord injury in rats and monkeys. Nat Med. 1997;3:73–76. doi: 10.1038/nm0197-73. [DOI] [PubMed] [Google Scholar]
- 6.Fitch MT, Silver J. CNS injury, glial scars, and inflammation: Inhibitory extracellular matrices and regeneration failure. Exp Neurol. 2008;209:294–301. doi: 10.1016/j.expneurol.2007.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fauad K, Tetzlaff W. Rehabilitative training and plasticity following spinal cord injury. Exp Neurol. 2012;235:91–99. doi: 10.1016/j.expneurol.2011.02.009. [DOI] [PubMed] [Google Scholar]
- 8.Strickland ER, Hook MA, Balaraman S, Huie JR, Grau JW, Miranda RC. MicroRNA dysregulation following spinal cord contusion: Implications for neural plasticity and repair. Neuroscience. 2011;186:146–160. doi: 10.1016/j.neuroscience.2011.03.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gingras AC, Raught B, Sonenberg N. mTOR signaling to translation. Curr Top Microbiol Immunol. 2004;279:169–197. doi: 10.1007/978-3-642-18930-2_11. [DOI] [PubMed] [Google Scholar]
- 10.Hay N, Sonenberg N. Upstream and downstream of mTOR. Genes Dev. 2004;18:1926–1945. doi: 10.1101/gad.1212704. [DOI] [PubMed] [Google Scholar]
- 11.Jaworski J, Sheng M. The growing role of mTOR in neuronal development and plasticity. Mol Neurobiol. 2006;34:205–219. doi: 10.1385/MN:34:3:205. [DOI] [PubMed] [Google Scholar]
- 12.Abe N, Borson SH, Gambello MJ, Wang F, Cavalli V. Mammalian target of rapamycin (mTOR) activation increases axonal growth capacity of injured peripheral nerves. J Biol Chem. 2010;285:28034–28043. doi: 10.1074/jbc.M110.125336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Park KK, Liu K, Hu Y, Smith PD, Wang C, Cai B, Xu B, Connolly L, Kramvis I, Sahin M, He Z. Promoting axon regeneration in the adult CNS by modulation of the PTEN/mTOR pathway. Science. 2008;322:963–966. doi: 10.1126/science.1161566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ambros V. microRNAs: Tiny regulators with great potential. Cell. 2001;107:823–826. doi: 10.1016/S0092-8674(01)00616-X. [DOI] [PubMed] [Google Scholar]
- 15.Fineberg SK, Kosik KS, Davidson BL. MicroRNA potentiate neural development. Neuron. 2009;64:303–309. doi: 10.1016/j.neuron.2009.10.020. [DOI] [PubMed] [Google Scholar]
- 16.Genda Y, Arai M, Ishikawa M, Tanaka S, Okabe T, Sakamoto A. microRNA changes in the dorsal horn of the spinal cord of rats with chronic constriction injury: A TaqMan® low density array study. Int J Mol Med. 2013;31:129–137. doi: 10.3892/ijmm.2012.1163. [DOI] [PubMed] [Google Scholar]
- 17.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 18.Jee MK, Jung JS, Choi JI, Jang JA, Kang KS, Im YB, Kang SK. MicroRNA 486 is a potentially novel target for the treatment of spinal cord injury. Brain. 2012;135:1237–1252. doi: 10.1093/brain/aws047. [DOI] [PubMed] [Google Scholar]
- 19.Harkema SJ. Neural plasticity after human spinal cord injury: Application of locomotor training to the rehabilitation of walking. Neuroscientist. 2001;7:455–468. doi: 10.1177/107385840100700514. [DOI] [PubMed] [Google Scholar]
- 20.Ding Y, Kastin AJ, Pan W. Neural plasticity after spinal cord injury. Curr Pharm Des. 2005;11:1441–1450. doi: 10.2174/1381612053507855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ziu M, Fletcher L, Rana S, Jimenez DF, Digicaylioglu M. Temporal differences in microRNA expression patterns in astrocytes and neurons after ischemic injury. PLoS One. 2011;6:e14724. doi: 10.1371/journal.pone.0014724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Loohuis NF Olde, Kos A, Martens GJ, Van Bokhoven H, Kasri N Nadif, Aschrafi A. MicroRNA networks direct neuronal development and plasticity. Cell Mol Life Sci. 2012;69:89–102. doi: 10.1007/s00018-011-0788-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ye X, Luo H, Chen Y, Wu Q, Xiong Y, Zhu J, Dia Y, Wu Z, Miao J, Wan J. MicroRNAs 99b-5p/100-5p regulated by endoplasmic reticulum stress are involved in abeta-induced pathologies. Front Aging Neurosci. 2015;7:210. doi: 10.3389/fnagi.2015.00210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jin Y, Tymen SD, Chen D, Fang ZJ, Zhao Y, Dragas D, Dai Y, Marucha PT, Zhou X. MicroRNA-99 family targets AKT/mTOR signaling pathway in dermal wound healing. PLoS One. 2013;8:e64434. doi: 10.1371/journal.pone.0064434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Edgerton VR, de Leon RD, Tillakaratne N, Recktenwald MR, Hodgson JA, Roy RR. Use dependent plasticity in spinal stepping and standing. Adv Neurol. 1997;72:233–247. [PubMed] [Google Scholar]
- 26.De Leon RD, Hodgson JA, Roy RR, Edgerton VR. Retention of hindlimb stepping ability in adult spinal cats after the cessation of step training. J Neurophysiol. 1999;81:85–94. doi: 10.1152/jn.1999.81.1.85. [DOI] [PubMed] [Google Scholar]
- 27.Dobkin BH. Activity-dependent learning contributes to motor recovery. Ann Neurol. 1998;44:158–160. doi: 10.1002/ana.410440204. [DOI] [PubMed] [Google Scholar]
- 28.Martinez M, Rossignol S. A dual spinal cord lesion paradigm to study spinal locomotor plasticity in the cat. Ann N Y Acad Sci. 2013;1279:127–134. doi: 10.1111/j.1749-6632.2012.06823.x. [DOI] [PubMed] [Google Scholar]