Abstract
In recent years, oncolytic viruses have attracted increasing interest due to their potent antitumor effects. Luteolin, a natural product, has additionally been observed to exhibit various pharmacological antitumor activities. Previously, a novel dual-targeting oncolytic adenovirus, complement decay-accelerating factor (CD55)-tumor necrosis factor ligand superfamily member 10 (TRAIL), was constructed, which exhibited significant growth inhibitory effects in various types of tumor cell. The present study investigated whether the combination of luteolin and CD55-TRAIL was able to exert a synergistic antitumor effect in colorectal carcinoma (CRC) cells. The cytotoxicity and tumor cell apoptosis mediated by combination treatment in CRC cells were detected via an MTT assay, Hoechst staining and western blotting, respectively. Tumor growth in vivo was examined in a CRC mouse xenograft model following various treatments. The results demonstrated that the addition of luteolin enhanced oncolytic adenovirus-mediated enhanced green fluorescent protein, early region 1A and TRAIL expression. The combination of CD55-TRAIL with luteolin synergistically inhibited tumor growth and promoted CRC cellular apoptosis in vitro and in vivo. Additionally, the combination of CD55-TRAIL with luteolin significantly decreased cytotoxicity in lung/bronchial normal epithelial cells, compared with single treatment. Therefore, the combination of CD55-TRAIL with luteolin may be a novel efficient therapeutic strategy for CRC in the future.
Keywords: oncolytic adenovirus, luteolin, tumor necrosis factor ligand superfamily member 10, colorectal carcinoma, combined therapy
Introduction
Colorectal cancer (CRC) has become a major global threat to health. According to a 2012 report, 1.4 million people were diagnosed with colorectal cancer worldwide; these cases accounted for 9.7% of the total number of cancer cases globally (1). Therefore, it is of importance that a novel and effective therapy for CRC be developed.
As a promising therapy, oncolytic viruses, including conditional replication oncolytic adenovirus (OA) and the OncoPox, replicate in and eventually lyse tumor cells (2). Cancer-targeting gene-viro-therapy (CTGVT), using oncolytic viral vectors carrying an antitumor gen, has been demonstrated to enhance the antitumor effects of oncolytic viruses (3–7).
A CTGVT using the carcinoembryonic antigen (CEA) promoter to regulate early region 1A (E1A) and carrying tumor necrosis factor ligand superfamily member 10 (TRAIL) was previously developed. TRAIL belongs to the tumor necrosis factor superfamily, which display antitumor effects by activating the death receptors in tumor cells to induce apoptosis (8,9). Oncolytic virus-mediated TRAIL has displayed antitumor effects in hepatocellular carcinoma (10,11), CRC (12–15), lung adenocarcinoma (16) and gastric carcinoma (17). Based on the tumor specific carcinoembryonic antigen (CEA), Ad·CEA·E1A·E1B (Δ55)-TRAIL [complement decay-accelerating factor (CD55)-TRAIL] was constructed, which used the CEA promoter instead of the native E1A promoter to improve tumor cell targeting and carried the TRAIL gene (11).
Luteolin (3′,4′,5,7-tetrahydroxyflavone) is a natural substance obtained from fruits, vegetables and certain medicinal plants (18). Previous studies have reported that luteolin is able to suppress tumor growth by inducing tumor cell apoptosis, cell cycle arrest, and anti angiogenesis (19–24). Luteolin may specifically inhibit tumor growth in CRC by activating nuclear factor erythroid 2-related factor 2 signaling (25). The combination of luteolin and TRAIL has been shown to promote apoptosis in bladder (26) and cervical cancer cells (27).
In the present study, the antitumor effect of co-treatment with luteolin and CD55-TRAIL were examined in CRC cells, and it was observed that the combination markedly decreased CRC progression in vitro and in vivo.
Materials and methods
Cells and culture
293 cells were obtained from Microbix Biosystems, Inc. (Mississauga, ONT, Canada). The human colorectal cancer cell lines HT-29, SW620 and SW480, and the human normal lung/bronchial normal epithelial cells Beas-2B, were acquired from the Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China). 293 cells and Beas-2B cells were cultured in Dulbecco's modified Eagle's medium (DMEM; Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA), supplemented with 10% heat-inactivated fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc.). The other cell lines, HT-29, SW620 and SW480, were cultured in DMEM supplemented with 5% FBS. All cells were cultured in a 5% CO2 humidified incubator at 37°C.
Generation and purification of the oncolytic virus
The OA CD55-TRAIL was constructed in a previous study (11). The CD55-TRAIL was amplified by infecting 293 cell cultures. The virus was collected by CsCl gradient centrifugation, followed by dialysis. Viral titer was determined by 50% tissue culture infective dose (TICD50) analysis in 293 cells (11).
Cell viability assay
Cells (5,000) were seeded into 96-well plates and cultured for 24 h. The culture medium was subsequently replaced with medium containing CD55-TRAIL [5 multiplicity of infection (MOI), 10 MOI, 15 MOI], luteolin (25 µM) (Beyotime Institute of Biotechnology, Haimen, China), or a combination of CD55-TRAIL and luteolin. Following incubation for approximately 24, 48, 72 and 96 h, MTT (0.5 mg/ml) was added to each well. Following 4 h incubation, the cell supernatant was completely removed and 150 ml dimethyl sulfoxide was added to each well. Following mixing thoroughly, the 96-well plates were read by a microplate reader at a wavelength of 490 nm (Tecan Group, Ltd., Mannedorf, Switzerland). The effect of combination treatment was evaluated using CalcuSyn version 2.1 (Biosoft, Cambridge, UK).
Detection of green fluorescent protein
A total of 5×104 cells were seeded into 6-well plates and were left to attach overnight. The cells were treated with CD55-EGFP (15 MOI) or a combination of CD55-TRAIL and luteolin (25 µM). Following 48 h incubation, the cells were subsequently observed under a fluorescence microscope (0.2 mm fields; magnification, ×200; IX71-22FL/PH, Olympus Corporation, Japan).
Apoptotic cell staining
A total of 5×104 cells were seeded into 6-well plates and were left to attach overnight. The cells were treated with CD55-TRAIL (15 MOI), luteolin (25 µM), or a combination of CD55-TRAIL and luteolin. Following 48 h incubation, the cells were incubated in Hoechst 33342 (Beyotime Institute of Biotechnology) for 10 min, and washed twice with PBS. The cells were subsequently observed under a fluorescence microscope (0.2 mm fields; magnification, ×200; IX71-22FL/PH, Olympus Corporation, Japan).
Western blot analysis
A total of 5×105 cells were seeded into 6-well plates and were left to attach overnight. The cells were treated with CD55-TRAIL (15 MOI), luteolin (25 µM), or a combination of CD55-TRAIL and luteolin. After 72 h, the cells were removed and lysed with radioimmunoprecipitation assay buffer (Beyotime Institute of Biotechnology, Shanghai, China) containing protease inhibiters to collect protein. The concentration of proteins collected was determined using the Pierce bicinchoninic acid protein assay kit (Thermo Fisher Scientific, Inc.). Subsequently, the protein (20 µg) was separated using 6% SDS-PAGE to detect cleaved poly-ADP-ribose polymerase (PARP; cat. no. 5625) and PARP and the protein (20 µg) were separated using 12% SDS-PAGE to detect GAPDH (cat. no. 51332), procaspase-9 (cat. no. 9508), cleaved caspase-9 (cat. no. 7237), E3 ubiquitin-protein ligase XIAP (XIAP; cat. no. 14334), E1A (cat. no. sc374663) and TRAIL (cat. no. 3219) and transferred onto a polyvinylidene difluoride membrane. The membranes were blocked with 5% non-fat dried milk at room temperature for 2 h and incubated in primary antibodies [GAPDH, procaspase-9, cleaved caspase-9, cleaved poly-ADP-ribose polymerase (PARP), PARP, E3 ubiquitin-protein ligase XIAP (XIAP), E1Aand TRAIL] overnight at 4°C. The working dilution of all primary antibodies was 1:1,000. The membranes of GAPDH, procaspase-9 and E1A were subsequently incubated in the corresponding mouse secondary antibody (cat. no. 10230269) and the membranes of cleaved caspase-9 cleaved PARP, PARP and TRAIL were subsequently incubated in corresponding rabbit secondary antibody (cat. no. 10245169). the working dilution of all secondary are the ratio of 1:5,000 and the all membranes are incubated at room temperature for 2 h. Protein expression was detected using the Odyssey infrared imaging system (LI-COR Biosciences, Lincoln, NE, USA). All antibodies, except the E1A antibodies, were purchased from Cell Signaling Technology, Inc. (Danvers, MA, USA). The E1A antibodies were obtained from Santa Cruz Biotechnology, Inc. (Dallas, TX, USA). The mouse and rabbit secondary antibodies were obtained from PerkinElmer, Inc. (Waltham, MA, USA).
Flow cytometric analysis
A total of 5×105 cells were seeded into 6-well plates and were left to attach overnight. The cells were treated with CD55-TRAIL (15 MOI), luteolin (25 µM), or a combination of CD55-TRAIL and luteolin. After 48 h, the cells were trypsinized and harvested. Aliquots of cells were resuspended in 500 ml binding buffer and stained with annexin V fluorescein isothiocyanate/propidium iodide (BD Biosciences, San Jose, CA, USA), according to the manufacturer's protocol. The cells were immediately identified using fluorescence-activated cell sorting (BD Biosciences).
Animals
Animal studies followed the regulations and standards set by the US Department of Agriculture and the National Institutes of Health. A total of 24 female BALB/c nude mice (4 weeks old; mean body weight 18 g) were purchased from the Zhejiang Chinese Medical University (Hangzhou, China) and housed at 28°C, 40–60% humidity and a 12-h light/dark cycle. The experimental protocol was approved by the Animal Care and Welfare Committee of Zhejiang Chinese Medical University (Hangzhou, China).
Tumor xenograft
A total of 8×106 HT-29 cells were injected subcutaneously into the right flank of nude mice. When the tumor had grown to a diameter of 400–600 mm3, the mice were divided randomly into 4 groups and were injected with PBS (vehicle), luteolin (50 mg/kg) alone, CD55-TRAIL (2×109 plaque-forming units) alone or CD55-TRAIL and luteolin. Subsequent to the injection, the tumor size was measured using a Vernier calliper every 5 days.
Histopathology, immunohistochemistry (IHC) and terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) assay
The liver, kidney, spleen and tumor tissues were harvested and fixed in 5% paraformaldehyde at room temperature for 48 h, dehydrated with gradient increasing ethanol concentrations and embedded in paraffin wax, and were cut into 5-mm sections. The sections were stained with hematoxylin and eosin at room temperature for 5 min for histological analysis. The sections were blocked with 5% BSA at room temperature for 20 min and subsequently incubated in anti-TRAIL antibodies (3219; Cell Signaling Technology, Inc.) overnight at 4°C, followed by avidin-biotin-peroxidase complex reagents (Vector Laboratories, Inc., Burlingame, CA, USA) for the IHC analysis. The working dilution of anti-TRAIL antibodies was 1:300. Hematoxylin was used as a counterstain at room temperature for 5 min. The in-situ apoptosis detection kit (Sino-American Biotechnology Co., Luoyang, China) was used to stain apoptotic cell tumor tissue sections, according to the manufacturer's protocol, for the TUNEL assay. All sections were counterstained with hematoxylin at room temperature for 5 min. All sections were subsequently observed under a microscope (0.2 mm fields; magnification, ×200; IX71-22FL/PH, Olympus Corporation, Japan).
Statistical analysis
All experiments were repeated three times and data are presented as the mean ± standard deviation. Statistical analysis was performed using GraphPad Prism 6 version is 6.01 (GraphPad Software, Inc., La Jolla, CA, USA) for a Student's t test. P<0.05 was considered to indicate a statistically significant difference.
Results
CD55-TRAIL with luteolin reduces the cell viability of CRC cells
The oncolytic virus CD55-TRAIL was constructed by inserting a TRAIL gene-expressing cassette into CD55. TRAIL gene expression was regulated by the human cytomegalovirus immediate-early promoter. CEA protein expression in HT-29 and SW620 cells was detected via western blotting to ensure the replication of CD55-TRAIL.
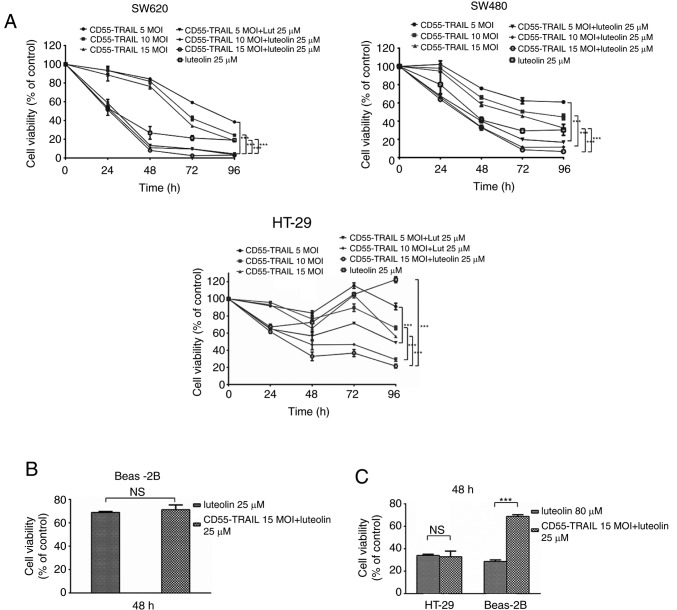
In order to evaluate the antitumor effect of CD55-TRAIL and luteolin in vitro, CRC cell lines SW620, SW480 and HT-29 were infected with CD55-TRAIL (at various concentrations) and luteolin (25 µM) (Fig. 1A) for 48 h. It was observed that, following treatment, cell viability significantly decreased in a time-dependent manner. At 96 h post-infection, cell viability was <30%. Treatment with CD55-TRAIL and luteolin was similar to luteolin alone, illustrating the decreased toxicity of CD55-TRAIL in normal BEAS-2B cells (Fig. 1B) at 48 h. An increased concentration of luteolin (80 µM), and the combination of CD55-TRAIL (15 MOI) with luteolin (25 µM), was used to treat HT-29 and BEAS-2B cells; in HT-29 CRC cells, the same cytotoxic effects were observed leading to a viability of <35%, although the combination treatment induced less toxicity in normal Beas-2B cells (>65% viability) (Fig. 1C). The results of the present study suggested that the combination of CD55-TRAIL with luteolin suppressed CRC cell proliferation with minimal toxic impact to normal cells compared with luteolin alone.
Figure 1.
Cytopathic effect of the combining CD55-TRAIL with luteolin in colorectal cancer and normal cells. (A) Cell viability was examined via an MTT assay. (B) Beas-2B cells were treated with luteolin (25 µM) or the combination of CD55-TRAIL (15 MOI) with luteolin (25 µM). (C) HT-29 cells and Beas-2B cells were treated with luteolin (80 µM) or the combination of CD55-TRAIL (15 MOI) with luteolin (25 µM). All data are presented as the mean ± standard deviation. n=3. **P<0.01; ***P<0.001. NS, not significant; CD55, complement decay-accelerating factor; TRAIL, tumor necrosis factor ligand superfamily member 10; MOI, multiplicity of infection.
Luteolin enhances CD55-mediated transgene expression in CRC cells
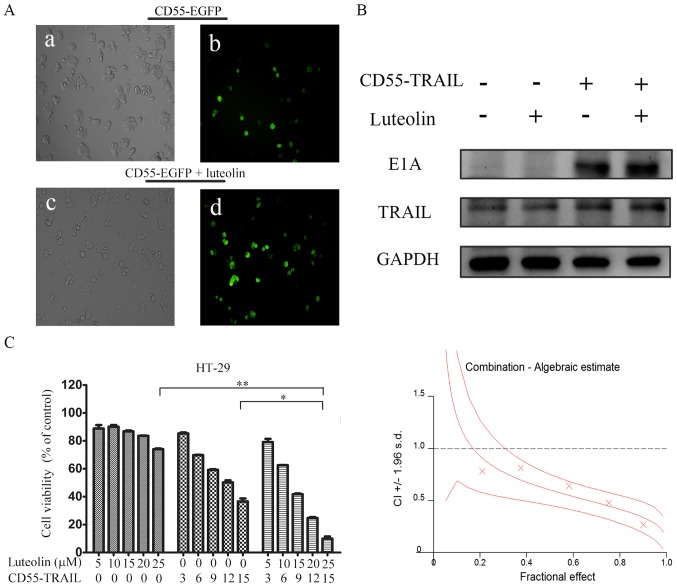
In order to detect whether the luteolin was able to enhance oncolytic adenovirus CD55-mediated gene expression, HT-29 cells were treated with CD55-enhanced green fluorescent protein (EGFP) (15 MOI) or CD55-EGFP (15 MOI) and luteolin (25 µM). After 24 h, it was observed that the percentage of EGFP fluorescent cells was increased in HT-29 cells treated with CD55-EGFP and luteolin compared with CD55-EGFP alone (Fig. 2A), suggesting that luteolin promoted CD55-EGFP replication and EGFP expression. In addition, E1A and TRAIL expression was detected in CD55-TRAIL-infected HT-29 cells. The results demonstrated that combination treatment with CD55-TRAIL and luteolin led to increased E1A and TRAIL expression compared with CD55-TRAIL alone or luteolin alone (Fig. 2B), indicating that luteolin was able to enhance CD55-mediated associated protein expression.
Figure 2.
Luteolin promotes CD55-TRAIL replication and associated gene expression. (A) Colorectal cancer HT-29 cells were treated with CD55-Trail (15 MOI; a and b), or with CD55-Trail (15 MOI) plus luteolin (25 µM; c and d). Images a and c were captured using a light microscope (0.2 mm fields; magnification, ×200); b and d were captured using a fluorescence microscope (0.2 mm fields; magnification, ×200). (B) E1A and TRAIL expression was detected by western blot analysis. (C) Cell viability was detected via an MTT assay. The quantitated data were processed using CalcuSyn software, and the CI values of the treatment groups are represented by X-marks. All data are presented as the mean ± standard deviation. n=3. *P<0.05, **P<0.01. CD55, complement decay-accelerating factor; TRAIL, tumor necrosis factor ligand superfamily member 10; MOI, multiplicity of infection; CI, combination index; E1A, early region 1A; EGFP, enhanced green fluorescent protein.
In order to determine whether the antitumor effect of the combination therapy was synergistic or additive, the effect of combination treatment was evaluated using CalcuSyn. HT-29 cells were treated with CD55-TRAIL alone at various concentrations, luteolin alone or a combination at the constant ratio of 3:5 for 48 h. The X-marks on the graph (Fig. 2C) were used as the combination index (CI) values. All the experimental CI values were <1, illustrating that the combination treatment had a synergistic effect (Fig. 2C).
Combination of CD55-TRAIL with luteolin induces apoptosis in cancer cells
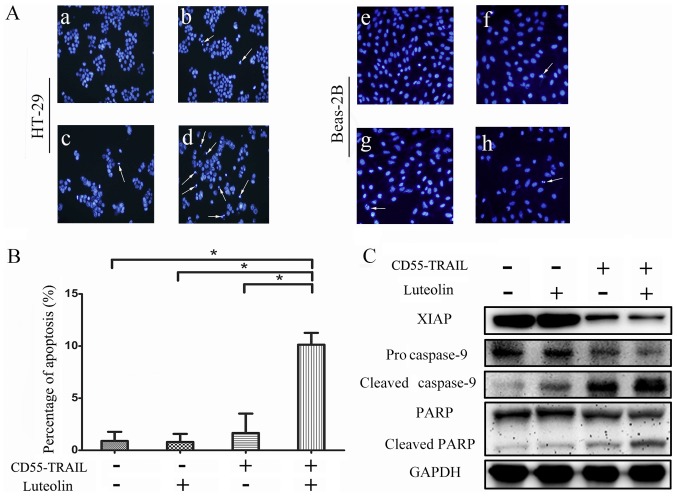
In order to determine whether the antitumor effects of the combination therapy were induced by an enhancement of apoptosis in cancer cells, Hoechst 33342 analysis was used to examine morphological alterations indicative of apoptosis in HT-29 cells. The results indicated that compared with CD55-TRAIL or luteolin alone, the combination-treated cells displayed greater chromatin condensation, nuclear fragmentation and apoptotic body formation (Fig. 3A). Notably, normal Beas-2B cells exhibited few signs of apoptosis (Fig. 3A). The examination of cell death by flow cytometry of treated HT-29 cells revealed that compared with 1.7% for luteolin and 3.8% for CD55-TRAIL, treatment with CD55-TRAIL with luteolin resulted in a significantly increased percentage of apoptotic cells at 9.8% (Fig. 3B). Therefore, combination treatment induced apoptosis to a greater degree compared with each treatment alone.
Figure 3.
Inducing apoptosis via the combination of CD55-TRAIL and luteolin. (A) After 48 h, nuclear fragmentation (arrows) was observed in HT-29 (a-d) and Beas-2B (e-h) cells using Hoechst staining under an inverted fluorescence microscope (0.2 mm fields; magnification, ×200). The a and e were untreated. The b and f were treated with luteolin. The c and g were treated with CD55-TRAIL. The d and h were treated with the combination of CD55-TRAIL with luteolin. (B) The percentage of apoptotic cells was detected using flow cytometric analysis. The data are presented as the mean ± standard deviation. n=3. *P<0.05. (C) Apoptosis-associated protein expression was detected by western blotting. GAPDH was used as the internal control. CD55, complement decay-accelerating factor; TRAIL, tumor necrosis factor ligand superfamily member 10; XIAP, E3 ubiquitin-protein ligase XIAP; PARP; poly-ADP-ribose polymerase.
In order to further elucidate the underlying mechanism of cellular apoptosis, expression of the caspase signaling pathway proteins was detected by western blot analysis. Procaspase-9, cleaved caspase-9, PARP, cleaved PARP and XIAP were detected in HT-29 cells treated for 48 h (Fig. 3C). There was a significant decrease in the expression of anti-apoptosis proteins procaspase-9 and PARP, and an increase in the pro-apoptosis proteins cleaved caspase-9 and cleaved PARP, in cells treated with the combination therapy compared with the other treatments. The decreased expression of XIAP with the combination treatment compared with other apoptosis treatment significantly increased the effect of apoptosis. The results of the present study suggested that the combination treatment enhanced cellular apoptosis via activation of the caspase apoptosis pathway.
Antitumor efficacy of the combination of CD55-TRAIL with luteolin in vivo
In order to further examine their antitumor efficacy in vivo, the HT-29 xenograft model was established in nude mice. Treatment with luteolin, CD55-TRAIL or a combination of the two for 48 h was applied subcutaneously to the right flank of the mice. As solid tumors may not exceed 20 mm diameter for ethical reasons, animals were sacrificed at day 30. The results demonstrated that the tumor volume in the groups receiving PBS, luteolin or CD55-TRAIL reached ~3,761 mm3, ~3,415 mm3 and ~3,072 mm3, respectively, within 30 days; the combination of CD55-TRAIL with luteolin significantly impeded the growth of the tumor, with a tumor volume of ~1,780 mm3 representing a 58% inhibition of tumor growth (Fig. 4A). Additionally, after 25 days, the tumor volume in the combination treatment was decreased, suggesting that the combination treatment has better anti-tumor effect.
Figure 4.
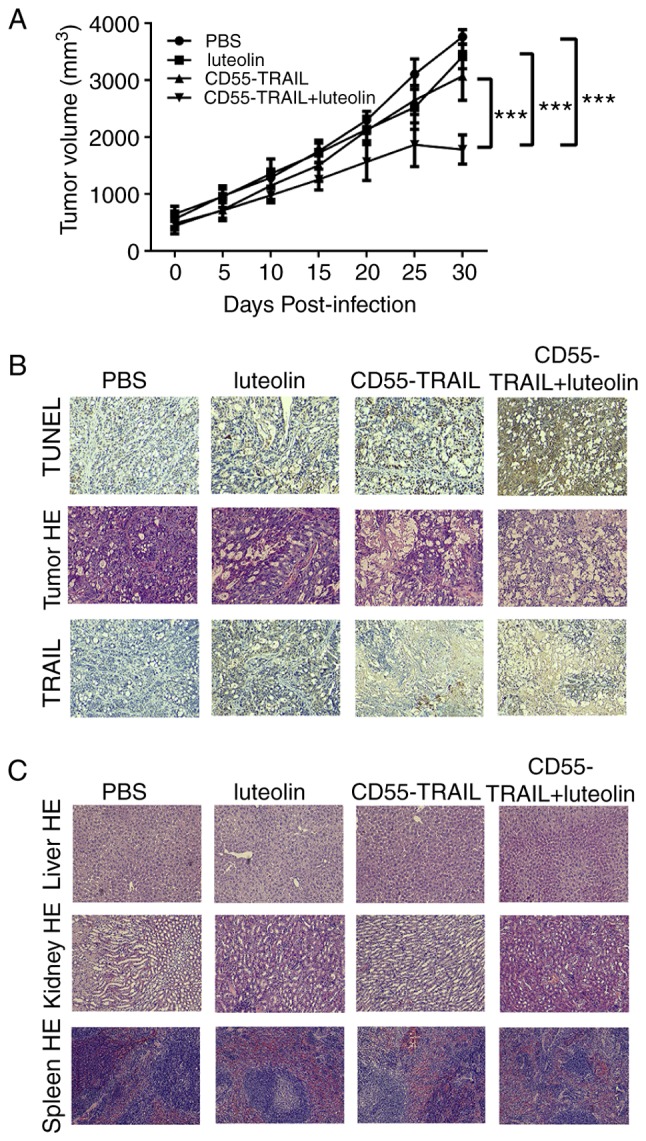
Antitumor efficacy of the combination of CD55-TRAIL with luteolin in vivo. (A) The tumor volume was measured every 5 days using the formula V (mm3)=1/2xlength xwidth2. Data are presented as the mean ± standard deviation, n=5. ***P<0.001. (B) The apoptosis of tumor sections was assayed using TUNEL staining. The cellular necrotic areas in the tumors were detected using HE staining. The TRAIL expression in tumor tissues was detected by immunohistochemical staining. (C) The toxicity to liver, kidney and spleen tissues was detected by HE staining. Magnification, ×200. HE, hematoxylin and eosin; CD55, complement decay-accelerating factor; TRAIL, tumor necrosis factor ligand superfamily member 10; TUNEL, terminal deoxynucleotidyl transferase dUTP nick end labeling.
Examination of TRAIL and cell death-associated protein TUNEL expression by IHC analysis of tumor tissues 20 days subsequent to drug treatment revealed that the expression of TUNEL was increased in animals receiving combination treated cells, compared with other mice (Fig. 4B). Hematoxylin and eosin staining demonstrated that the combination of CD55-TRAIL with luteolin caused more severe cytopathic effects in tumor tissues compared with each therapy alone (Fig. 4B). In addition, the liver, kidney and spleen tissues without toxic effects were indicated by hematoxylin and eosin staining, implying that little to no cell damage or apoptosis had occurred in these tissues (Fig. 4C). IHC staining demonstrated that the TRAIL protein was highly expressed in tumor tissue following treatment with the combination of CD55-TRAIL and luteolin (Fig. 4B).
Discussion
Luteolin functions as a tumor suppressor by inducing tumor cell apoptosis, cell cycle arrest, and antiangiogenesis (19–24). However, these effects are accompanied by severe side effects which may cause damage to normal cells. In addition, long-term use of chemotherapy drugs may cause drug resistance in the tumor, leading to an increase in the dose of the drug, thereby resulting in more serious side effects. A number of combined therapeutic strategies have demonstrated fewer negative effects (28,29). Therefore, a combination strategy which may markedly decrease these severe side effects may provide a potentially promising treatment for CRC.
The present study revealed the potent antitumor effect of oncolytic adenovirus CD55-TRAIL with luteolin on CRC cells in vitro and in vivo, while minimizing cytotoxicity to normal cells. The combination therapy significantly and synergistically inhibited CRC cell proliferation. This inhibition was induced in part by the enhancement of CD55-mediated TRAIL expression in CRC cells by luteolin. The combination of CD55-TRAIL with luteolin additionally decreased the toxicity of luteolin in lung/bronchial normal epithelial cells. It was demonstrated that the combination of CD55-TRAIL with luteolin reduced the dose of luteolin required to elicit the same antitumor effect as luteolin alone.
The results of the present study demonstrated that the combination treatment induced antitumor effects by enhancing apoptosis, evidenced by an increase in chromatin condensation, nuclear fragmentation, the formation of apoptotic bodies and the expression of caspase apoptotic pathway proteins in CRC cells. In addition, compared with each treatment individually, the combination of CD55-TRAIL with luteolin decreased the expression of the caspase inhibitor gene XIAP in order to activate the caspase apoptotic pathway. This finding was consistent with a previous report showing that the combination of TRAIL with luteolin inhibited the expression of the XIAP gene to activate the caspase apoptotic pathway in HeLa cells (30). The distinct molecular mechanism of the combination treatment strategy remains to be further elucidated in future studies.
Progress has been made in the field of OAs, including the CTGVT strategy and the use of specific promoters instead of the E1A promoter to improve targeting. However, OAs, due to their heterogeneity, may induce an inflammatory response following infection of the tumor tissue and elicit antiviral effects. In addition, the bioavailability of oncolytic adenoviruses is low when administered by intravenous injection. Further studies are required to resolve these issues. Thus, OAs have been considered less as an individual treatment for tumors, and more as a combination therapy, including OAs combined with nanomaterials (31,32), immune cells (33,34) and chemotherapy drugs (30,31), which have exhibited an improved therapeutic effect compared with oncolytic viruses alone. Additionally, previous studies have focused on the identification of tumor biomarkers based on microRNAs (35,36), which may provide the possibility of improving the antitumor effect of oncolytic virotherapy.
The results of the present study demonstrated that the combined use of luteolin and CD55-TRAIL had a potent antitumor effect, and that the antitumor effect of CD55-TRAIL was synergized by luteolin in CRC cells with reduced toxicity to normal epithelial cells. Additionally, this combination resulted in a strong apoptotic effect in CRC cells in vitro and in vivo. In conclusion, the combination of CD55-TRAIL with luteolin provided a potentially effective clinical therapy for CRC.
Acknowledgements
The present study was supported by the Zhejiang Provincial Natural Science Foundation of China (grant nos. LY16H160056 and LZ13H160004), the Grant for 521 Talent Project of Zhejiang Sci-Tech University and the Innovation Project of Zhejiang Sci-Tech University (grant no. YCX15035).
References
- 1.Global battle against cancer won't be won with treatment alone-effective prevention measures urgently needed to prevent cancer crisis. Cent Eur J Public Health. 2014;22:23–28. [PubMed] [Google Scholar]
- 2.Breitbach CJ, Burke J, Jonker D, Stephenson J, Haas AR, Chow LQ, Nieva J, Hwang TH, Moon A, Patt R, et al. Intravenous delivery of a multi-mechanistic cancer-targeted oncolytic poxvirus in humans. Nature. 2011;477:99–102. doi: 10.1038/nature10358. [DOI] [PubMed] [Google Scholar]
- 3.Liu XY. A new anticancer strategy-gene-virotherapy of cancer. Chin J Cancer Biother. 2001;8:1. [Google Scholar]
- 4.Zhang ZL, Zou WG, Luo CX, Li BH, Wang JH, Sun LY, Qian QJ, Liu XY. An armed oncolytic adenovirus system, ZD55-gene, demonstrating potent antitumoral efficacy. Cell Res. 2003;13:481–489. doi: 10.1038/sj.cr.7290191. [DOI] [PubMed] [Google Scholar]
- 5.Wang Y, Liu T, Huang P, Zhao H, Zhang R, Ma B, Chen K, Huang F, Zhou X, Cui C, Liu X. A novel Golgi protein (GOLPH2)-regulated oncolytic adenovirus exhibits potent antitumor efficacy in hepatocellular carcinoma. Oncotarget. 2015;6:13564–13578. doi: 10.18632/oncotarget.3769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lei W, Liu HB, Wang SB, Zhou XM, Zheng SD, Guo KN, Ma BY, Xia YL, Tan WS, Liu XY, Wang YG. Tumor suppressor in lung cancer-1 (TSLC1) mediated by dual-regulated oncolytic adenovirus exerts specific antitumor actions in a mouse model. Acta Pharmacol Sin. 2013;34:531–540. doi: 10.1038/aps.2012.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.He G, Lei W, Wang S, Xiao R, Guo K, Xia Y, Zhou X, Zhang K, Liu X, Wang Y. Overexpression of tumor suppressor TSLC1 by a survivin-regulated oncolytic adenovirus significantly inhibits hepatocellular carcinoma growth. J Cancer Res Clin Oncol. 2012;138:657–670. doi: 10.1007/s00432-011-1138-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ashkenazi A, Pai RC, Fong S, Leung S, Lawrence DA, Marsters SA, Blackie C, Chang L, McMurtrey AE, Hebert A, et al. Safety and antitumor activity of recombinant soluble Apo2 ligand. J Clin Invest. 1999;104:155–162. doi: 10.1172/JCI6926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pan G, O'Rourke K, Chinnaiyan AM, Gentz R, Ebner R, Ni J, Dixit VM. The receptor for the cytotoxic ligand TRAIL. Science. 1997;276:111–113. doi: 10.1126/science.276.5309.111. [DOI] [PubMed] [Google Scholar]
- 10.Ma H, Liu Y, Liu S, Kung HF, Sun X, Zheng D, Xu R. Recombinant adeno-associated virus-mediated TRAIL gene therapy suppresses liver metastatic tumors. Int J Cancer. 2005;116:314–321. doi: 10.1002/ijc.20982. [DOI] [PubMed] [Google Scholar]
- 11.Zhang R, Zhang X, Ma B, Xiao B, Huang F, Huang P, Ying C, Liu T, Wang Y. Enhanced antitumor effect of combining TRAIL and MnSOD mediated by CEA-controlled oncolytic adenovirus in lung cancer. Cancer Gene Ther. 2016;23:168–177. doi: 10.1038/cgt.2016.11. [DOI] [PubMed] [Google Scholar]
- 12.Zhao L, Dong A, Gu J, Liu Z, Zhang Y, Zhang W, Wang Y, He L, Qian C, Qian Q, Liu X. The antitumor activity of TRAIL and IL-24 with replicating oncolytic adenovirus in colorectal cancer. Cancer Gene Ther. 2006;13:1011–1022. doi: 10.1038/sj.cgt.7700969. [DOI] [PubMed] [Google Scholar]
- 13.Hang ZQ, Zheng MF, Huang JH. Detection and diagnostic value of serum carcinoembryonic antigen and cytokeratin 19 fragment in lung cancer patients. Zhonghua Zhong Liu Za Zhi. 2011;33:847–849. (In Chinese) [PubMed] [Google Scholar]
- 14.Verberne CJ, Nijboer CH, de Bock GH, Grossmann I, Wiggers T, Havenga K. Evaluation of the use of decision-support software in carcino-embryonic antigen (CEA)-based follow-up of patients with colorectal cancer. BMC Med Inform Decis Mak. 2012;12:14. doi: 10.1186/1472-6947-12-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang YR, Yan JX, Wang LN. The diagnostic value of serum carcino-embryonic antigen, alpha fetoprotein and carbohydrate antigen 19-9 for colorectal cancer. J Cancer Res Ther. 2014;10(Suppl):S307–S309. doi: 10.4103/0973-1482.151538. [DOI] [PubMed] [Google Scholar]
- 16.Shi J, Zheng D, Liu Y, Sham MH, Tam P, Farzaneh F, Xu R. Overexpression of soluble TRAIL induces apoptosis in human lung adenocarcinoma and inhibits growth of tumor xenografts in nude mice. Cancer Res. 2005;65:1687–1692. doi: 10.1158/0008-5472.CAN-04-2749. [DOI] [PubMed] [Google Scholar]
- 17.Lai H, Jin Q, Lin Y, Mo X, Li B, He K, Chen J. Combined use of lysyl oxidase, carcino-embryonic antigen, and carbohydrate antigens improves the sensitivity of biomarkers in predicting lymph node metastasis and peritoneal metastasis in gastric cancer. Tumour Biol. 2014;35:10547–10554. doi: 10.1007/s13277-014-2355-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ross JA, Kasum CM. Dietary flavonoids: Bioavailability, metabolic effects, and safety. Annu Rev Nutr. 2002;22:19–34. doi: 10.1146/annurev.nutr.22.111401.144957. [DOI] [PubMed] [Google Scholar]
- 19.Huang YT, Hwang JJ, Lee PP, Ke FC, Huang JH, Huang CJ, Kandaswami C, Middleton E, Jr, Lee MT. Effects of luteolin and quercetin, inhibitors of tyrosine kinase, on cell growth and metastasis-associated properties in A431 cells overexpressing epidermal growth factor receptor. Br J Pharmacol. 1999;128:999–1010. doi: 10.1038/sj.bjp.0702879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ko WG, Kang TH, Lee SJ, Kim YC, Lee BH. Effects of luteolin on the inhibition of proliferation and induction of apoptosis in human myeloid leukaemia cells. Phytother Res. 2002;16:295–298. doi: 10.1002/ptr.871. [DOI] [PubMed] [Google Scholar]
- 21.Kim JH, Jin YR, Park BS, Kim TJ, Kim SY, Lim Y, Hong JT, Yoo HS, Yun YP. Luteolin prevents PDGF-BB-induced proliferation of vascular smooth muscle cells by inhibition of PDGF beta-receptor phosphorylation. Biochem Pharmacol. 2005;69:1715–1721. doi: 10.1016/j.bcp.2005.04.002. [DOI] [PubMed] [Google Scholar]
- 22.Lee HJ, Wang CJ, Kuo HC, Chou FP, Jean LF, Tseng TH. Induction apoptosis of luteolin in human hepatoma HepG2 cells involving mitochondria translocation of Bax/Bak and activation of JNK. Toxicol Appl Pharmacol. 2005;203:124–131. doi: 10.1016/j.taap.2004.08.004. [DOI] [PubMed] [Google Scholar]
- 23.Leung WC, Wu CH, Lin CH, Lee HZ. Luteolin induced DNA damage leading to human lung squamous carcinoma CH27 cell apoptosis. Eur J Pharmacol. 2005;508:77–83. doi: 10.1016/j.ejphar.2004.12.032. [DOI] [PubMed] [Google Scholar]
- 24.Bagli E, Stefaniotou M, Morbidelli L, Ziche M, Psillas K, Murphy C, Fotsis T. Luteolin inhibits vascular endothelial growth factor-induced angiogenesis; inhibition of endothelial cell survival and proliferation by targeting phosphatidylinositol 3′-kinase activity. Cancer Res. 2004;64:7936–7946. doi: 10.1158/0008-5472.CAN-03-3104. [DOI] [PubMed] [Google Scholar]
- 25.Pandurangan AK, Sadagopan SK Ananda, Dharmalingam P, Ganapasam S. Luteolin, a bioflavonoid inhibits Azoxymethane-induced colorectal cancer through activation of Nrf2 signaling. Toxicol Mech Methods. 2014;24:13–20. doi: 10.3109/15376516.2013.843111. [DOI] [PubMed] [Google Scholar]
- 26.Park HS, Choi YH. Induction of Apoptosis by Combination Treatment with Luteolin and TRAIL in T24 Human Bladder Cancer Cells. J Korean Society Food Sci Nut. 2013;42:1363–1369. doi: 10.3746/jkfn.2013.42.9.1363. [DOI] [Google Scholar]
- 27.Horinaka M, Yoshida T, Shiraishi T, Nakata S, Wakada M, Nakanishi R, Nishino H, Sakai T. The combination of TRAIL and luteolin enhances apoptosis in human cervical cancer HeLa cells. Biochem Biophys Res Commun. 2005;333:833–838. doi: 10.1016/j.bbrc.2005.05.179. [DOI] [PubMed] [Google Scholar]
- 28.Ma BY, Wang Y, Zhou X, Huang P, Zhang R, Liu T, Cui C, Liu X, Wang Y. Synergistic suppression effect on tumor growth of hepatocellular carcinoma by combining oncolytic adenovirus carrying XAF1 with cisplatin. J Cancer Res Clin Oncol. 2015;141:419–429. doi: 10.1007/s00432-014-1835-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cheng PH, Lian S, Zhao R, Rao XM, McMasters KM, Zhou HS. Combination of autophagy inducer rapamycin and oncolytic adenovirus improves antitumor effect in cancer cells. Virol J. 2013;10:293. doi: 10.1186/1743-422X-10-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shi RX, Ong CN, Shen HM. Protein kinase C inhibition and x-linked inhibitor of apoptosis protein degradation contribute to the sensitization effect of luteolin on tumor necrosis factor-related apoptosis-inducing ligand-induced apoptosis in cancer cells. Cancer Res. 2005;65:7815–7823. doi: 10.1158/0008-5472.CAN-04-3875. [DOI] [PubMed] [Google Scholar]
- 31.Wang L, Yao B, Li Q, Mei K, Xu JR, Li HX, Wang YS, Wen YJ, Wang XD, Yang HS, et al. Gene therapy with recombinant adenovirus encoding endostatin encapsulated in cationic liposome in coxsackievirus and adenovirus receptor-deficient colon carcinoma murine models. Hum Gene Ther. 2011;22:1061–1069. doi: 10.1089/hum.2011.014. [DOI] [PubMed] [Google Scholar]
- 32.Nigatu AS, Vupputuri S, Flynn N, Neely BJ, Ramsey JD. Evaluation of cell-penetrating peptide/adenovirus particles for transduction of CAR-negative cells. J Pharm Sci. 2013;102:1981–1993. doi: 10.1002/jps.23556. [DOI] [PubMed] [Google Scholar]
- 33.Zhang SN, Choi IK, Huang JH, Yoo JY, Choi KJ, Yun CO. Optimizing DC vaccination by combination with oncolytic adenovirus coexpressing IL-12 and GM-CSF. Mol Ther. 2011;19:1558–1568. doi: 10.1038/mt.2011.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang Z, Zhang Q, Xu K, Shan J, Shen J, Liu L, Xu Y, Xia F, Bie P, Zhang X, et al. Combined therapy with cytokine-induced killer cells and oncolytic adenovirus expressing IL-12 induce enhanced antitumor activity in liver tumor model. PLoS One. 2012;7:e44802. doi: 10.1371/journal.pone.0044802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang F, Zheng Z, Guo J, Ding X. Correlation and quantitation of microRNA aberrant expression in tissues and sera from patients with breast tumor. Gynecol Oncol. 2010;119:586–593. doi: 10.1016/j.ygyno.2010.07.021. [DOI] [PubMed] [Google Scholar]
- 36.Ding X, Ding J, Ning J, Yi F, Chen J, Zhao D, Zheng J, Liang Z, Hu Z, Du Q. Circulating microRNA as a potential biomarker for liver injury. Mol Med Rep. 2012;5:1428–1432. doi: 10.3892/mmr.2012.838. [DOI] [PubMed] [Google Scholar]