Abstract
Structure-specific recognition protein 1 (SSRP1) has been considered as a potential biomarker, since aberrant high expression of SSRP1 has been detected in numerous malignant tumors. However, the correlation between the expression level of SSRP1 and glioma remains unclear. The present study attempted to investigate the role of SSRP1 in the pathogenesis of glioma. In the present study, our data revealed that SSRP1 overexpression was detected in glioma tissues at both the mRNA and protein levels using quantitative real-time RT-PCR and immunohistochemical analysis. We also demonstrated that the upregulated expression of SSRP1 was correlated with the World Health Organization (WHO) grade of glioma. The knockdown of SSRP1 by siRNA not only resulted in the inhibition of cell proliferation, but also significantly inhibited glioma cell migration and invasion. Mechanistic analyses revealed that SSRP1 depletion suppressed the activity of the phosphorylation of the MAPK signaling pathway. In conclusion, the present study indicated that SSRP1 regulated the proliferation and metastasis of glioma cells via the MAPK signaling pathway.
Keywords: glioma, SSRP1, GEO data, proliferation, metastasis, MAPK pathway
Introduction
Cancer of the brain and the central nervous system is one of the 10 most common types of cancer in China, and its incidence rate has evidently increased from 2000 to 2011 (1). Glioma is the most common malignant primary brain tumor, and accounts for the vast majority of all intracranial tumors (2). At present, standard therapeutic options include maximal safe resection, followed by radiation therapy and adjuvant chemotherapy, which are not particularly effective (3). Despite the improvement of treatment modalities, the majority of patients with malignant glioma may have poor prognosis within 2 years of diagnosis (4).
Structure-specific recognition protein 1 (SSRP1) is a subunit of the facilitates chromatin transcription (FACT) complex, which attaches to the nucleosome and reassembles the nucleosome (5). SSRP1 was initially identified in 1991 as a high-mobility group protein 1 (HMG1)-related DNA binding protein, and the capacity of SSRP1 for biological functions can be attributed to its HMG domain (6). As an important histone chaperone, it plays a role as a transcription factor in the regulation of several targets to modulate cellular processes such as DNA replication, DNA damage repair, apoptosis and cell cycle regulation (7–11). It has been documented that the expression of SSRP1 was associated with the stage of cellular differentiation. High SSRP1 levels were observed in stem or less-differentiated cells, while low SSRP1 levels were observed in more differentiated cells (12). The high expression of SSRP1 was detected in multiple human types of cancer, including non-small-cell lung cancer, renal cell carcinoma, pancreatic ductal and colorectal adenocarcinoma, and hepatocellular carcinoma (13–15). In addition, it is a potential marker and target of aggressive types of cancer (13,16,17).
The mitogen-activated protein kinase (MAPK) signaling pathway is a highly conserved module that regulates various cellular functions, including proliferation, differentiation and malignant transformation (18). The MAPK pathway has been reported to be activated in over 88% of gliomas (19). The suppression of MAPK signaling synergizes the cytotoxicity of receptor tyrosine kinase inhibitors in glioma tumor-initiating cells (20). Research on the function of SSRP1 has focused on directing nucleosome reorganization as the histone chaperone, while the expression and potential molecular mechanism of SSRP1 in glioma remains unknown.
In the present study, we evaluated the expression of SSRP1 in human patient samples and investigated its function in the progression of glioma. In addition, we explored the effect of SSRP1 silencing on glioma cell proliferation and invasion. The results revealed that SSRP1 may function as a regulator, and promote cell proliferation and invasion by enhancing the activity of the MAPK signaling pathway. Therefore, the present study revealed that SSRP1 played a role in the progression of glioma, and also provided valuable information for understanding the mechanism of glioma.
Materials and methods
Tissue samples
Human glioma tissues were obtained during surgery at the Department of Neurosurgery, Renmin Hospital of Wuhan University (Wuhan, China), from 2010 to 2016. These control non-glioma human brain tissues were collected from unmatched patients undergoing surgery for intracranial hypertension. A total of 83 paraffin-embedded tissue samples, which included 77 glioma tissues, and 6 normal brain (NB) tissues, were histopathologically diagnosed by two neuropathologists. For real-time quantitative reverse transcription-polymerase chain reaction (RT-PCR), 30 frozen (stored in liquid nitrogen) glioblastoma and 10 NB tissues were evaluated. All tissues were classified according to the 2016 World Health Organization (WHO) classification of tumors of the central nervous system. Prior patient consent and approval from the Ethics Committee of Wuhan University were obtained for the use of these clinical materials. The authors assert that all experiments complied with the ethical standards of relevant national and institutional guidelines as well as the laws of the People's Republic of China. The detailed demographics of patients are presented in Table I.
Table I.
Correlation between the clinicopathological characteristics and expression of SSRP1 protein in glioma.
| SSRP1 expression | ||||
|---|---|---|---|---|
| Characteristics | No. of patients | High | Low | P-value |
| Age (years) | ||||
| ≥50 | 42 | 28 | 14 | 0.223 |
| <50 | 35 | 21 | 14 | |
| Sex | ||||
| Male | 39 | 27 | 12 | 0.535 |
| Female | 38 | 22 | 16 | |
| WHO grade | ||||
| I+II | 11 | 3 | 8 | 0.009 |
| III+IV | 66 | 46 | 20 | |
SSRP1, structure-specific recognition protein 1; WHO, World Health Organization.
Cells and cell culture
Human glioblastoma-derived cancer cell lines U118 and U251 were purchased from the Cell Bank Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China). The American Type Culture Collection (ATCC; Manassas, VA, USA) suspects that U118 is a contaminated cell line which has similar cytogenetics and similar origin with glioblastoma cell line U138MG, since there is no doubt that U118 is a cell line derived from malignant gliomas, we thus used U118 as experimental cells. These cells were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS) (both from Gibco Grand Island, NY, USA), 100 U/ml of penicillin and 100 µg/ml of streptomycin (Sigma-Aldrich, St. Louis, MO, USA) at 37°C with 5% CO2.
Gene expression profiles
SSRP1 expression datasets were obtained from the Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geo/). Three datasets were retrieved for the study: GSE50161 (13 NB, 15 pilocytic astrocytoma, 46 ependymoma, 22 medulloblastoma and 34 glioblastoma tissues were collected for microarray analysis), GSE4290 (23 NB and 157 glioma tissues were analyzed; glioma tissues comprised of 45 grade II, 31 grade III and 81 grade IV cases), and GSE3185 (10 astrocytomas and 10 glioblastoma tissues were analyzed). The expression values of SSRP1 were transformed to relative expression.
Immunohistochemical staining
The paraffin-embedded tissues were placed in xylene 3 times for 15 min at room temperature, hydrated in a series of 100, 95, 90, 80, 70 and 60% ethanol solutions, and washed in phosphate-buffered saline (PBS). The sections were treated with 3% H2O2 and subjected to antigen retrieval by citrate buffer (10 mmol/l, pH 6.0) for 15 min. Then, the sections were incubated with primary rabbit anti-SSRP1 polyclonal antibody (1:200; GeneTex, Inc., Irvine, CA,. USA) overnight at 4°C. After washing with PBS, the slides were incubated with poly-HRP goat anti-rabbit antibody (Maixin Bio, Fujian, China) for 30 min, and incubated with diaminobenzidine for 5 min. Next, the sections were counterstained with hematoxylin, dehydrated in ethanol and coverslips were placed on the slides. Images were captured using an Olympus BX40 microscope and the CC-12 Soft Imaging System (Olympus, Tokyo, Japan).
Immunohistochemical evaluation
SSRP1-positive cells displayed brownish yellow granules on the cytoplasm. These results were evaluated through immunohistochemical scores (IHC scores), according to the intensity of the staining and the percentage of immunoreactive cells. The estimate of the staining intensity was scored as follows: 0, no staining; 1, weak staining; 2, moderate staining; and 3, strong staining. The percentage was rated on a scale of 0–4, as follows: 0, <5%; 1, 5–25%; 2, 26–50%; 3, 51–75%; and 4, 76–100%. The IHC score was obtained by multiplying the percentage and intensity score. These IHC scores ranged from 0 to 12, in which scores 0–4 were considered as low expression, while scores 5–12 were considered as high expression. These scores were independently determined by two independent senior pathologists.
Western blot analysis
Cells were lysed in ice-cold RIPA buffer (50 mM of Tris-HCl pH 7.4, 150 mM of NaCl, 1% Triton X-100, 0.25% deoxycholate, 1.5 mM of MgCl2, 1 mM of EGTA, 1 mM of phenylmethylsulfonyl fluoride, 10 mM of ZnAF, 10 mM of pervanadate, 10 µg/ml of leupeptin and 10 µg/ml of aprotinin) and incubated for 30 min. The protein concentration was determined using the BCA method. The cell lysate was heated at 100°C for 10 min after being mixed with sample loading buffer. The protein samples were equally loaded on 10% SDS-PAGE, and transferred onto nitrocellulose membranes. After being blocked with 5% non-fat milk in Tris-buffer, the membrane strips were incubated with a primary antibody overnight at 4°C, followed by Alex Fluor 680/790-labeled goat anti-rabbit or goat anti-mouse IgG (Li-COR Biosciences, Lincoln, NE, USA). The strips were visualized using the LI-COR Odyssey Infrared Imaging System (Li-COR Biosciences).
The primary antibodies were as follows: p38, phospho-p38, ERK, phospho-ERK, JNK and phospho-JNK (1:1,000; Cell Signaling Technology, Danvers, MA, USA); MMP2, VEGF, EGFR, cyclin D and E (1:1,000; Abcam, Cambridge, MA, USA); p65, Bcl2, Snail, c-MYC, SSRP1 and GAPDH (1:1,000; GeneTex, Inc.).
RNA isolation and RT-PCR
Total RNA was isolated from U118 and U251 cell lines, and extracted from NB and glioma tissues with TRIzol reagents (Invitrogen, Carlsbad, CA, USA). Then, cDNA was prepared from 2 µg of total RNA using the PrimeScript RT reagent kit with gDNA Eraser (Takara, Tokyo, Japan) according to the manufacturer's instructions. The primers were as follows: SSRP1 forward, 5′-GGATTGAAAGAGGGCATGAA-3′ and reverse, 5′-AGAGGCGTTGCTGTCAAACT-3′; and GAPDH forward, 5′-ACAACTTTGGTATCGTGGAAGG-3′ and reverse, 5′-GCCATCACGCCACAGTTTC-3′. RT-PCR was performed to quantify the mRNA expression using the SYBR-Green PCR Master Mix (Takara). GAPDH was used for SSRP1 normalization.
Transient transfection with siRNAs
Small interfering RNAs (siRNAs) were designed and synthesized by Suzhou GenePharma, Inc. (Shanghai, China). Two siRNAs targeting the SSRP1 gene were designed and purchased, and the most effective siRNAs were identified using both RT-PCR and western blotting. The sequences of the siRNAs against SSRP1 were as follows: siRNA1 sense, 5′-GCCAUGUCUACAAGUAUGATT-3′ and antisense, 5′-UCAUACUUGUAGACAUGGCTT-3′; siRNA2 sense, 5′-CCCAGAAUGGUGUUGUCAAATT-3′ and antisense, 5′-UUUGACAACACAUUCUGGGTT-3′. The sequence of the negative control siRNA was as follows: sense, 5′-UUCUCCGAACGUGUCACGUTT-3′ and antisense, 5′-ACGUGACACGUUCGGAGAATT-3′. The siRNA sequences were derived from another research group at our laboratory, and the specificity had been verified (14). Glioma cells were plated onto a 6- or a 96-well plate at 40–50% confluency. After 24 h, siRNA transfections were conducted using HyperFect (Qiagen, Hilden, Germany), according to the manufacturer's protocol. Cells were harvested after 48–72 h. All siRNAs were used at a final concentration of 20 nM.
Cell proliferation assay
Cell proliferation was analyzed using Cell Counting Kit-8 (CCK-8; Dojindo, Tokyo, Japan). Cells were seeded in 96-well plates on day 0, and the cell growth was assessed at days 1, 2 and 3 after culturing, according to the manufacturer's instructions. On average, 5 replicates for each time-point were statistically analyzed.
5-Ethynyl-2′-deoxyuridine (EdU) incorporation assay
Cells were seeded into 96-well plates containing complete medium, and were allowed to attach overnight. Then, the cell growth was determined using the CellLight Edu imaging detecting kit (RiboBio, Guangzhou, China) according to the manufacturer's protocol. In brief, the cells were cultured in medium with 10 µM of EdU for 24 h, and fixed with 4% paraformaldehyde for 20 min. Subsequently, the cells were incubated in an Apollo reaction cocktail for 30 min, and the cell nuclei were stained with 5 µg/ml of Hoechst 33342 for 30 min. The cells were visualized under a fluorescence microscope (Olympus BX51; Olympus, Tokyo, Japan). Experiments were performed in triplicate.
Cell cycle analysis
Cells were harvested with 0.25% trypsin and washed twice with ice-cold PBS. After centrifugation, the cells were suspended with 75% methanol overnight at −20°C. Then, the cells were washed twice with PBS, incubated in PBS with 1 mg/ml of RNAase at 37°C, and stained with 50 mg/ml of propidium iodide (PI) in PBS. For each experiment, 2.5×104 cells were analyzed using BD FACSAria (BD Biosciences, Franklin Lakes, NJ, USA). Six replicates for each time-point were statistically analyzed.
Apoptosis analysis
Cells in each group were plated in 6-well plates, and harvested at 48 h after transient transfection with siRNA. Then, the cells were stained with ApoScreen Annexin V and PI, according to the manufacturer's instructions (BD Biosciences). For each experiment, 2.5×104 cells were analyzed using BD FACSAria (BD Biosciences). Six replicates for each time-point were statistically analyzed.
Cell migration and invasion assays
The cell migration assay was conducted using 8-µm pore size Transwell chambers (Corning, Corning, NY, USA). Cells in each group were suspended in serum-free DMEM. Then, 5×104 cells in 100 µl of DMEM were plated into the upper chamber, and 600 µl of DMEM containing 10% FBS was added to the lower chamber. These chambers were cultured at 37°C with 5% CO2 for 24 h. The cells in the upper chamber of the insert were collected using a cotton swab. Cells that had adhered to the lower surface were fixed with 4% paraformaldehyde, stained with 0.1% crystal violet and counted under a microscope (Olympus BX51). The invasion assay was the same, except that the Transwell chambers were precoated with Matrigel (R&D Systems, Minneapolis, MN, USA), and the cells were cultured for 36 h.
Statistical analysis
All statistical analyses are presented as the average of at least triplicate samples and as the mean ± standard deviation. SPSS 19.0 (SPSS, Inc., Chicago, IL, USA) and GraphPad Prism 6.0 software (GraphPad Software, Inc., La Jolla, CA, USA) were used for statistical analysis. A P-value <0.05 was considered statistically significant.
Results
SSRP1 mRNA and protein are overexpressed in glioma tissues
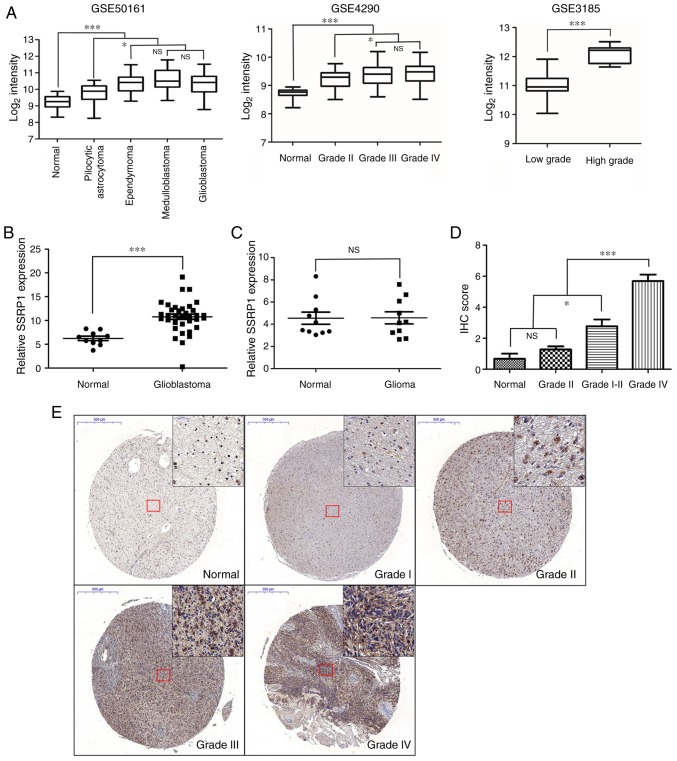
In order to observe the mRNA expression level of SSRP1 in glioma, 3 independent GEO datasets (GSE50161, GSE4290 and GSE3185) were analyzed. As shown in Fig. 1A, the expression of SSRP1 was upregulated in tumor tissues, compared to NB tissues; and the expression levels of SSRP1 were significantly higher in the glioblastoma multiform group, compared with the low-grade glioma group. The mRNA expression of SSRP1 in 10 NB and 30 glioblastoma tissues were examined by RT-PCR. Compared with NB tissues, glioblastoma tissues exhibited higher SSRP1 mRNA expression levels (P<0.001; Fig. 1B). Next, we attempted to elucidate the clinicopathological significance of the mRNA expression of SSRP1 in the peripheral blood of glioma patients, and found that the difference between glioma patients and normal controls was not statistically significant (P=0.97; Fig. 1C). In addition, the protein expression levels of SSRP1 were assessed in an independent cohort of 83 cases, which included 6 NB cases, 3 grade I, 8 grade II, 22 grade III and 44 grade IV cases, using immunohistochemical staining. The staining density of SSRP1 in the high-grade group exhibited stronger coloring and broader distribution, compared to that in the low-grade group (Fig. 1E). As shown in Table I, among the 11 cases of glioma tissues at grades I–II, 3 (27.3%) cases revealed positive staining, while 11 out of 22 (50.0%) cases of glioma tissues at grade III and 35 out of 44 (79.5%) cases of glioma tissues at grade IV were detected with positive staining of SSRP1 via immunohistochemical analysis. Quantitative analysis also revealed that the average score of SSRP1 staining increased from histologic grade I to grade IV (P<0.05; Fig. 1D). However, it was found that there was no significant association between the expression of SSRP1 and the age and sex of these patients (Table I).
Figure 1.
Expression of structure-specific recognition protein 1 (SSRP1) in human glioma tissues. (A) The mRNA expression level of SSRP1 in 3 datasets from the GEO repository (GSE50161, GSE4290 and GSE3185). The expression of the SSRP1 gene was higher in glioma tissues than in normal brain (NB) tissues. (B) SSRP1 mRNA expression was analyzed in 30 glioblastoma and 10 NB tissues by RT-PCR. (C) The fold change of SSRP1 expression in blood samples of 10 patients with glioblastoma and 10 normal control subjects. (D) Graphical representation of the IHC scores of the SSRP1 protein in 83 paraffin-embedded tissue samples. (E) The protein expression of SSRP1 in 83 paraffin-embedded tissue samples was assessed by immunohistochemical (IHC) staining. Representative images of the IHC staining for the SSRP1 protein is shown (magnification, ×100), the top right corner are the microscopic images captured at a magnification of ×400. Each bar is presented as the mean ± standard deviation (SD); *P<0.05, ***P<0.001. NS, no significance.
Downregulated SSRP1 expression suppresses cell proliferation in vitro
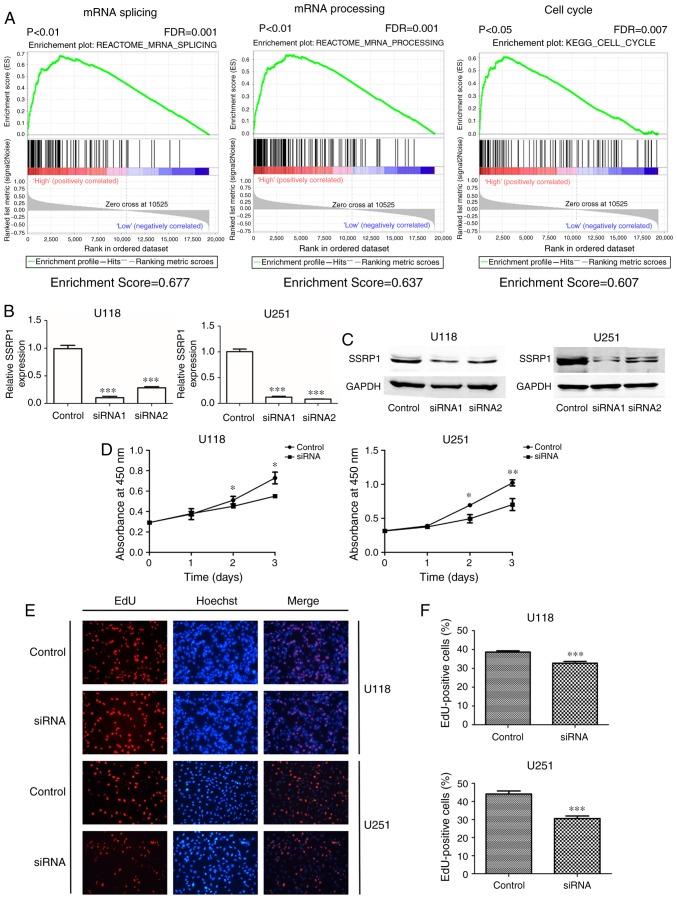
Gene set enrichment analysis (GSEA) was performed to identify biological processes potentially modulated by SSRP1. As shown in Fig. 2A, the GSEA results from gene profiling data (GSE50161) demonstrated that a mass of gene sets were enriched by SSRP1, and the enriched expression of gene sets included mRNA splicing, mRNA processing and the cell cycle (Fig. 2A).
Figure 2.
Downregulation of structure-specific recognition protein 1 (SSRP1) inhibits the proliferation in glioma cells. (A) The gene set enrichment analysis (GSEA) of GSE50161 revealed that SSRP1 may be involved in the progression of mRNA splicing, mRNA processing and the cell cycle. (B and C) The knockdown of SSRP1 by siRNA in U118 and U251 cells at both the mRNA and protein levels is shown. (D) Silencing of SSRP1 inhibited cell growth from day two as determined by CCK-8 assay in U118 and U251 cells. (E and F) EdU incorporation assay was performed to assess the DNA replication of U118 and U251 cells. Nuclei were stained with Hoechst (blue), and the proliferative cells were dyed red with EdU. A graphical representation of the percentage of EdU-positive cells is presented; *P<0.05, **P<0.01, ***P<0.001. Data is presented as the mean ± standard deviation (SD).
A plasmid pcDNA3.1-SSRP1 was constructed to increase the expression of SSRP1 in glioma cells. U251 cells transfected with pcDNA3.1-SSRP1 plasmid (U251-SSRP1) has a higher mRNA expression level, while the protein level was only 1.20-fold. The average percentage of EdU-positive U251-SSRP1 cells was similar with that in the U251 cells transfected with empty vector (38.30 and 39.51%; P=0.502). Flow cytometric analysis revealed that the average percentage of U251-SSRP1 cells in the S + G2/M phase was 52.17%, which had no significant differences with the control group (P=0.102). Overexpression of SSRP1 could not exert its biological effects in U251, and the reason may be attributed to the high expression of exogenous expression of SSRP1. In addition, further increased expression of SSRP1 was at variance with objective reality. Thus, we did not perform a functional study of the overexpression SSRP1.
siRNA was used to specifically knockdown the expression of SSRP1 in U118 and U251 cell lines, which were established from high-grade glioma tumors. The efficient knockdown of SSRP1 expression was assessed through both RT-PCR and western blotting, and siRNA1 revealed more interference efficiency (P<0.001; Fig. 2B and C). Therefore, siRNA1 was selected to knockdown the expression of SSRP1 in U118 and U251.
Subsequently, we verified the effect of the downregulated expression of SSRP1 in glioma cell growth in vitro. A CCK-8 assay was used to evaluate U118 and U251 cell growth at 24, 48 and 72 h after siRNA transfection. The results revealed that the cell growth was significantly decreased in cells transfected with si-SSRP1 compared with the cells transfected with scramble siRNA (negative control) (P<0.05; Fig. 2D). In addition, cell proliferation was also assessed using the EdU incorporation assays. As shown in Fig. 2E and F, the average percentage of EdU-positive cells in the control group and the SSRP1 knockdown group were 38.60 and 32.73% in the U118 cells, and 44.1 and 30.6% in the U251 cells (P<0.001). The knockdown of SSRP1 expression led to a marked decrease in the percentage of EdU-positive cells, as compared with the control group (P<0.001).
All the aforementioned results revealed that the downregulation of SSRP1 suppressed the proliferation of glioma cells in vitro.
siRNA-mediated knockdown of SSRP1 arrests the cell cycle and causes apoptosis in glioma cells
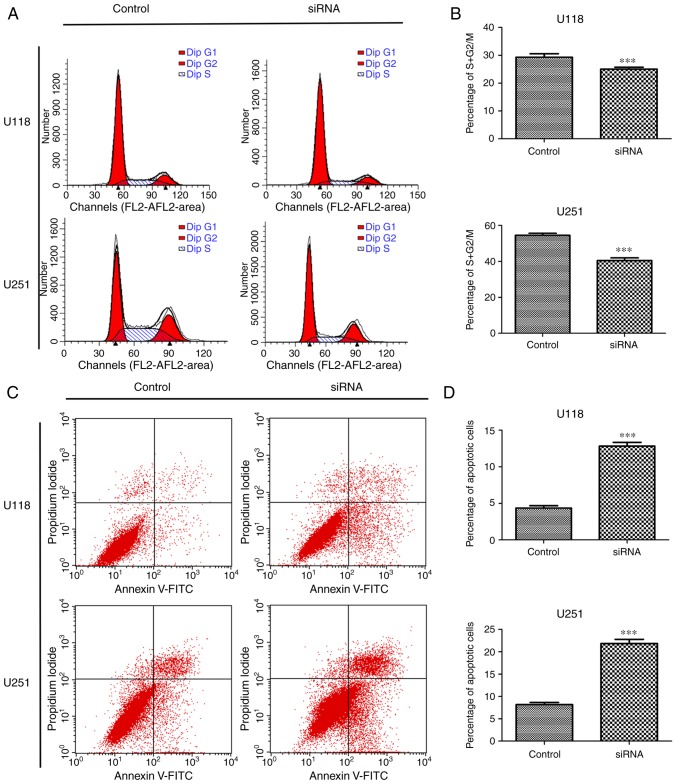
In order to explore whether the effect of SSRP1 depletion on cell proliferation was related to the cell cycle and apoptosis, flow cytometric analysis was first performed. Cells transfected with si-SSRP1 exhibited a significant decrease in the percentage of cells in the S + G2/M phase compared with the cells transfected with scramble siRNA in the U118 (29.31 vs. 24.05%) and U251 cell lines (54.53 vs. 40.47%) (Fig. 3A and B). Moreover, the function of SSRP1 in apoptosis was also investigated. After transfection with si-SSRP1 or scramble siRNA for 48 h, the average percentage of apoptotic U118 cells treated with si-SSRP1 and the control group was 4.36 and 12.83%, respectively (P<0.001; Fig. 3C and D). Experiments using U251 yielded similar results, and the average percentage of apoptotic cells was significantly increased after treatment with si-SSRP1. The percentage of apoptotic U251 cells in the two groups was 8.18 and 21.83% (P<0.001; Fig. 3C and D). These results revealed that SSRP1 was involved in the cell cycle process and apoptosis of glioma cells.
Figure 3.
Effect of structure-specific recognition protein 1 (SSRP1) on the cell cycle and apoptosis. (A and B) Silencing of SSRP1 arrested the cell cycle in both U118 and U251 cells as dtermined by flow cytometric analysis. A graphical representation of the mean ± standard deviation (SD) data revealed the percentage of cells in the S + G2/M phase. (C) The apoptosis rate of cells was assessed with Annexin V-FITC/propidium iodide by flow cytometry. (D) The quantitative analysis of apoptotic rates; ***P<0.001. Data are presented as the mean ± SD.
Decrease of SSRP1 by siRNA inhibits glioma cell migration and invasion
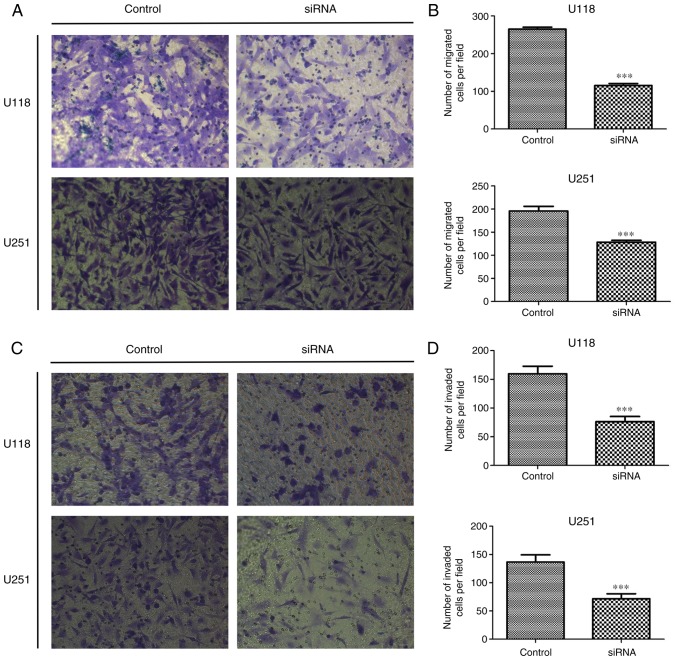
In order to investigate the role of SSRP1 in glioma metastasis, we examined whether SSRP1 played a role in cell migration and invasion using the Transwell apparatus. After 24 h of incubation, the number of migrated cells in both the U118 and U251 cells transfected with si-SSRP1 was significantly lower than that in the negative control group (P<0.001, for each; Fig. 4A and B). The Transwell matrix penetration assay was performed to evaluate the effects of SSRP1 on cell invasion. Compared with the control group, the cells with downregulated SSRP1 expression by siRNA demonstrated a significantly decreased invasion in both U118 and U251 cell lines (P<0.001, for both; Fig. 4C and D). All these results demonstrated that SSRP1 modulates glioma cell migration and invasion in vitro.
Figure 4.
Knockdown of structure-specific recognition protein 1 (SSRP1) decreases cell migration and invasion. (A and B) Knockdown of SSRP1 decreases the migration ability of U118 and U251 cells. The representative images of migrated cells are shown (magnification, ×400), and the migrated cells of each image are presented as the mean ± standard deviation (SD). (C and D) Suppressed SSRP1 inhibited the invasion of U118 and U251 cells. Data are presented as the mean ± SD of at least 3 independent experiments; ***P<0.001.
SSRP1 controls the expression of proliferation-and migration-associated genes in glioma
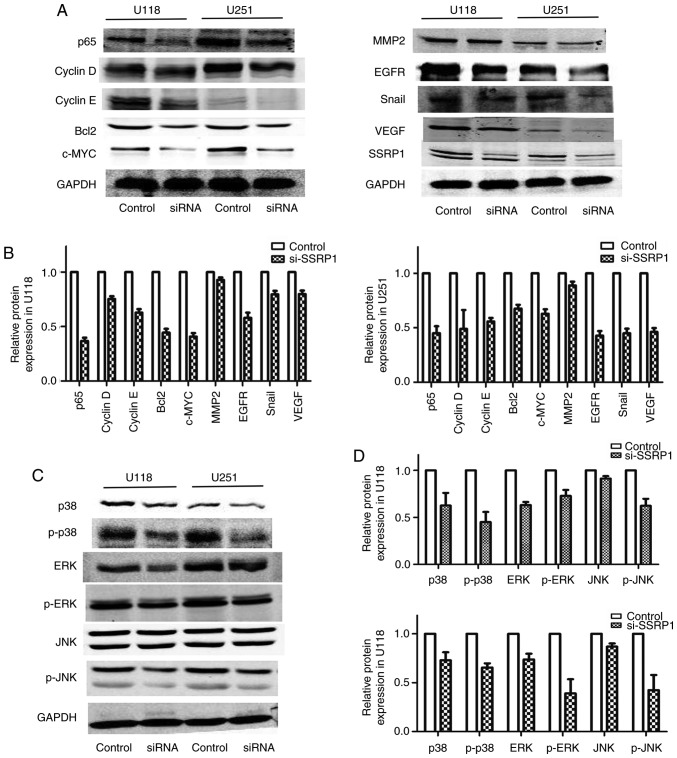
In order to explore the mechanism by which SSRP1 modulated the progression of cell proliferation and metastasis, the proteins involved in the cell cycle, migration and invasion in U118 and U251 cells were analyzed. The downregulated expression of SSRP1 by si-SSRP1 suppressed the protein levels of p65, c-myc and the cell cycle regulators including cyclin D and E (Fig. 5A). Compared to the negative control group, the protein levels of epithelial-mesenchymal transition and metastasis, including EGFR, VEGF and Snail, decreased in the group transfected with si-SSRP1, but MMP2 did not decrease (Fig. 5A).
Figure 5.
Structure-specific recognition protein 1 (SSRP1) regulates the expression of proliferation- and migration-associated genes in glioma via the MAPK pathway. (A and B) Knockdown of SSRP1 expression decreased the expression of proliferation-associated genes, including p65, c-myc, cyclin D and E. The expression of EGFR, VEGF and Snail, proteins involved in migration, were also suppressed. (C and D) The downregulated expression of SSRP1 significantly decreased the total and phosphorylated protein levels of p38 and ERK, and only the phosphorylated protein levels of JNK.
SSRP1 regulates the MAPK signaling pathway
The MAPK signaling pathway has an important role in regulating the proliferation and migration of glioblastoma cells. In order to evaluate the underlying mechanisms of SSRP1 in regulating glioma cell proliferation and migration, western blot assay was carried out to detect the alterations in the main proteins in the MAPK pathway. As shown in Fig. 5B, the downregulated expression of SSRP1 significantly decreased the phosphorylation of p38, ERK and JNK. Furthermore, the total p38 and ERK protein expression was also decreased in the group transfected with si-SSRP1, but not the total JNK protein level (Fig. 5B). These results revealed that SSRP1 is an upstream factor that modulated the MAPK signaling pathway in glioma.
Discussion
SSRP1 has been reported as a subunit of histone chaperone protein FACT, which causes the disruption of nucleosome and histone replacements (21,22). It has been reported to be involved in the progression of DNA replication, DNA damage repair, cell proliferation and cell apoptosis (14,15,23). Previous studies have focused on the molecular mechanism directing transcription elongation. However, few studies have reported the correlation between SSRP1 and cancer, and no study has focused on the clinical significance and oncogenicity effect of SSRP1 in glioma.
Our studies revealed that both the mRNA and protein levels of SSRP1 were upregulated in glioma tissues, compared with those in NB tissues, and that the SSRP1 protein level was higher in the high-grade glioma patients than in the low-grade glioma patients, which were identical to the results of the GEO datasets analyses. These findings suggest that SSRP1 may play an important role in the development of human glioma, and that the overexpression level may predict high-grade glioma.
The overexpression of SSRP1 in glioblastoma predicted the oncogenic role of SSRP1 in glioma tumorigenesis. First, we used multiple methods, including the CCK-8 and EdU incorporation assays, and FACS flow cytometric analysis, to identify the role of SSRP1 in GBM cell growth progression. Furthermore, we demonstrated that the viability and proliferation abilities were significantly decreased in glioma cells with the silencing of SSRP1 by siRNA transfection. In addition, we also ascertained the biological function of SSRP1 on GBM cell motility, and detected that the knockdown of SSRP1 by siRNA inhibited glioma cell migration and invasion. Glioblastoma is the most common aggressive phenotype of glioma, and is identified by the characteristics of cellular proliferation, diffuse infiltration, necrosis, angiogenesis and intense resistance to apoptosis (24). The results of the present study reflected that the decreased expression of SSRP1 inhibited the ability of cell proliferation, anti-apoptosis and metastasis, which were the hallmark features of glioblastoma.
In order to further confirm the function of SSRP1, we examined some cell growth regulators and metastasis markers. It was observed that the protein levels of proliferation-associated genes, including p65, c-myc, cyclin D and E, were inhibited after SSRP1 downregulation in glioma U118 and U251 cells. Furthermore, we demonstrated that SSRP1 knockdown suppressed the expression of metastasis-associated genes such as EGFR, VEGF and Snail.
The MAPK pathway regulated multiple cellular programs including embryogenesis, proliferation, apoptosis and differentiation, based on cues derived from the surface and metabolic state of the cells (25). The 3 major MAPK pathways are ERKs, JNKs and the p38 families (26). In the present study, we also demonstrated that the downregulated expression of SSRP1 significantly decreased the phosphorylation of p38, ERK and JNK, and that total p38 and ERK protein expression was also decreased, but not the total JNK protein level. Hence, the present study revealed that SSRP1 may be involved in tumor progression via the MAPK pathway.
In conclusion, the present study indicated that SSRP1 may regulate the function of the MAPK pathway to promote cell proliferation and metastasis in gliomas. SSRP1 overexpression in glioma specimens could be used for the diagnosis of glioma, and our results revealed the potential role for SSRP1 in glioma therapy. In addition, further research to clarify the molecular mechanism of SSRP1 involved in glioma tumorigenesis is warranted.
Acknowledgements
The present study was supported by grants from the National Science Foundation of China (nos. 81502175 and 81572489).
References
- 1.Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 2.Ohgaki H, Kleihues P. Genetic pathways to primary and secondary glioblastoma. Am J Pathol. 2007;170:1445–1453. doi: 10.2353/ajpath.2007.070011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baumert BG, Hegi ME, Van den Bent MJ, von Deimling A, Gorlia T, Hoang-Xuan K, Brandes AA, Kantor G, Taphoorn MJ, Hassel MB, et al. Temozolomide chemotherapy versus radiotherapy in high-risk low-grade glioma (EORTC 22033–26033): A randomised, open-label, phase 3 intergroup study. Lancet Oncol. 2016;17:1521–1532. doi: 10.1016/S1470-2045(16)30313-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Norden AD, Drappatz J, Wen PY. Novel anti-angiogenic therapies for malignant gliomas. Lancet Neurol. 2008;7:1152–1160. doi: 10.1016/S1474-4422(08)70260-6. [DOI] [PubMed] [Google Scholar]
- 5.Belotserkovskaya R, Oh S, Bondarenko VA, Orphanides G, Studitsky VM, Reinberg D. FACT facilitates transcription-dependent nucleosome alteration. Science. 2003;301:1090–1093. doi: 10.1126/science.1085703. [DOI] [PubMed] [Google Scholar]
- 6.Röttgers K, Krohn NM, Lichota J, Stemmer C, Merkle T, Grasser KD. DNA-interactions and nuclear localisation of the chromosomal HMG domain protein SSRP1 from maize. Plant J. 2000;23:395–405. doi: 10.1046/j.1365-313x.2000.00801.x. [DOI] [PubMed] [Google Scholar]
- 7.Birch JL, Tan BC, Panov KI, Panova TB, Andersen JS, Owen-Hughes TA, Russell J, Lee SC, Zomerdijk JC. FACT facilitates chromatin transcription by RNA polymerases I and III. EMBO J. 2009;28:854–865. doi: 10.1038/emboj.2009.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kumari A, Mazina OM, Shinde U, Mazin AV, Lu H. A role for SSRP1 in recombination-mediated DNA damage response. J Cell Biochem. 2009;108:508–518. doi: 10.1002/jcb.22280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mason PB, Struhl K. The FACT complex travels with elongating RNA polymerase II and is important for the fidelity of transcriptional initiation in vivo. Mol Cell Biol. 2003;23:8323–8333. doi: 10.1128/MCB.23.22.8323-8333.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tan BC, Liu H, Lin CL, Lee SC. Functional cooperation between FACT and MCM is coordinated with cell cycle and differential complex formation. J Biomed Sci. 2010;17:11. doi: 10.1186/1423-0127-17-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang W, Zeng F, Liu Y, Shao C, Li S, Lv H, Shi Y, Niu L, Teng M, Li X. Crystal structure of human SSRP1 middle domain reveals a role in DNA binding. Sci Rep. 2015;5:18688. doi: 10.1038/srep18688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garcia H, Fleyshman D, Kolesnikova K, Safina A, Commane M, Paszkiewicz G, Omelian A, Morrison C, Gurova K. Expression of FACT in mammalian tissues suggests its role in maintaining of undifferentiated state of cells. Oncotarget. 2011;2:783–796. doi: 10.18632/oncotarget.340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garcia H, Miecznikowski JC, Safina A, Commane M, Ruusulehto A, Kilpinen S, Leach RW, Attwood K, Li Y, Degan S, et al. Facilitates chromatin transcription complex is an ‘accelerator’ of tumor transformation and potential marker and target of aggressive cancers. Cell Reports. 2013;4:159–173. doi: 10.1016/j.celrep.2013.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ding Q, He K, Luo T, Deng Y, Wang H, Liu H, Zhang J, Chen K, Xiao J, Duan X, et al. SSRP1 contributes to the malignancy of hepatocellular carcinoma and is negatively regulated by miR-497. Mol Ther. 2016;24:903–914. doi: 10.1038/mt.2016.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dermawan JK, Gurova K, Pink J, Dowlati A, De S, Narla G, Sharma N, Stark GR. Quinacrine overcomes resistance to erlotinib by inhibiting FACT, NF-κB, and cell-cycle progression in non-small cell lung cancer. Mol Cancer Ther. 2014;13:2203–2214. doi: 10.1158/1535-7163.MCT-14-0013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koman IE, Commane M, Paszkiewicz G, Hoonjan B, Pal S, Safina A, Toshkov I, Purmal AA, Wang D, Liu S, et al. Targeting FACT complex suppresses mammary tumorigenesis in Her2/neu transgenic mice. Cancer Prev Res. 2012;5:1025–1035. doi: 10.1158/1940-6207.CAPR-11-0529. [DOI] [PubMed] [Google Scholar]
- 17.Carter DR, Murray J, Cheung BB, Gamble L, Koach J, Tsang J, Sutton S, Kalla H, Syed S, Gifford AJ, et al. Therapeutic targeting of the MYC signal by inhibition of histone chaperone FACT in neuroblastoma. Sci Transl Med. 2015;7:312ra176. doi: 10.1126/scitranslmed.aab1803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lake D, Corrêa SA, Müller J. Negative feedback regulation of the ERK1/2 MAPK pathway. Cell Mol Life Sci. 2016;73:4397–4413. doi: 10.1007/s00018-016-2297-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Z, Guo Q, Wang R, Xu G, Li P, Sun Y, She X, Liu Q, Chen Q, Yu Z, et al. The D Domain of LRRC4 anchors ERK1/2 in the cytoplasm and competitively inhibits MEK/ERK activation in glioma cells. J Hematol Oncol. 2016;9:130. doi: 10.1186/s13045-016-0355-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shingu T, Holmes L, Henry V, Wang Q, Latha K, Gururaj AE, Gibson LA, Doucette T, Lang FF, Rao G, et al. Suppression of RAF/MEK or PI3K synergizes cytotoxicity of receptor tyrosine kinase inhibitors in glioma tumor-initiating cells. J Transl Med. 2016;14:46. doi: 10.1186/s12967-016-0803-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Takahata S, Yu Y, Stillman DJ. FACT and Asf1 regulate nucleosome dynamics and coactivator binding at the HO promoter. Mol Cell. 2009;34:405–415. doi: 10.1016/j.molcel.2009.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tsunaka Y, Fujiwara Y, Oyama T, Hirose S, Morikawa K. Integrated molecular mechanism directing nucleosome reorganization by human FACT. Genes Dev. 2016;30:673–686. doi: 10.1101/gad.274183.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gao XJ, Feng JX, Zhu S, Liu XH, Tardieux I, Liu LX. Protein phosphatase 2C of toxoplasma gondii interacts with human SSRP1 and negatively regulates cell apoptosis. Biomed Environ Sci. 2014;27:883–893. doi: 10.3967/bes2014.130. [DOI] [PubMed] [Google Scholar]
- 24.Furnari FB, Fenton T, Bachoo RM, Mukasa A, Stommel JM, Stegh A, Hahn WC, Ligon KL, Louis DN, Brennan C, et al. Malignant astrocytic glioma: Genetics, biology, and paths to treatment. Genes Dev. 2007;21:2683–2710. doi: 10.1101/gad.1596707. [DOI] [PubMed] [Google Scholar]
- 25.Raman M, Chen W, Cobb MH. Differential regulation and properties of MAPKs. Oncogene. 2007;26:3100–3112. doi: 10.1038/sj.onc.1210392. [DOI] [PubMed] [Google Scholar]
- 26.Owens DM, Keyse SM. Differential regulation of MAP kinase signalling by dual-specificity protein phosphatases. Oncogene. 2007;26:3203–3213. doi: 10.1038/sj.onc.1210412. [DOI] [PubMed] [Google Scholar]