Abstract
Recent studies have demonstrated that microRNA-154 (miR-154) is involved in tumorigenesis, progression, invasion and metastasis in several types of human cancer. However, whether it plays a role in bladder cancer (BC) is unclear. The aim of the present study was to determine miR-154 levels in human BC tissues and investigate the correlation between miR-154 levels and clinicopathological characteristics as well as patient outcome. Using RT-qPCR, we found that the expression levels of miR-154 were significantly lower in BC tissues compared to adjacent normal tissues. We also demonstrated that downregulation of miR-154 was associated with advanced clinicopathological features and worse prognoses for patients with BC. Using a variety of integrated approaches, we demonstrated that both runt-related transcription factor 2 (RUNX2) and remodeling and spacing factor 1 (RSF1) were miR-154 targets. Notably, there was an inverse correlation between RSF1, RUNX2 and miR-154 expression in BC tissues. The biological functions of miR-154 were examined in vitro using Cell Counting Kit-8 (CCK-8), wound healing, and Transwell assays with T24 human bladder carcinoma cells transfected with miR-154 mimics or negative controls. These assays demonstrated that miR-154 significantly suppressed proliferation, migration and invasion of T24 cells (P<0.05). Furthermore, overexpression of RSF1 and RUNX2 rescued miR-154-induced inhibition of these aggressive behaviors. Our results indicated that miR-154, and its downstream targets RSF1 and RUNX2, are promising options for future BC therapies.
Keywords: bladder cancer, miR-154, prognosis, runt-related transcription factor 2, remodeling and spacing factor 1
Introduction
Bladder cancer (BC) is one of the most common malignancies worldwide, with a global age-standardized incidence rate of 9.0 for men and 2.2 for women (per 100,000 person-years) (1,2). Approximately 25% of newly diagnosed cases of BC present with muscle invasion and require radical surgery or radiotherapy, although these patients often have poor outcomes despite systemic therapy (3). BC affects the urinary lining of the bladder through a complicated, multifactorial etiology that involves both genetic and environmental factors (4). Novel prognostic markers and effective treatment strategies are therefore essential to improve outcomes for patients with BC.
MicroRNAs (miRNAs) are small non-coding RNAs that are ~22 nucleotides in length. They play pivotal regulatory roles in post-transcriptional suppression (5) and have been reported to be involved in many cellular processes, such as differentiation, morphogenesis and tumorigenesis (6–8). Aberrant miRNA expression is found in a variety of cancers, including BC, suggesting that they may have roles as oncogenes or tumor-suppressor genes (9,10). For example, miR-497 was significantly downregulated in bladder transitional cell carcinoma cells and tissues. This led to the upregulation of the transcription factor E2F3 and suppression of cell proliferation and invasion (11). Similarly, miR-106a had an inhibitory effect on the proliferation of BC cells through the modulation of MAPK signaling (12). Previous studies have also shown that miR-154, located in the human imprinted 14q32 domain, acts as a tumor suppressor in various types of human cancer, including prostate (13), colorectal (14) and lung cancer (15). Moreover, low miR-154 levels in glioma (16) and colorectal cancer (17) were significantly associated with poor prognosis. Nevertheless, the levels of miR-154 and their clinical significance in human BC have not yet been evaluated.
In our analysis, we first investigated the levels of miR-154 in multiple BC tissues. We found that miR-154 was downregulated in BC tissues and lower miR-154 expression levels were strongly associated with worse overall survival (OS) rates in patients with BC. Further study suggested that this was due to decreased inhibition of BC cell proliferation, migration and invasion mediated by miR-154. Subsequent bioinformatics analysis revealed that both runt-related transcription factor 2 (RUNX2) and remodeling and spacing factor 1 (RSF1) were targets of miR-154 and were involved in miR-154-induced inhibition of the tumorigenic properties of BC cells.
Materials and methods
Patients and tissue samples
In total, 86 fresh BC tissue specimens, along with adjacent normal tissue specimens, were collected from 2010 to 2013 at the Department of Urology, Peking Union Medical College Hospital (Beijing, China). None of the patients had received any treatment prior to surgery. The pathological type of BC was observed and microscopically validated by an independent pathologist. Tumor grading and staging were carried out according to the 6th AJCC TNM staging system and graded according to Fuhrman's nuclear grading system. The exclusion criteria included incomplete clinical or follow-up information, recurrent tumors, other chronic system diseases, and an unwillingness to participate in the present study. Clinical characteristics and follow-up information for individual patients were collected from their medical records. OS was defined as the time from inclusion to death of the patient, for any reason. The study was approved by the Research Ethics Committee of the Peking Union Medical College Hospital and written informed consent was provided by all patients.
Real-time quantitative PCR (RT-qPCR)
Total RNA was isolated from tissues and cells using TRIzol reagent, according to the manufacturer's protocol (Invitrogen, Carlsbad, CA, USA). For the detection of miR-154, complementary DNAs were synthesized using a TaqMan MicroRNA Reverse Transcription kit and a stem-loop RT primer. Amplification was performed using a TaqMan miRNA assay (Applied Biosystems, Foster City, CA, USA) on an ABI PRISM 7500 Real-Time PCR System with the following conditions: 95°C for 2 min, followed by 40 cycles at 94°C (15 sec), 60°C (60 sec) and 72°C (30 sec). For mRNA detection, complementary DNAs were synthesized via a PrimeScript RT-PCR kit (Takara, Otsu, Japan) and quantification was performed using RT Real-Time SYBR-Green assays (Bio-Rad Laboratories, Berkeley, CA, USA). Each sample was examined in triplicate and the relative miRNA and mRNA expression levels were determined using the 2−ΔΔCt method and normalized to the housekeeping genes U6 and GAPDH. Primers used for qPCR are shown in Table I.
Table I.
The forward and reverse primers for real-time PCR.
| Name | Sequence | |
|---|---|---|
| has-miR-154 | RT | GTCGTATCCAGTGCGTGTCG |
| primer | TGGAGTCGGCAATTGCACTGGATACGACCGAAGG | |
| Forward | GTGGTACTTGAAGATAGGTT | |
| Reverse | TTGGTACTGAAAAATAGGTC | |
| RSF1 | Forward | GAGGAGGATGCCGATACTATGC |
| Reverse | TGCTTTCAGGAGTGCAAGAGTC | |
| RUNX2 | Forward | AAGTGAGGTTAGGGCGAAATG |
| Reverse | AAGGTAGTTGATTGCCAACGAA | |
| U6 | Forward | GAGCGGTAGCACCATTTGAA |
| Reverse | GTGCAGGGTCCGAGG | |
| GAPDH | Forward | GAAGGTGAAGGTCGGAGTC |
| Reverse | GAAGATGGTGATGGGATTTC |
miR-154, microRNA-154; RSF1, remodeling and spacing factor 1; RUNX2, runt-related transcription factor 2.
Cell culture and transfection
The T24 human BC cell line was purchased from the American Type Culture Collection (ATCC; Manassas, VA, USA) and cultured in Dulbeccos modified Eagles medium (DMEM) (Mediatech, Manassas, VA, USA) supplemented with 10% fetal bovine serum (FBS), and 1% U/ml penicillin and streptomycin (both from Invitrogen). All cells were maintained at 37°C in a humidified atmosphere of air containing 5% CO2. Cells were transfected with miR-154 mimics (sense, 5′UAGGUUAUCCGUGUUGCCUUCG3′ and antisense, 5′AAGGCAACACGAUAACCUAUU3′) and negative controls (scrambled oligos, miR-NC) (Shanghai GenePharma, Shanghai, China) using Lipofectamine 2000 (Invitrogen) at a final concentration of 100 nM, following the manufacturer's instructions. Overexpression of RSF1 and RUNX2 was achieved using RSF1 and RUNX2 ORF expression clones (Vector Gene Technology of Beijing, Beijing, China) and a pcDNA3.1 empty vector was used as a negative control.
Bioinformatics analysis and luciferase reporter assay
The downstream targets of miR-154 were predicted using miRBase, miTarget and TargetScan. Putative complementary sequences for miR-154 were identified in the 3′-untranslated regions (3′-UTRs) of RSF1 and RUNX2 mRNA. The 3′-UTRs of RSF1 and RUNX2, containing the miR-154 target sequence, were synthesized and inserted into the XbaI and FseI sites of a pGL3 control vector (Promega, Madison, WI, USA) to generate a overall survival an RSF1-wild-type and RUNX2-wild-type luciferase reporter. Mutation of the binding site was performed using site-directed mutagenesis (Promega) to create the pGL3-RSF1- and pGL3-RUNX2-mutant type plasmids. For detection, T24 cells were cultured in 96-well plates and co-transfected with miR-1545 mimics (or miR-NC) and reporter plasmids using Lipofectamine 2000. After 48 h, the cells were assayed with a Dual-Luciferase Reporter Assay System (Promega). Firefly luciferase activity for each sample was normalized to Renilla luciferase activity.
Cell proliferation assay
Cell Counting Kit-8 (CCK-8; Dojindo, Kumamoto, Japan) assays were used for analysis of cell proliferation. T24 cells were seeded into 96-well plates at 1×104 cells/well and incubated in 10% CCK-8 solution at 37°C until color conversion had occurred. The absorbance was assessed at 450 nm using a microplate reader (Multiskan; Devices, Menlo Park, CA, USA) 24, 48, 72 and 96 h after transfection.
Wound-healing and Transwell invasion assays
T24 cells were seeded in 24-well plates. When cells were 80–90% confluent, a sterile pipette tip was used to manually scratch the monolayer. After three washes with phosphate-buffered saline (PBS), cell migration was observed using an inverted microscope. The cells were then incubated at 37°C for a further 48 h before cell migration was assessed again. For the invasion assays, transfected cells were transferred to the top of Matrigel-coated invasion chambers (24-well plates, 8 µm pore size; Becton-Dickinson, San Jose, CA, USA) with serum-free medium. Medium containing 10% FBS was used as the chemoattractant in the lower chambers. After 24 h of incubation, the media were removed and the wells were washed twice with PBS. Invasive cells that were attached to the lower surface were fixed with 4% paraformaldehyde, stained with 0.1% crystal violet, and transferred to a microscope slide. Microphotographs were collected in six representative fields using an inverted microscope at a magnification of ×200 (Olympus BX53; Olympus, Tokyo, Japan).
Western blot analyses
T24 cells and human BC specimens were lysed in 1% RIPA lysis buffer (Beyotime, Jiangsu, China), and supplemented with protease inhibitors at 4°C for 1 h. Protein quantification was performed using the BCA method (Beyotime). Equal amounts of protein were ran on 10% sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE) gels, and transferred to polyvinylidene fluoride (PVDF) membranes (Millipore, Billerica, MA, USA). After blocking with 1% bovine serum albumin (BSA) for 30 min, the membranes were incubated with primary antibodies against either RSF1 (ab109002), RUNX2 (ab23981) or GAPDH (ab9485) (all from Abcam, Cambridge, UK) at 4°C overnight. The membranes were then washed three times with Tris-buffered saline with Tween-20 (TBST), followed by incubation with horseradish peroxidase-conjugated (HRP) goat anti-rabbit secondary antibody (Sigma, St. Louis, MO, USA) for 1 h at room temperature. The blots were incubated with an enhanced chemiluminescence substrate (ECL; Plus) and visualized on a G:BOX Chemi XR5 imaging system (Syngene, Cambridge, UK).
Statistical analysis
All values are expressed as the median and range, or mean ± standard deviation (SD), where appropriate. Data analysis was performed using GraphPad Prism software 6.0 (GraphPad Software, Inc., La Jolla, CA, USA). Comparisons between two groups were performed using unpaired two-tailed Student's t-tests or χ2 test and comparisons among >2 groups were carried out with one-way ANOVA plus post hoc Bonferroni or post hoc Dunnett tests. Assessment of any association between miR-154 expression and various clinicopathological parameters were made using Pearson's χ2 tests and SPSS version 19.0 (SPSS, Inc., Chicago, IL, USA). Spearman's correlation analysis was performed to compare mRNAs and miR-154 levels. Analysis of OS was performed according to the Kaplan-Meier method and comparisons were carried out using log-rank tests. P<0.05 was considered statistically significant.
Results
miR-154 is specifically downregulated in BC tissues and is a predictor of poor outcome
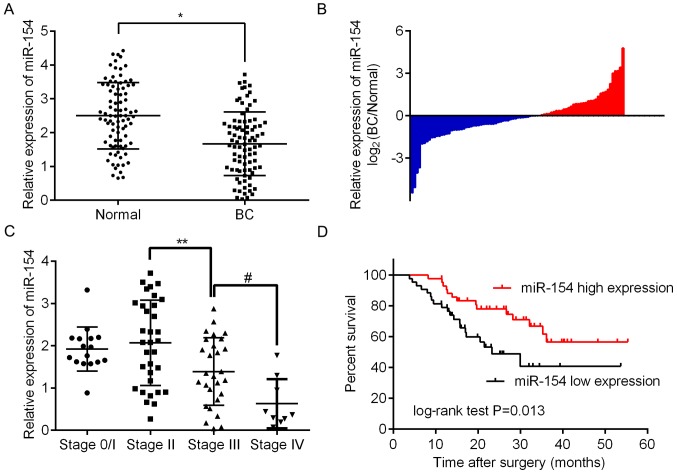
To assess the expression of miR-154 in BC tissue, we analyzed 86 paired BC/normal tissue samples by RT-qPCR. The results revealed that there was significantly less miR-154 in BC tissues relative to adjacent normal tissue (Fig. 1A and B; P<0.05). To assess any correlation between miR-154 and the clinical or pathological features of BC, patients were assigned to two groups, one with high expression relative to the median and the other with low expression. As shown in Table II, it was determined that low miR-154 expression was significantly associated with more advanced tumor staging (P=0.020), higher histologic grades (P=0.041) and lymph node metastasis (P=0.024). However, there were no significant associations between miR-154 expression and age, sex or tumor size. These findings revealed that miR-154 may play an important role in the progression of BC. This supports results shown in Fig. 1C demonstrating that there is a trend for miR-154 to be lower in more advanced tumor, node and metastasis (TNM) stages, with the lowest expression found in stage 4. Furthermore, Kaplan-Meier curves revealed that low levels of miR-154 had a statistically significant association with shorter OS in patients when compared to those with higher expression levels (P<0.01; Fig. 1D).
Figure 1.
Low expression of miR-154 in human bladder cancer (BC) tissue and its correlation with poor prognosis. (A) The relative expression of miR-154 in 86 paired BC/normal tissues analyzed by RT-qPCR; *P<0.05 vs. the normal group. (B) The levels of miR-154 expression, reported as the log2-fold difference between BC and adjacent normal tissues. (C) The expression profile of miR-154 in patients with BC in different TNM stages; **P<0.05 vs. stage 2, #P<0.05 vs. stage 3. (D) A Kaplan-Meier curve demonstrating that downregulation of miR-154 in BC was correlated with poor overall survival; P=0.013, obtained by log-rank test.
Table II.
Correlation between miR-154 expression and clinicopathological parameters of baldder cancer.
| miR-154 expression | |||
|---|---|---|---|
| Clinical characteristics | Low | High | P-valuea |
| Age (years) | 0.914 | ||
| <60 | 19 | 17 | |
| ≥60 | 27 | 23 | |
| Sex | 0.279 | ||
| Male | 20 | 23 | |
| Female | 26 | 17 | |
| Tumor size (cm) | 0.363 | ||
| <3 | 28 | 29 | |
| ≥3 | 18 | 11 | |
| T stage | 0.020 | ||
| Ta/T1 | 8 | 8 | |
| T2 | 12 | 21 | |
| T3 | 18 | 10 | |
| T4 | 8 | 1 | |
| Histologic grade | 0.041 | ||
| G1 | 12 | 20 | |
| G2 | 17 | 13 | |
| G3 | 17 | 7 | |
| Lymph node metastasis | 0.024 | ||
| N0/1 | 19 | 28 | |
| N2 | 20 | 10 | |
| N3 | 7 | 2 | |
Pearson χ2 test was used to derive P-values. miR-154, microRNA-154.
RSF1 and RUNX2 are directly targeted by miR-154 in BC cells
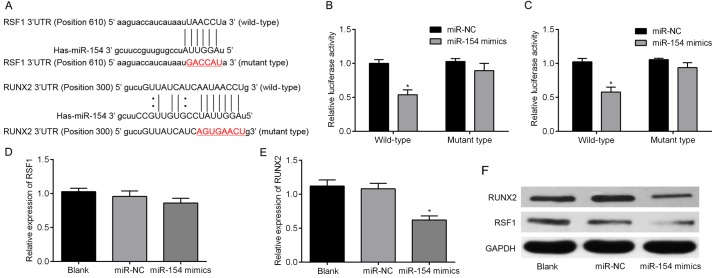
Three algorithms, miRBase, TargetScan and miRanda, were used to predict the putative targets of miR-154. Two genes, RSF1 and RUNX2, were then selected for further validation since they both occurred in all three algorithms. As shown in Fig. 2A, the 3′-UTRs of RSF1 and RUNX2 were inserted into a reporter plasmid downstream of luciferase. Matching mutants were also created, with changes underlined in the presented sequences. T24 cells were then transiently co-transfected with miR-154 mimics (or miR-NC) and individually constructed plasmids. Luciferase reporter assays were performed to detect luciferase expression. These revealed that overexpression of miR-154 resulted in downregulation of luciferase when fused to the RSF1 and RUNX2 3′-UTRs in T24 cells (Fig. 2B and C). To determine whether miR-154 suppressed endogenous RSF1 and RUNX2, miR-154 was overexpressed using mimic transfection into T24 cells. Notably, upregulation of miR-154 only downregulated RUNX2 mRNA, but failed to suppress endogenous expression of RSF1 mRNA (Fig. 2D and E). For further validation, cells were collected after transfection and analyzed by western blotting. The results revealed that both RSF1 and RUNX2 protein were decreased by miR-154 overexpression (Fig. 2F). Collectively, our results revealed that RSF1 and RUNX2 are both target genes of miR-154.
Figure 2.
miR-154 directly targets RSF1 and RUNX2 in BC cells. (A) Several bioinformatics tools were used to predict miR-154 binding sites. This identified putative sites in the 3′-UTRs of the RSF1 and RUNX2 genes. (B) The relative luciferase activities of cells co-transfected with either wild WT or MUT 3′-UTR constructs containing RSF1 and either miR-154 mimics or miR-NC. (C) The relative luciferase activity of cells transfected with WT/MUT 3′-UTR constructs containing RUNX2 and either miR-154 mimics or miR-NC. (D) The expression levels of RSF1 mRNA in T24 cells transfected with miR-154 or miR-NC. (E) RUNX2 was significantly lower in cells transfected miR-154 mimics compared to the miR-NC group, assessed by RT-qPCR. (F) miR-154 expression affected both RSF1 and RUNX2 protein expression in T24 cells; *P<0.05 vs. the miR-NC group. RSF1, remodeling and spacing factor 1; RUNX2, runt-related transcription factor 2; BC, bladder cancer; Wt, wild-type; MUT, mutant-type.
RSF1 and RUNX2 are highly expressed in BC tissues and are negatively correlated with miR-154 levels
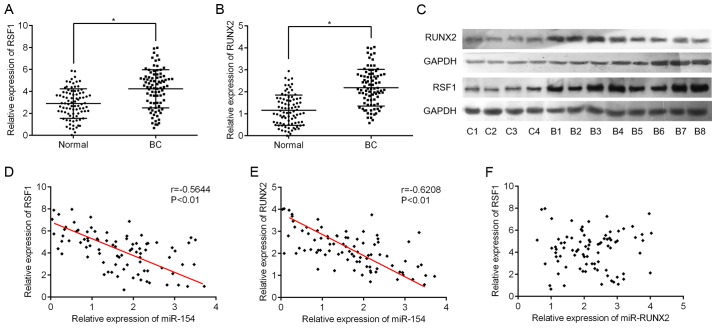
Next, we used RT-qPCR to assess the expression levels of RSF1 and RUNX2 in BC tissues. As expected, RSF1 and RUNX2 mRNA expression was significantly higher in BC tissues compared to adjacent normal tissues (Fig. 3A and B). Supporting the RT-qPCR data, the levels of RSF1 and RUNX2 protein also exhibited the same pattern in eight BC and four normal samples (Fig. 3C). Furthermore, we found that RSF1 gene expression was negatively correlated with miR-154 levels (Pearson correlation r=-0.5644, P<0.001; Fig. 3D). A similar correlation was also observed between RUNX2 and miR-154 (r=-0.6208, P<0.001; Fig. 3E). However, no linear relationship was found between RSF1 and RUNX2 expression in BC tissues (r=0.016, P=0.893; Fig. 3F).
Figure 3.
An inverse correlation between RSF1, RUNX2 and miR-154 expression in BC tissues. (A) The expression of RSF1 mRNA was significantly increased in BC tissues when compared to corresponding normal tissue; *P<0.05 vs. normal tissue. (B) The expression of RUNX2 mRNA was also increased in BC tissue compared to normal tissue; *P<0.05 vs. normal tissue. (C) Western blot analyses showing the expression levels of RSF1 and RUNX2 protein in four normal (C1-4) and eight BC samples (B1-8). (D) The relationship between miR-154 and RSF1 mRNA in BC tissue was assessed by Pearson correlation; r=0.5644, P<0.001. (E) The relationship between miR-154 and RUNX2 mRNA in BC tissue; r=0.6208, P<0.001. (F) The relationship between RSF1 and RUNX2 mRNA in BC tissue; r=0.016, P=0.893. RSF1, remodeling and spacing factor 1; RUNX2, runt-related transcription factor 2; BC, bladder cancer.
miR-154 suppresses proliferation, migration and invasion of BC cells
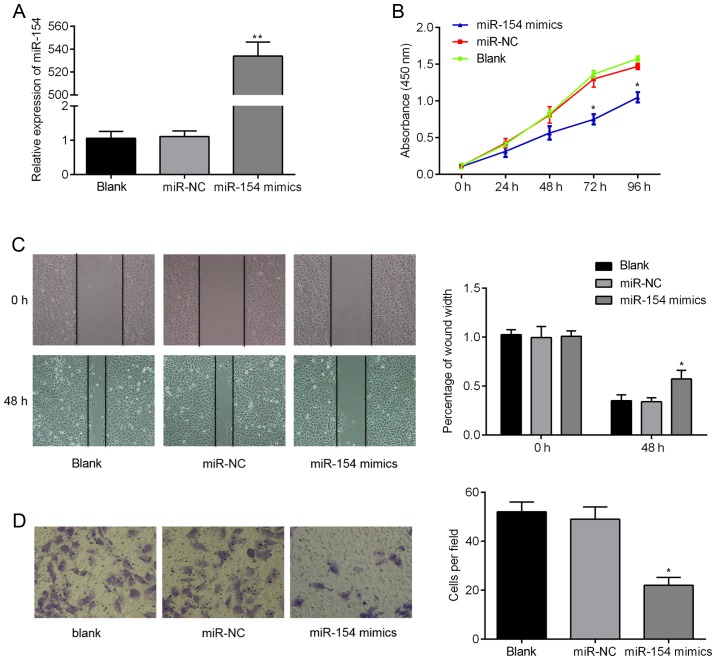
We next assessed the in vitro biological functions of miR-154 in BC cells in more detail. RT-qPCR analysis confirmed that miR-154 mimics were successfully transfected into T24 cells and could be used for subsequent analysis. As shown in Fig. 4A, miR-154 expression levels were >500-fold higher in the miR-154 mimic group compared to controls (P<0.01). We then examined the effects that miR-154 had on cell proliferation, migration and invasion. We found using CCK-8 assays that ectopic miR-154 expression suppressed proliferation of T24 cells. Additionally, the migratory capacity of cells transfected with miR-154 mimics was lower 48 h after wound creation, when compared to the control cells (Fig. 4C; P<0.05). Finally, Transwell migration assays over 24 h revealed that there were twice as many migratory cells transfected with miR-NC relative to cells transfected with miR-154 mimics (Fig. 4D; P<0.05). Combined, these findings suggest that overexpression of miR-154 suppresses the tumorigenic behavior of BC cells in vitro.
Figure 4.
miR-154 inhibits T24 cell proliferation, migration and invasion. (A) The relative expression of miR-154 in cells transfected with either miR-154 mimics, miR-NC or neither. (B) Cell proliferation was determined in T24 cells transfected with miR-154 mimic or miR-NC using a CCK-8 assay. (C) The migratory capacity of cells in different groups was determined using a wound healing assay. (D) The invasive capacity of T24 cells between groups was investigated using a Transwell invasion assay; *P<0.05 vs. miR-NC, **P<0.01 vs. miR-NC.
miR-154 inhibits proliferation, migration and invasion in BC cells by regulating RSF1 and RUNX2
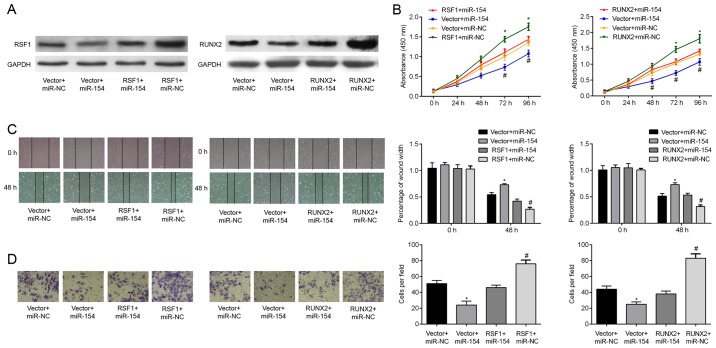
We finally determined whether overexpression of RSF1 or RUNX2 could counteract the inhibitory effects of miR-154 on T24 cells. To achieve this, cells were transfected with either RSF1 or RUNX2 inserted into a pcDNA3.1 overexpression plasmid. Successful transfection was ascertained by western blotting, demonstrating that the relative expression of RSF1 and RUNX2 protein was increased relative to the controls. Significantly, increasing RSF1 and RUNX2 protein expression in BC cells reversed the cell proliferation induced by miR-154 overexpression (Fig. 5B). Wound healing assays revealed that the decrease in T24 cell migration caused by miR-154 mimics was attenuated by co-transfection with RSF1 and RUNX2 (Fig. 5C; P<0.05). Similarly, upregulation of RSF1 and RUNX2 resulted in a greater cell invasive capacity and rescued miR-154-induced suppression of cell invasion (Fig. 5D; all P<0.05). Collectively, these data support the hypothesis that miR-154 inhibits T24 cell proliferation, migration, and invasion by negatively targeting the RSF1 and RUNX2 genes.
Figure 5.
miR-153 suppresses proliferation, migration and the invasive potential of BC cells by downregulating RSF1 and RUNX2. (A) RT-qPCR was used to assess the expression of RSF1 and RUNX2 mRNA after transfection with a RSF1 or RUNX2 ORF expression clone, or an empty pcDNA3.1 vector, and either miR-154 mimics or miR-NC. (B) Cell proliferation between different groups was investigated using a CCK-8 assay (C) Cell migration was assessed using a wound-healing assay and T24 cells. (D) Transwell invasion assays were used to measure the cell invasive capacity between groups; *P<0.05 vs. vector + miR-NC; #P<0.05 vs. RSF1 + miR-154 or RUNX2 + miR-154. BC, bladder cancer; RSF1, remodeling and spacing factor 1; RUNX2, runt-related transcription factor 2.
Discussion
A growing body of evidence has suggested that miRNAs play a fundamental role in the proliferation, invasion and metastasis of malignant human carcinomas (18). Therefore, targeting miRNAs may be an effective approach for the treatment of advanced cancer. Recently, aberrant miRNAs involved in tumorigenesis and cancer progression were identified in bladder cancer (BC) (19,20). In the present study, we demonstrated that miR-154 was downregulated in BC tissues compared to adjacent normal tissues. We also found that miR-154 levels were strongly associated with the aggressive clinicopathological features of BC, such as progression to advanced TNM stages, high histologic grades and lymph node status. In addition, survival analysis revealed that patients with low miR-154 expression had shorter OS relative to those with high miR-154 expression. Finally, we found that miR-154 could suppress BC cell proliferation, migration and invasion in vitro. To the best of our knowledge, this is the first study confirming the clinical significance and functional attributes of miR-154 in BC.
Previous studies have identified miR-154 as a tumor-suppressor involved in several types of cancer. For example Xu et al (2016) revealed that miR-154 is frequently downregulated in breast cancer tissue and cell lines where it is involved in the proliferation, migration and invasion of breast cancer cells by directly targeting E2F transcription factor 5 (E2F5) (21). In addition, miR-154 was identified as a suppressor of epithelial mesenchymal transition by downregulating ZEB2 (22) and HMGA2 (23). A recent study revealed by Lin et al (2016) also revealed that ZEB2 overexpression can reverse miR-154-induced inhibition of migration and invasion in lung cancer (15). Building on this research, the present study demonstrated that overexpression of miR-154 decreased RSF1 and RUNX2 protein expression levels in T24 cells. This is likely through miR-154 binding the 3′-UTR of RUNX2 mRNA, promoting its degradation. There was however no significant effect on RSF1 mRNA levels, which may be due to incomplete complementation with the 3′-UTR.
RSF1, also known as hepatitis B X-antigen-associated protein (HBXAP), is a member of the ATP-dependent chromatin remodeling factor family (24). RSF1 interacts with its partner, sucrose non-fermenting protein 2 homologous (hSNF2H), to form the RSF-1/hSNF2H complex. This complex responds to a variety of growth signals and environmental cues to remodel chromatin and is essential for transcriptional activation/suppression and cell cycle progression (25–27). There is also evidence to suggest that expression of the RSF1 gene is increased in various cancers and its upregulation is critical to tumor development (28). Ren et al (2013) revealed that high-expression of RSF1 was associated with larger tumors and more advanced TNM staging in primary breast cancer (29). A study by Liang et al (2012) also found that increased levels of RSF1 were associated with poor outcomes for patients with BC, suggesting that the protein is involved in cell survival and growth (30). In the present study, we found that RSF1 was directly targeted by miR-154. Furthermore, it played an important role in controlling the malignant behavior of BC cells, explaining the clinical significance of the RSF1. However, the exact molecular mechanisms associated with the effects of RSF1 protein, such as NF-κB signaling (31), in BC still remain undefined in the present study.
In addition, we also identified another target gene of miR-154, RUNX2. This is a member of the mammalian RUNX family of transcription factors that have been well-established to be involved in development and cancer (32). For example, RUNX2, a key regulator of osteoblast differentiation, has been implicated in the evolution of breast and prostate cancer metastasis (33, 34). Consistent with our findings, Abdelzaher et al (2016) reported that RUNX2 is frequently upregulated in urothelial carcinomas and is likely involved in both carcinogenesis and tumor aggressiveness (35). There are also numerous studies examining the oncogenic role of RUNX2 through its regulatory effects on various tumorigenesis-associating genes and pathways. For example, Lim et al (2010) demonstrated that RUNX2 regulated survivin (BIRC5) expression in prostate cancer and protected cancer cells against apoptosis (36). Another study concluded that RUNX2 could act as a negative regulator for p53-dependent apoptosis and may therefore be an attractive therapeutic target for cancer (37). Our own study revealed an association between upregulation of RUNX2 in malignant epithelial BC carcinoma tissues, although we demonstrated that this was regulated by miR-154. We therefore suggest that RUNX2 promotes cell proliferation, migration, and invasion in BC and that these factors may be controlled by miR-154. However, it is still unclear whether RUNX2 affects cell apoptosis in BC cells and more studies are required to better understand the molecular mechanisms and biological pathways mediated by RUNX2. Since miRNAs can regulate various genes simultaneously, targeting miRNAs could influence multiple critical molecules involved in tumorigenesis (38). iTRAQ proteomics and bioinformatics (39) could be further used to identify more miR-154 targets.
In summary, the present study found that miR-154 downregulation in BC was correlated with a poor survival rate. Additionally, miR-154 inhibited BC cell proliferation, migration, and invasion by regulating RSF1 and RUNX2 expression. Our data revealed that the miR-154-RSF1/RUNX2 interaction in BC may be an attractive target for future cancer treatment.
Glossary
Abbreviations
- BC
bladder cancer
- miR-154
microRNA-154
- RUNX2
runt-related transcription factor 2
- RSF1
remodeling and spacing factor 1
References
- 1.Babjuk M, Böhle A, Burger M, Capoun O, Cohen D, Compérat EM, Hernández V, Kaasinen E, Palou J, Rouprêt M, et al. EAU Guidelines on Non-Muscle-invasive Urothelial Carcinoma of the Bladder: Update 2016. Eur Urol. 2017;71:447–461. doi: 10.1016/j.eururo.2016.05.041. [DOI] [PubMed] [Google Scholar]
- 2.Burger M, Catto JW, Dalbagni G, Grossman HB, Herr H, Karakiewicz P, Kassouf W, Kiemeney LA, La Vecchia C, Shariat S, et al. Epidemiology and risk factors of urothelial bladder cancer. Eur Urol. 2013;63:234–241. doi: 10.1016/j.eururo.2012.07.033. [DOI] [PubMed] [Google Scholar]
- 3.Stenzl A, Cowan NC, De Santis M, Kuczyk MA, Merseburger AS, Ribal MJ, Sherif A, Witjes JA. European Association of Urology (EAU): Treatment of muscle-invasive and metastatic bladder cancer: Update of the EAU guidelines. Eur Urol. 2011;59:1009–1018. doi: 10.1016/j.eururo.2011.03.023. [DOI] [PubMed] [Google Scholar]
- 4.Ye F, Wang L, Castillo-Martin M, McBride R, Galsky MD, Zhu J, Boffetta P, Zhang DY, Cordon-Cardo C. Biomarkers for bladder cancer management: Present and future. Am J Clin Exp Urol. 2014;2:1–14. [PMC free article] [PubMed] [Google Scholar]
- 5.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 6.Chang SJ, Weng SL, Hsieh JY, Wang TY, Chang MD, Wang HW. MicroRNA-34a modulates genes involved in cellular motility and oxidative phosphorylation in neural precursors derived from human umbilical cord mesenchymal stem cells. BMC Med Genomics. 2011;4:65. doi: 10.1186/1755-8794-4-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cui X, Kong C, Zhu Y, Zeng Y, Zhang Z, Liu X, Zhan B, Piao C, Jiang Z. miR-130b, an onco-miRNA in bladder cancer, is directly regulated by NF-κB and sustains NF-κB activation by decreasing Cylindromatosis expression. Oncotarget. 2016;7:48547–48561. doi: 10.18632/oncotarget.10423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Song T, Zhang X, Yang G, Song Y, Cai W. Decrement of miR-199a-5p contributes to the tumorigenesis of bladder urothelial carcinoma by regulating MLK3/NF-κB pathway. Am J Transl Res. 2015;7:2786–2794. [PMC free article] [PubMed] [Google Scholar]
- 9.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 10.Lee JY, Ryu DS, Kim WJ, Kim SJ. Aberrantly expressed microRNAs in the context of bladder tumorigenesis. Investig Clin Urol. 2016;57(Suppl 1):S52–S59. doi: 10.4111/icu.2016.57.S1.S52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang Y, Zhang Z, Li Z, Gong D, Zhan B, Man X, Kong C. MicroRNA-497 inhibits the proliferation, migration and invasion of human bladder transitional cell carcinoma cells by targeting E2F3. Oncol Rep. 2016;36:1293–1300. doi: 10.3892/or.2016.4923. [DOI] [PubMed] [Google Scholar]
- 12.Shin SS, Park SS, Hwang B, Kim WT, Choi YH, Kim WJ, Moon SK. MicroRNA-106a suppresses proliferation, migration, and invasion of bladder cancer cells by modulating MAPK signaling, cell cycle regulators, and Ets-1-mediated MMP-2 expression. Oncol Rep. 2016;36:2421–2429. doi: 10.3892/or.2016.5015. [DOI] [PubMed] [Google Scholar]
- 13.Zhu C, Shao P, Bao M, Li P, Zhou H, Cai H, Cao Q, Tao L, Meng X, Ju X, et al. miR-154 inhibits prostate cancer cell proliferation by targeting CCND2. Urol Oncol. 2014;32:31.e9–31.e16. doi: 10.1016/j.urolonc.2012.11.013. [DOI] [PubMed] [Google Scholar]
- 14.Xin C, Zhang H, Liu Z. miR-154 suppresses colorectal cancer cell growth and motility by targeting TLR2. Mol Cell Biochem. 2014;387:271–277. doi: 10.1007/s11010-013-1892-3. [DOI] [PubMed] [Google Scholar]
- 15.Lin X, Yang Z, Zhang P, Liu Y, Shao G. miR-154 inhibits migration and invasion of human non-small cell lung cancer by targeting ZEB2. Oncol Lett. 2016;12:301–306. doi: 10.3892/ol.2016.4577. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 16.Wang L, Wu L, Wu J. Downregulation of miR-154 in human glioma and its clinicopathological and prognostic significance. J Int Med Res. 2016;44:994–1001. doi: 10.1177/0300060516649487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kai Y, Qiang C, Xinxin P, Miaomiao Z, Kuailu L. Decreased miR-154 expression and its clinical significance in human colorectal cancer. World J Surg Oncol. 2015;13:195. doi: 10.1186/s12957-015-0607-5. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 18.Baranwal S, Alahari SK. miRNA control of tumor cell invasion and metastasis. Int J Cancer. 2010;126:1283–1290. doi: 10.1002/ijc.25014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xiong F, Liu K, Zhang F, Sha K, Wang X, Guo X, Huang N. MiR-204 inhibits the proliferation and invasion of renal cell carcinoma by inhibiting RAB22A expression. Oncol Rep. 2016;35:3000–3008. doi: 10.3892/or.2016.4624. [DOI] [PubMed] [Google Scholar]
- 20.Zhou J, Dai W, Song J. miR-1182 inhibits growth and mediates the chemosensitivity of bladder cancer by targeting hTERT. Biochem Biophys Res Commun. 2016;470:445–452. doi: 10.1016/j.bbrc.2016.01.014. [DOI] [PubMed] [Google Scholar]
- 21.Xu H, Fei D, Zong S, Fan Z. MicroRNA-154 inhibits growth and invasion of breast cancer cells through targeting E2F5. Am J Transl Res. 2016;8:2620–2630. [PMC free article] [PubMed] [Google Scholar]
- 22.Pang X, Huang K, Zhang Q, Zhang Y, Niu J. miR-154 targeting ZEB2 in hepatocellular carcinoma functions as a potential tumor suppressor. Oncol Rep. 2015;34:3272–3279. doi: 10.3892/or.2015.4321. [DOI] [PubMed] [Google Scholar]
- 23.Zhu C, Li J, Cheng G, Zhou H, Tao L, Cai H, Li P, Cao Q, Ju X, Meng X, et al. miR-154 inhibits EMT by targeting HMGA2 in prostate cancer cells. Mol Cell Biochem. 2013;379:69–75. doi: 10.1007/s11010-013-1628-4. [DOI] [PubMed] [Google Scholar]
- 24.Shih IeM, Sheu JJ, Santillan A, Nakayama K, Yen MJ, Bristow RE, Vang R, Parmigiani G, Kurman RJ, Trope CG, et al. Amplification of a chromatin remodeling gene, Rsf-1/HBXAP, in ovarian carcinoma; Proc Natl Acad Sci USA; 2005; pp. 14004–14009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cosma MP, Tanaka T, Nasmyth K. Ordered recruitment of transcription and chromatin remodeling factors to a cell cycle- and developmentally regulated promoter. Cell. 1999;97:299–311. doi: 10.1016/S0092-8674(00)80740-0. [DOI] [PubMed] [Google Scholar]
- 26.Shamay M, Barak O, Doitsh G, Ben-Dor I, Shaul Y. Hepatitis B virus pX interacts with HBXAP, a PHD finger protein to coactivate transcription. J Biol Chem. 2002;277:9982–9988. doi: 10.1074/jbc.M111354200. [DOI] [PubMed] [Google Scholar]
- 27.Maeda D, Chen X, Guan B, Nakagawa S, Yano T, Taketani Y, Fukayama M, Wang TL, Shih IeM. Rsf-1 (HBXAP) expression is associated with advanced stage and lymph node metastasis in ovarian clear cell carcinoma. Int J Gynecol Pathol. 2011;30:30–35. doi: 10.1097/PGP.0b013e3181e9a319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sheu JJ, Choi JH, Guan B, Tsai FJ, Hua CH, Lai MT, Wang TL, Shih IeM. Rsf-1, a chromatin remodelling protein, interacts with cyclin E1 and promotes tumour development. J Pathol. 2013;229:559–568. doi: 10.1002/path.4147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ren J, Chen QC, Jin F, Wu HZ, He M, Zhao L, Yu ZJ, Yao WF, Mi XY, Wang EH, et al. Overexpression of Rsf-1 correlates with pathological type, p53 status and survival in primary breast cancer. Int J Clin Exp Pathol. 2014;7:5595–5608. [PMC free article] [PubMed] [Google Scholar]
- 30.Liang PI, Wu LC, Sheu JJ, Wu TF, Shen KH, Wang YH, Wu WR, Shiue YL, Huang HY, Hsu HP, et al. Rsf-1/HBXAP overexpression is independent of gene amplification and is associated with poor outcome in patients with urinary bladder urothelial carcinoma. J Clin Pathol. 2012;65:802–807. doi: 10.1136/jclinpath-2012-200897. [DOI] [PubMed] [Google Scholar]
- 31.Liu Y, Li G, Liu C, Tang Y, Zhang S. RSF1 regulates the proliferation and paclitaxel resistance via modulating NF-κB signaling pathway in nasopharyngeal carcinoma. J Cancer. 2017;8:354–362. doi: 10.7150/jca.16720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ito Y. RUNX genes in development and cancer: Regulation of viral gene expression and the discovery of RUNX family genes. Adv Cancer Res. 2008;99:33–76. doi: 10.1016/S0065-230X(07)99002-8. [DOI] [PubMed] [Google Scholar]
- 33.Akech J, Wixted JJ, Bedard K, van der Deen M, Hussain S, Guise TA, van Wijnen AJ, Stein JL, Languino LR, Altieri DC, et al. Runx2 association with progression of prostate cancer in patients: Mechanisms mediating bone osteolysis and osteoblastic metastatic lesions. Oncogene. 2010;29:811–821. doi: 10.1038/onc.2009.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pratap J, Wixted JJ, Gaur T, Zaidi SK, Dobson J, Gokul KD, Hussain S, van Wijnen AJ, Stein JL, Stein GS, et al. Runx2 transcriptional activation of Indian Hedgehog and a downstream bone metastatic pathway in breast cancer cells. Cancer Res. 2008;68:7795–7802. doi: 10.1158/0008-5472.CAN-08-1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Abdelzaher E, Kotb AF. High Coexpression of Runt-related transcription factor 2 (RUNX2) and p53 independently predicts early tumor recurrence in bladder urothelial carcinoma patients. Appl Immunohistochem Mol Morphol. 2016;24:345–354. doi: 10.1097/PAI.0000000000000193. [DOI] [PubMed] [Google Scholar]
- 36.Lim M, Zhong C, Yang S, Bell AM, Cohen MB, Roy-Burman P. Runx2 regulates survivin expression in prostate cancer cells. Lab Invest. 2010;90:222–233. doi: 10.1038/labinvest.2009.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ozaki T, Wu D, Sugimoto H, Nagase H, Nakagawara A. Runt-related transcription factor 2 (RUNX2) inhibits p53-dependent apoptosis through the collaboration with HDAC6 in response to DNA damage. Cell Death Dis. 2013;4:e610. doi: 10.1038/cddis.2013.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen Y, Gao DY, Huang L. In vivo delivery of miRNAs for cancer therapy: Challenges and strategies. Adv Drug Deliv Rev. 2015;81:128–141. doi: 10.1016/j.addr.2014.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tang Z, Qiu H, Luo L, Liu N, Zhong J, Kang K, Gou D. miR-34b modulates skeletal muscle cell proliferation and differentiation. J Cell Biochem. 2017 Apr 19; doi: 10.1002/jcb.26079. (Epub ahead of print). doi: 10.1002/jcb.26079. [DOI] [PubMed] [Google Scholar]