Abstract
Prostate cancer (PCa) cells expressing full-length androgen receptor (AR-FL) are susceptible to androgen deprivation therapy (ADT). However, outgrowth of castration-resistant prostate cancer (CRPC) can occur due to the expression of constitutively active (ligand-independent) AR splice variants, particularly AR-V7. We previously demonstrated that sulforaphane (SFN), an isothiocyanate phytochemical, can decrease AR-FL levels in the PCa cell lines, LNCaP and C4-2B. Here, we examined the efficacy of SFN in targeting both AR-FL and AR-V7 in the CRPC cell line, CWR22Rv1 (22Rv1). MTT cell viability, wound-heal assay, and colony forming unit (CFU) measurements revealed that 22Rv1 cells are resistant to the anti-androgen, enzalutamide (ENZ). However, co-exposure to SFN sensitized these cells to the potent anticancer effects of ENZ (P<0.05). Immunoblot analyses showed that SFN (5–20 µM) rapidly decreases both AR-FL and AR-V7 levels, and immunofluorescence microscopy (IFM) depicted decreased AR in both cytoplasm and nucleus with SFN treatment. SFN increased both ubiquitination and proteasomal activity in 22Rv1 cells. Studies using a protein synthesis inhibitor (cycloheximide) or a proteasomal inhibitor (MG132) indicated that SFN increases both ubiquitin-mediated aggregation and subsequent proteasomal-degradation of AR proteins. Previous studies reported that SFN inhibits the chaperone activity of heat-shock protein 90 (Hsp90) and induces the nuclear factor erythroid-2-like 2 (Nrf2) transcription factor. Therefore, we investigated whether the Hsp90 inhibitor, ganetespib (G) or the Nrf2 activator, bardoxolone methyl (BM) can similarly suppress AR levels in 22Rv1 cells. Low doses of G and BM, alone or in combination, decreased both AR-FL and AR-V7 levels, and combined exposure to G+BM sensitized 22Rv1 cells to ENZ. Therefore, adjunct treatment with the phytochemical SFN or a safe pharmaceutical combination of G+BM may be effective against CRPC cells, especially those expressing AR-V7.
Keywords: prostate cancer, castration-resistant prostate cancer, androgen receptor, AR-V7, sulforaphane, enzalutamide, ganetespib, bardoxolone-methyl, combination therapy, sensitization
Introduction
Prostate cancer (PCa) is the second leading cause of cancer-related deaths in men in the United States (1). Despite the initial promise of androgen deprivation therapy (ADT), relapse of aggressive and therapeutic resistant tumors is a principal cause of death among patients (1). This ADT resistant form of PCa is referred to as hormone refractory or castration-resistant prostate cancer (CRPC) (2), which is associated with continuous androgen receptor (AR) signaling even in the absence of androgen (3–7). Various mechanisms underlying the constitutive AR signaling may include AR gene amplification, ligand-independent AR activation by cytokines or kinases, both intracrine and/or intratumoral androgen production, overexpression of AR co-activators, and most importantly, the expression of constitutively active AR splice variants (AR-Vs) (8,9). These truncated forms of AR lack the C-terminal ligand binding domain (LBD) but retain the transactivating N-terminal domain (NTD), thus promoting transcriptional activation of AR target genes despite castrated hormone levels (8–10).
AR-V7 (also known as AR3) is a major splice variant of full-length AR (AR-FL) that encodes a functional protein (11–19). Increased AR-V7 levels are detected in tumor specimens (11) and in circulating tumor cells (18) from CRPC patients. Survey of primary tumor tissues before and after castration resistance clearly showed increased AR-V7 expression following the outgrowth of CRPC tumors (11–17). Furthermore, resistance to the potent second-generation anti-androgens, e.g. enzalutamide (ENZ) and abiraterone acetate (ABI) has been attributed to overexpression of AR-V7 (20,21). Studies also demonstrated a crucial role of AR-FL in regulating dimerization and transactivation function of AR-V7 (22), which is implicated in castration-resistant cell growth (23,24). Therefore, there is an urgent need to develop therapeutic strategies that effectively suppress the constitutive tumor promoting signals associated with AR-FL and AR-V7 action in CRPC.
Sulforaphane (SFN), an isothiocyanate phytochemical found in cruciferous vegetables (e.g. broccoli), is a promising anticancer agent with multiple cellular targets (25–27). Several studies have also implicated SFN as a promising agent for metastatic CRPC, especially since it shows specific toxicity towards transformed cells without significant adverse effect on primary prostate epithelial cells (28–31). At pharmacological doses, SFN has been shown to slow down the progression of PCa (32–34). A recent study has also documented the ability of SFN to target the cancer stem cell (CSC) phenotype (35,36). Mechanistic studies have reported SFN-induced cell death to be initiated by reactive oxygen species (ROS) (37,38) and the release of hydrogen sulfide (39). Therefore, SFN may partly display its effect via epigenetic modifications of Nrf2 gene leading to the activation of downstream anti-oxidative/detoxification stress pathway and also by suppression of Akt survival pathway (40–42). SFN has also been shown to inhibit AR-FL levels by destabilization of protein, primarily by inactivating HDAC6 and the subsequent suppression of the chaperone function of heat-shock proteins (Hsp90) (43–45). However, efficacy of SFN against CRPC cells that express both AR-FL and AR-V7 has not been tested.
Our previous study using both androgen-dependent (LNCaP) and androgen-independent (C4-2B) cell lines showed that SFN can potentiate the efficacy of ENZ by rapidly decreasing AR-FL levels (46). In the current study, we show that SFN can suppress the levels of both AR-FL and AR-V7 proteins in the 22Rv1 cells. Mechanistic studies demonstrated the efficacy of SFN in increasing both ubiquitination and proteasomal activity in 22Rv1 cells. Since the multimodal actions of SFN are known to decrease Hsp90 (43–45) and increase Nrf2 (40,41), we tested whether combined exposure to an Hsp90 inhibitor (ganetespib) and an Nrf2 activator (bardoxolone-methyl) can be similarly effective. Our findings showed that co-exposure to physiologically achievable doses of the above clinically approved drugs decreases both AR-FL and AR-V7 levels and sensitizes 22Rv1 cells to the anticancer effects of ENZ.
Materials and methods
Cell culture
CWR22Rv1 (an androgen-independent PCa cell line that expresses both AR-FL and AR-V7) and LNCaP (an androgen-dependent PCa cell line that expresses only AR-FL) were purchased from American Type Culture Collection (ATCC; Rockville, MD, USA). The C4-2B cell line (an androgen-independent sub-line of LNCaP that expresses AR-FL and low levels of AR-Vs) was a kind gift from Dr. Leland Chung (47). The cell lines were maintained in RPMI-1640 media supplemented with 10% fetal bovine serum (FBS) (Atlanta Biologicals; Lawrenceville, GA, USA) and 1% penicillin/streptomycin (CellGro; Manassas, VA, USA) in a humidified incubator containing 5% CO2 at 37°C. To mimic steroid hormone deprived conditions, experiments were carried out in phenol-red free media supplemented with 10% charcoal-stripped FBS (CS-FBS) (Atlanta Biologicals).
Reagents
Sulforaphane (SFN) and MTT [3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide] were purchased from Sigma-Aldrich (St. Louis, MO, USA). Ganetespib (G) and enzalutamide (ENZ) were obtained from ApexBio (Houston, TX, USA). Cycloheximide (CHX) was purchased from Cayman chemicals (Ann Arbor, MI, USA). MG132 was obtained from EMD Millipore (Billerica, MA, USA). Bardoxolone methyl was purchased from Selleckchem (Houston, TX, USA). All drugs were dissolved in 100% DMSO and diluted in media immediately before use. The final DMSO concentration used in experiments was less than 0.1%. The primary antibodies including rabbit polyclonal anti-AR (N-20) (sc-816) and mouse monoclonal anti-GAPDH (sc-47724) were purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA). A mouse monoclonal antibody against ubiquitinated proteins (FK2) (BML-PW8810) was obtained from Enzo Life Sciences (Farmingdale, NY, USA). The horseradish peroxidase (HRP)-conjugated goat anti-rabbit (A0545) and goat anti-mouse (A9044) secondary antibodies were purchased from Sigma-Aldrich. The goat anti-rabbit secondary antibody tagged with Texas red (T-2767) was purchased from Thermo Fisher Scientific (Rockford, IL, USA).
MTT assay
MTT assays were performed to determine cell viability post exposure to the drug(s). In brief, ~5,000 cells were seeded in 96-well culture plates and allowed to adhere overnight. Cells were then synchronized by overnight incubation in serum-free medium, and treated with desired concentrations of drug(s), alone or in different combinations for 24–72 h. Cell viability was determined by adding MTT solution (5 mg/ml) and incubating for 3 h at 37°C. The formazan crystals were then solubilized in DMSO and optical density (O.D.) was measured at 540 nm by using a µQuant spectrophotometric plate reader from Bio-Tek (Seattle, WA, USA). In each individual experiment, changes in cell survival following drug treatments are expressed as percent of untreated control.
Western immunoblot
Whole cell lysates were harvested at different time points post-treatment(s) using RIPA lysis buffer from Santa Cruz Biotechnology and total protein content was quantified using the bicinchoninic acid (BCA) protein assay reagent from Thermo Fisher Scientific. Briefly, 10 µg of protein lysate was electrophoresed in 10% SDS-PAGE gels followed by electrotransfer onto nitrocellulose membrane. After blocking nonspecific binding using 5% casein in TBS-T buffer (tris buffer saline with 0.1% Tween-20), membranes were incubated overnight at 4°C with the primary antibodies against AR (1:500 dilution), GAPDH (1:3,000 dilution) or ubiquitin (1:1,000 dilution). This was followed by incubation with the corresponding HRP-conjugated secondary antibodies (1:2,000 dilution) for 1 h. Membranes were developed using the SuperSignal West Femto Substrate (Thermo Fisher Scientific). Immunoblots were scanned using the ImageQuant LAS 500 scanner (GE Healthcare; Princeton, NJ, USA) and band intensities were quantified using the ImageJ software from NIH (Bethesda, MD, USA). Densitometric value for AR proteins (AR-FL and AR-V7) was normalized to GAPDH levels.
Isolation of Triton-soluble and -insoluble fractions
Cells were lysed in RIPA lysis buffer containing 1% Triton X-100 on ice and centrifuged at 16,000 × g for 15 min at 4°C. The supernatant was collected as Triton-soluble (TS) fraction and the pellets (Triton-insoluble fraction) (TI) were further solubilized in SDS buffer (2% SDS in 50 mM Tris-HCl) followed by boiling for 15 min (48). Approximately 10 µg of protein from both TS and TI fractions was electrophoresed in 10% SDS-PAGE gels and immunoblots were analyzed for AR protein levels.
Proteasomal activity assay
A Proteasome Assay kit (Cayman chemicals; cat # 10008041) was used to measure proteasomal activity of control (untreated) and SFN treated cells. The proteasome inhibitor, MG132 was used as a positive control. Briefly, cells were seeded in 96-well microtiter plates (1×105 cells per well) and allowed to adhere overnight. After appropriate drug treatments, plates were centrifuged at 500 × g for 5 min and culture media was aspirated. Assay buffer (200 µl) was added to each well followed by centrifugation at 500 × g for 5 min and the supernatant was aspirated. The lysis buffer (100 µl) was then added to each well followed by gentle shaking for 30 min. Plates were centrifuged at 1,000 × g for 10 min and 90 µl of the supernatant from each well was transferred to corresponding wells in a black (opaque) 96-well plate (Sigma-Aldrich). For sample activity measurements, assay buffer (10 µl) was added to these wells followed by the addition of proteasome substrate, SUC-LLVY-AMC (10 µl). Fluorescence intensity was measured using an FLx-800 fluorimeter (Bio-Tek) with the absorption (excitation) set at 360 nm and the emission set at 480 nm. Mean fluorescence intensities (MFI) were normalized to protein content in each sample.
Wound-healing assay
Wound-healing assays were carried out to measure the effect of drugs on the migratory phenotype of PCa cells, as previously described (49). Briefly, cells were seeded in 6-well plates (1×106 cells per well) and grown until they formed a confluent monolayer. The monolayers were scratched using a 200 µl pipette tip, wells were washed with PBS and images of the wound (0-time point) were captured using a Leica Microsystems microscope (Buffalo Grove, IL, USA). Growth media (CS-FBS) was returned to each culture and treatments were initiated. Change in wound width was captured after 48 h and cell migration (wound closure) was calculated by measuring the distance between 4–5 random points within the wound edges.
Colony forming units assay
Cells (500 cells/dish) were seeded in 60-mm petri dishes in 3 replicates and grown in medium supplemented with 2% FBS. The drugs, alone or in combination, were added after 48 h and replenished in the second week. After two weeks in culture, colonies were fixed with 100% ethanol and stained with 0.2% crystal violet in 20% methanol. The colony forming units (CFU) were enumerated using ImageJ software (NIH). Change in total CFUs were compared in both control (untreated) and drug-exposed cultures.
Immunofluorescence microscopy
Subcellular localization of AR post-treatment with SFN was visualized by immunofluorescence microscopy (IFM). Briefly, cells (3×104) were seeded in chamber slides (EMD Millipore) and allowed to adhere overnight. After treatment, cells were fixed in ice cold methanol followed by permeabilization with 0.1% Triton X-100 for 1 h. After blocking in 10% goat serum, slides were incubated overnight at 4°C with the primary antibody (1:300 dilution) followed by incubation with the corresponding Texas Red tagged secondary antibody (1:1,000 dilution) for 1 h. The Vectashield (Burlingame, CA) mounting medium containing the nuclear stain diamino-2-phenylindole (DAPI) was then added to the slides and cover slips were mounted. Images were captured using a fluorescent microscope from Leica Microsystems Inc.
Statistical analysis
Statistical analysis was carried out using GraphPad Prism (version 6) Software (San Diego, CA, USA). Results were expressed as the standard error of mean (± SEM). Significant changes from controls were determined by a two-tailed Student's t-test and P-values of <0.05 were considered significant. For synergy determination, the CompuSyn software (ComboSyn, Inc., Paramus, NJ, USA) was used and combination index (CI) was calculated based on the Chou-Talalay method, which quantitatively determines additive (CI=1), synergistic (CI <1) or antagonistic (CI >1) effects (50).
Results
The AR-V7 expressing 22Rv1 cells are resistant to androgen deprivation conditions and enzalutamide
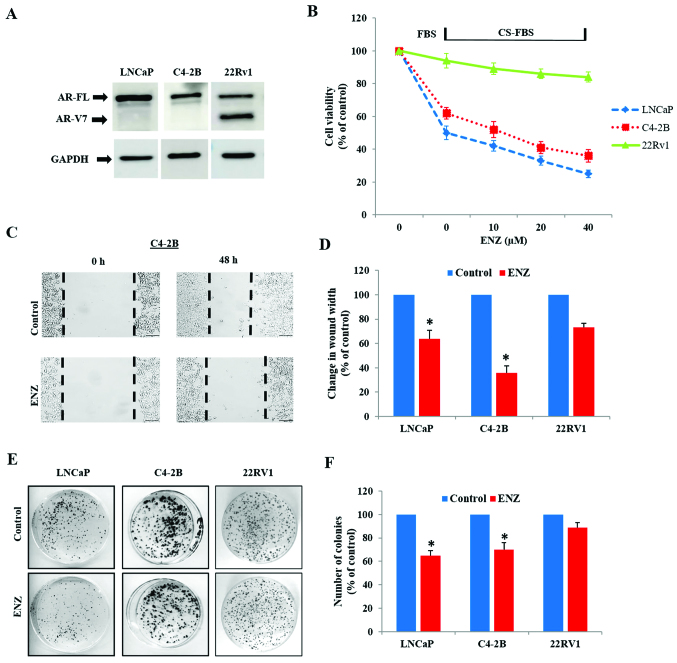
Immunoblot analysis showed that while the LNCaP and C4-2B cells express only AR-FL protein (110 kDa), 22Rv1 cells express AR-V7 (75 kDa) in addition to AR-FL (Fig. 1A). In order to confirm the inherent resistance of 22Rv1 to androgen deprivation and anti-androgen treatment, as compared to LNCaP and C4-2B, cells were cultured under normal (FBS media) and androgen-depleted conditions (CS-FBS media) in the absence or presence of enzalutamide (ENZ). Differences in cell viability, migratory behavior and clonogenic ability were compared in these three cell lines. In contrast to LNCaP and C4-2B cells, androgen depletion (CS-FBS) had no impact on the viability of 22Rv1 cells (Fig. 1B). Furthermore, although both LNCaP and C4-2B cells were susceptible to ENZ (10–40 µM), only a slight suppression in 22Rv1 cell viability was seen following 72 h exposure to ENZ. Wound-healing assays clearly showed that ENZ decreased the wound-closure ability (migration potential) in both LNCaP and C4-2B cells (Fig. 1C and D), but did not affect the migratory behavior of 22Rv1 cells (Fig. 2C and D). In addition, colony-forming unit (CFU) assays showed that long-term exposure to ENZ (0.4 µM) showed less suppressive effect on the clonogenic ability of 22Rv1 cells, compared to LNCaP and C4-2B cells (Fig. 1E and F).
Figure 1.
Anticancer effects of hormone-deprivation and enzalutamide in LNCaP, C4-2B and 22Rv1 cells. (A) A representative immunoblot showing the expression of only AR-FL in LNCaP and C4-2B cells, and both AR-FL and AR-V7 in 22Rv1 cells. (B) Cell viability was measured by MTT-assays. The LNCaP, C4-2B and 22Rv1 cells were exposed to either normal media (FBS) or steroid-hormone depleted media (CS-FBS) and increasing doses of ENZ (10–40 µM) for 72 h. Percentage change in cell viability is shown (n=3). (C and D) Quantification of cell migration was examined by wound-healing assays. (C) Representative images of scratch wound at the 0 and 48 h time points are shown in both untreated (control) and ENZ (20 µM) treated C4-2B cells. (D) Percentage changes in wound-width at 48 h are shown in LNCaP, C4-2B and 22Rv1 cells (n=2). (E and F) Quantification of clonogenic ability by colony forming unit (CFU) assays. (E) Representative images of CFUs in LNCaP, C4-2B and 22Rv1 cells in untreated (control) and after 14 day treatment with ENZ (0.4 µM). (F) Percentage changes in CFUs are shown. In all graphs, error bars represent standard error of mean (± SEM) and significant differences are shown as P-values (*P<0.05).
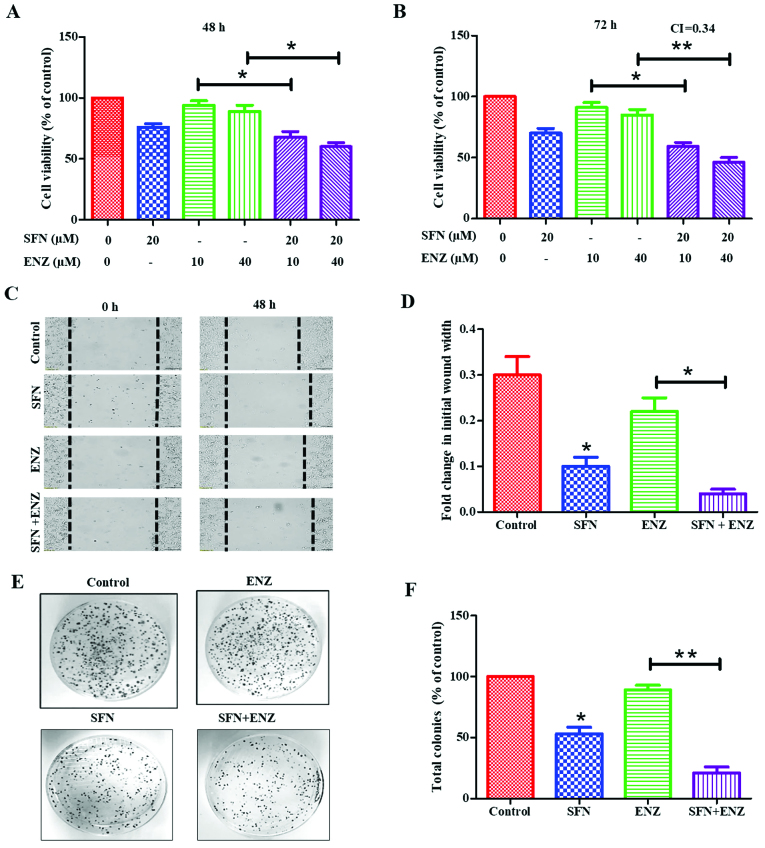
Figure 2.
Effect of SFN co-exposure on ENZ resistance of 22Rv1 cells. Cytotoxic effects of SFN (20 µM) or ENZ (10 or 40 µM), alone and in combination, are shown in 22Rv1 cells following (A) 48 h and (B) 72 h post-exposure (n=3) (C) Representative images of the effect of SFN and/or ENZ on migration of 22Rv1 cell are shown at 0 and 48 h time points. (D) Effects of SFN and/or ENZ on change in wound-width at 48 h, as compared to control (untreated) cells are shown (n=2). (E) Representative images of CFUs generated by 22Rv1 cells. (F) Percentage changes in CFUs in untreated (control) and in cells treated with SFN (0.2 µM) and/or ENZ (0.4 µM) are shown (n=2). In all graphs, error bars represent ± SEM and significant differences are shown as P-values (*P<0.05; **P<0.01).
SFN increases the anticancer efficacy of ENZ in 22Rv1 cells
Exposure to SFN alone showed a dose- and time-dependent suppressive effect on 22Rv1 proliferation, with ~40% suppression following 72 h exposure to 30 µM of SFN (data not shown). However, co-exposure to lower doses of SFN was able to sensitize these cells to ENZ (Fig. 2A and B). When ENZ (10 or 40 µM) was used in combination with sub-IC50 dose of SFN (20 µM), a significant (P<0.05) increase in cytotoxicity was observed within 48 h, which was more evident at 72 h. Combination index (CI) calculations demonstrated that this drug combination functions in a synergistic manner at 72 h post-exposure (CI=0.34) (Fig. 2B). Furthermore, co-exposure to SFN increased the ability of ENZ to suppress migration/motility of 22Rv1 cells (Fig. 2C and D). Wound-healing assays were carried out in the absence or presence of SFN (10 µM), alone and in combination with ENZ (20 µM). In control cultures, the wound-width decreased by approximately 30% after 48 h in culture, and exposure to ENZ alone did not significantly decrease the wound closure. However, exposure to SFN alone decreased wound-closure by almost 70% and co-exposure to SFN and ENZ decreased it by as much as 90%. Our investigations also showed that long-term exposure to SFN can suppress the clonogenic ability of 22Rv1 cells, and further increase the efficacy of ENZ in suppressing CFUs (Fig. 2E and F). To optimize our combination studies, we first determined the effect of each drug alone [data not shown] and then used doses that caused <50% decrease in CFUs. Although ENZ (0.4 µM) alone did not significantly decrease CFUs, exposure to SFN (0.2 µM) caused ~50–55% decrease and cells exposed to SFN and ENZ combination showed almost a complete suppression of CFUs (P<0.01) (Fig. 2F). Thus, SFN potentiates the efficacy of ENZ in suppressing the proliferation, migration and clonogenic ability of 22Rv1 cells.
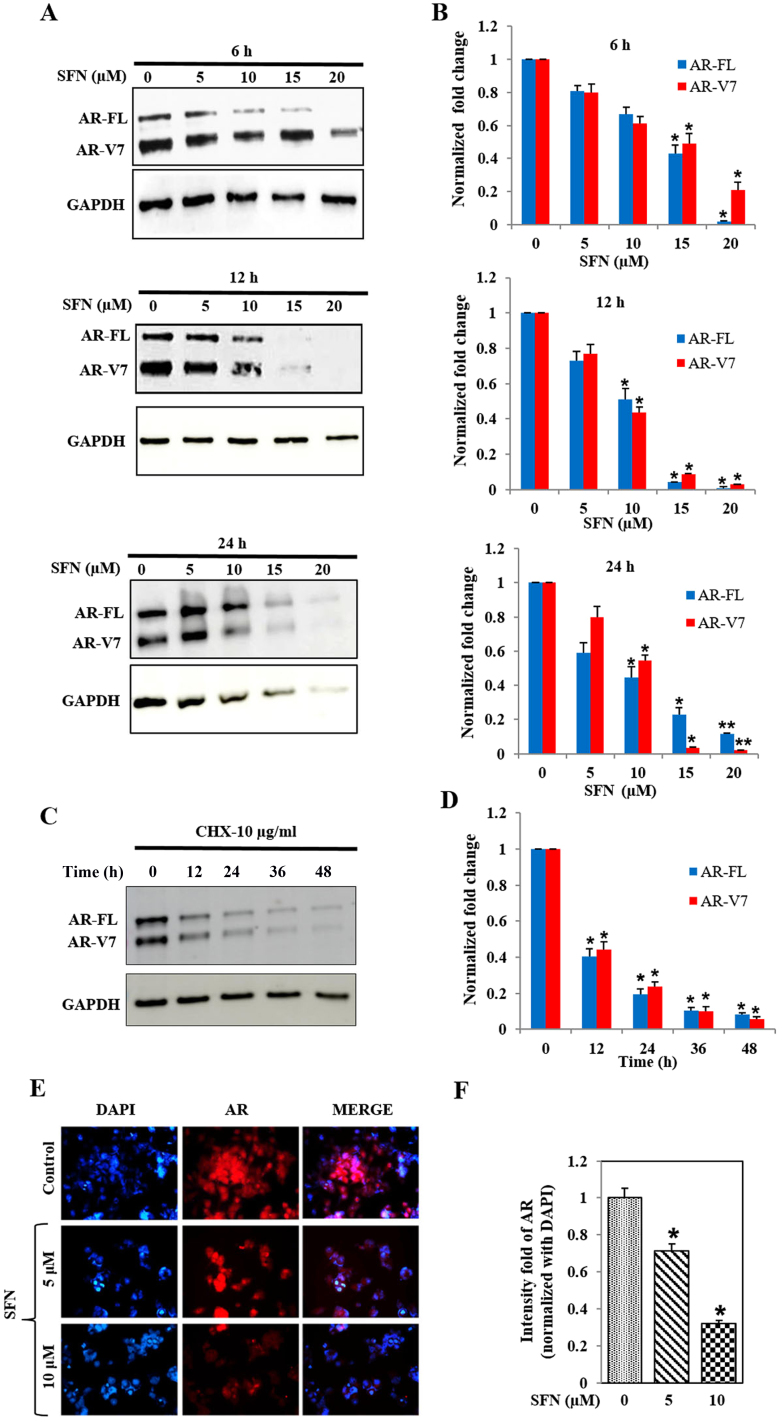
SFN decreases both AR-FL and AR-V7 levels in 22Rv1 cells
Immunoblot analysis showed that SFN treatment reduced protein levels of both AR-FL and AR-V7 in 22Rv1 cells, in a concentration- and time-dependent manner (Fig. 3A and B). High dose of SFN (20 µM) abrogated AR protein levels within 6 h, whereas 50% reduction was observed with 15 µM SFN at this time point. At 12 and 24 h, significant (P<0.05) suppression of both AR-FL and AR-V7 was evident even with the lower dose of SFN (10 µM). The reduction in AR-FL protein levels was more pronounced than the concomitant reduction in AR-V7 levels, suggesting that SFN may differentially affect the stability of these two AR proteins. Studies using the translation inhibitor, cycloheximide (10 µg/ml) clearly showed that the inherent stability of these two AR proteins is much greater in the 22Rv1 cells (Fig. 3C and D) and clearly indicated that SFN increases the rate of degradation of both AR-FL and AR-V7. Immunofluorescence microscopy (IFM) studies demonstrated that the treatment of 22Rv1 cells with SFN (5 and 10 µM) can significantly reduce total AR protein levels within 24 h (Fig. 3E and F). Comparative analysis demonstrated that SFN suppresses both cytoplasmic and constitutively expressed nuclear AR levels in 22Rv1 cells.
Figure 3.
Effect of SFN on AR protein levels in 22Rv1 cells. (A) Immunoblot analysis of AR-FL and AR-V7 levels in 22Rv1 cells exposed to SFN (0–20 µM) for 6–24 h. Representative immunoblots of AR-FL, AR-V7 and GAPDH levels are shown. (B) Bar graphs show dose- and time-dependent effects on AR-FL and AR-V7 levels. Data were normalized to GAPDH in corresponding samples and fold change from controls are presented (n=2). (C and D) Effect of CHX (10 µg/ml) on AR-FL and AR-V7 protein levels in 22Rv1 cells. (C) A representative immunoblot is shown. (D) Bar graphs depict the normal rate of AR degradation following inhibition of protein synthesis by CHX (n=2). (E and F) Immunofluorescence microscopy (IFM) of AR levels in 22Rv1 cells treated with SFN (5 and 10 µM) for 24 h. (E) A representative image of IFM is shown. Left panels depict DAPI stained nuclei (blue), middle panels show AR immunoreactivity, and merged images are in the right panels. (F) Quantitation of staining intensity, showing fold change in AR following normalization to DAPI (n=2). Data are expressed as mean ± SEM and significant differences are shown as P-values (*P<0.05; **P<0.01).
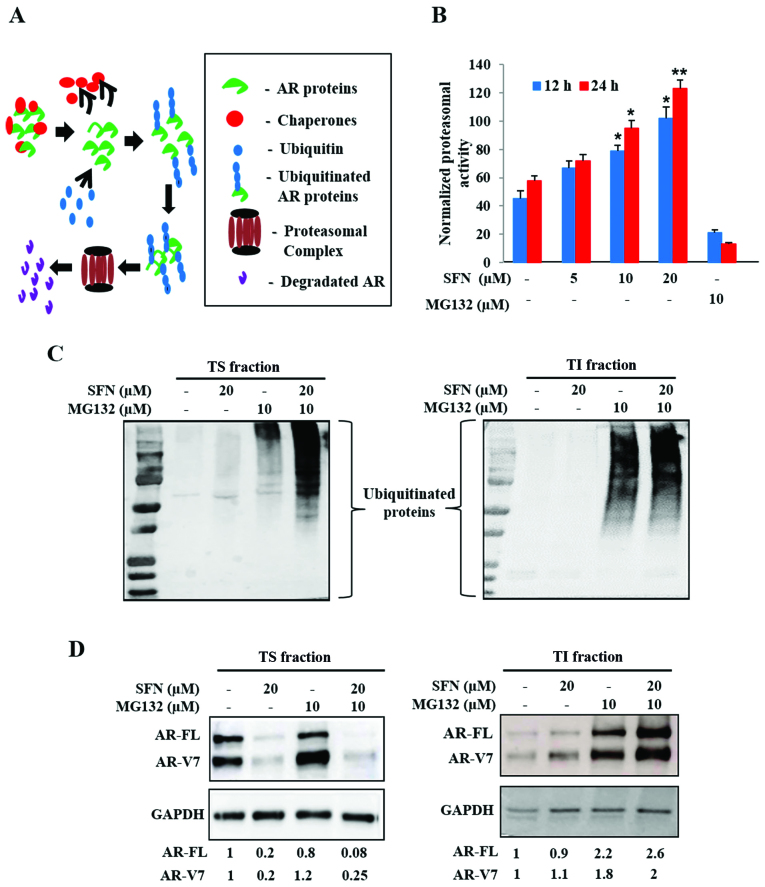
SFN increases proteasomal activity, protein ubiquitination and aggregation of both AR-FL and AR-V7
Intracellular proteins are continually degraded to their constituent amino acids via the ubiquitin-proteasome system (UPS) and are often protected from degradation via heat shock protein (Hsp) chaperones. Stability of AR proteins is similarly maintained via chaperone binding, ubiquitination and transit to the 26S proteasomal complex (Fig. 4A). Since our studies implicated that SFN may decrease the stability of both AR-FL and AR-V7, we tested if SFN treatment resulted in increased proteasomal degradation of AR. Exposure to SFN significantly increased proteasomal activity in 22Rv1 cells in both dose (5–20 µM) and time (12, 24 h) dependent manner, an effect validated by using MG132, a pharmacological proteasomal inhibitor (Fig. 4B). These findings suggested that SFN-mediated increase in proteasomal activity may be responsible for degradation of both AR-FL and AR-V7.
Figure 4.
Effect of SFN on ubiquitin-proteasome pathway mediated degradation of AR proteins. (A) Schematic of AR protein degradation via the ubiquitin-proteasome system (UPS). Legends are included in the box. (B) Effect of SFN (5–20 µM) on proteasomal activity in 22Rv1 cells following 12 and 24 h post-exposure. The proteasomal inhibitor, MG132 (10 µM) was used as a positive control. Exposure to SFN increases proteasomal activity of 22Rv1 cells (n=3). (C) Effect of SFN (20 µM) alone and in combination with MG132 (10 µM) on ubiquitinated protein levels. Representative immunoblots from 22Rv1 cell lysates, showing ubiquitinated proteins in both Triton soluble (TS, left) and triton insoluble (TI, right) fractions are shown. (D) Effect of SFN and/or MG132 on AR protein levels in TS (left) and TI (right) fractions. Different exposure times (TS, 20 sec and TI, 60 sec) enabled optimal detection of ubiquitinated proteins and AR bands. Fold changes in AR-FL and AR-V7, following normalization to GAPDH, are shown. Bar graph data are expressed as mean ± SEM and significant differences are shown as P-values (*P<0.05; **P<0.01).
To document AR-FL and AR-V7 levels in both the soluble cytosolic fraction and in ubiquitinated aggresomal fraction, we isolated both Triton-soluble (TS) and Triton-insoluble (TI) fractions, respectively. Immunoblot analyses of TS (20 sec exposure) and TI (60 sec exposure) indicated that proteasomal blockade via MG132 increases ubiquitinated protein levels in both TS and TI fractions (Fig. 4C). Although exposure to SFN alone showed negligible increase in ubiquitination, a striking enhancement of the ubiquitinated protein bands was evident upon co-exposure to MG132, in both TS fraction (Fig. 4C, left) and TI fraction (Fig. 4C, right). This indicated that SFN mediates ubiquitination and aggregation of cellular proteins.
An augmented UPS may be responsible for rapid suppression of both AR-FL and AR-V7 in SFN treated 22Rv1 cells. Therefore, immunoblot analyses of AR-FL and AR-V7 levels were carried out in both TS and TI fractions (Fig. 4D). Negligible quantities of AR proteins were present in insoluble aggregates (TI fraction) under basal conditions. Inhibition of proteasomal degradation by MG132 slightly increased AR levels in the TS fraction; however, it significantly augmented both AR-FL and AR-V7 levels in the TI aggregates. Although exposure to SFN (20 µM) reduced both AR-FL and AR-V7 levels in the TS fraction (Fig. 4D, left), some of these AR proteins were consistently found in the TI fraction (Fig. 4D, right). Co-exposure to MG132 considerably increased the amount of aggregated AR proteins in the TI fraction. These findings implicated that the multimodal actions of SFN increase ubiquitination, aggregation and proteasomal degradation to enable a rapid and profound decrease in both AR-FL and AR-V7 protein levels in 22Rv1 cells.
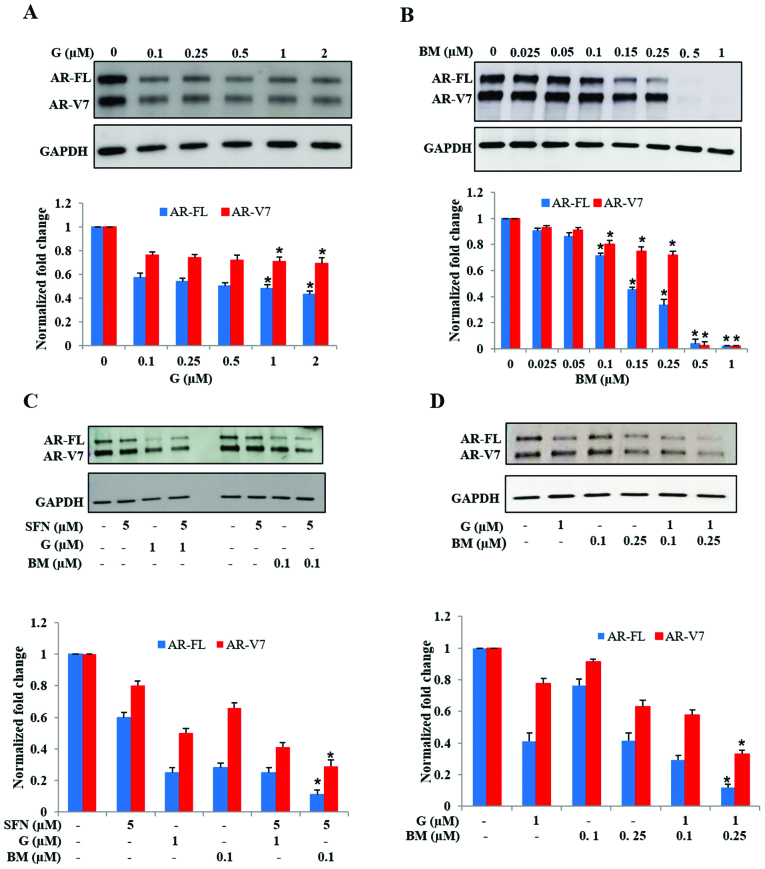
Inhibition of Hsp90 or activation of Nrf2 reduces AR protein levels in 22Rv1 cells
Several past studies have reported that SFN functions by inhibiting the chaperone activity of Hsp90 (43–45) and by decreasing oxidative stress via increased Nrf2 function (40,41). Therefore, we investigated whether the AR suppressive effects of SFN can be replicated by targeting the above two metabolic pathways. Treatment of 22Rv1 cells with the Hsp90 inhibitor, ganetespib (G) decreased both AR-FL and AR-V7 protein levels (Fig. 5A). Notably, exposure to G did not show a dose-dependent effect in suppression of AR-FL and AR-V7 levels. However, exposure to the Nrf2 activator, bardoxolone methyl (BM) showed a pronounced effect in abrogating both AR-FL and AR-V7 levels (Fig. 5B). Treatment with BM significantly reduced both AR-FL and AR-V7 protein levels in a dose-dependent manner. We observed a precipitous decrease in AR levels at the highest doses of BM used (0.5 and 1.0 µM) which totally eliminated both AR-FL and AR-V7 proteins within 24 h. These findings indicated that both Hsp90 inhibition and Nrf2 induction, two pathways targeted by SFN, are involved in suppressing AR protein levels in 22Rv1 cells, but may have different potency.
Figure 5.
Effect of Hsp90 inhibition or Nrf2-induction on AR-FL and AR-V7 protein levels in 22Rv1 cells. Cells were exposed to increasing concentrations of either (A) Ganetespib (G; 0–2000 nM) or (B) Bardoxolone methyl (BM; 0–1000 nM) and AR protein levels were measured. Top panels show representative immunoblots depicting AR-FL, AR-V7 and GAPDH levels. Bottom panels show fold change in AR proteins, normalized to GAPDH levels. (C) The effect of G or BM co-exposure with SFN on AR-FL and AR-V7 protein levels is shown. (D) The effect of combined treatment with G and BM on AR-FL and AR-V7 protein levels is shown. In both groups, top panels show representative immunoblots and bottom panels show normalized fold changes in AR proteins (n=2). Bar graphs show mean ± SEM and P-values (*P<0.05).
We investigated whether augmentation of the Hsp90 inhibitory activity or the Nrf2 inductive effect of SFN will enhance its efficacy, by using low dose SFN (5 µM) in presence of either G (1 µM) or BM (0.1 µM) (Fig. 5C). Immunoblot studies indicated that increasing the Hsp90 inhibitory function of SFN, via the addition of G, did not significantly enhance its potency in suppressing AR proteins. However, co-exposure to SFN and low-dose BM showed significantly increased suppression, as compared to SFN or BM alone. In addition, we observed that, even in the absence of SFN, combined treatment with low-dose G (1 µM) and BM (0.1 or 0.25 µM) was able to reduce AR protein levels, which was much more than with either drug alone (Fig. 5D). These findings indicated that the knowledge obtained from the multimodal actions of SFN can be utilized to use a pharmaceutical drug combination that suppresses both AR-FL and AR-V7 in 22Rv1 cells.
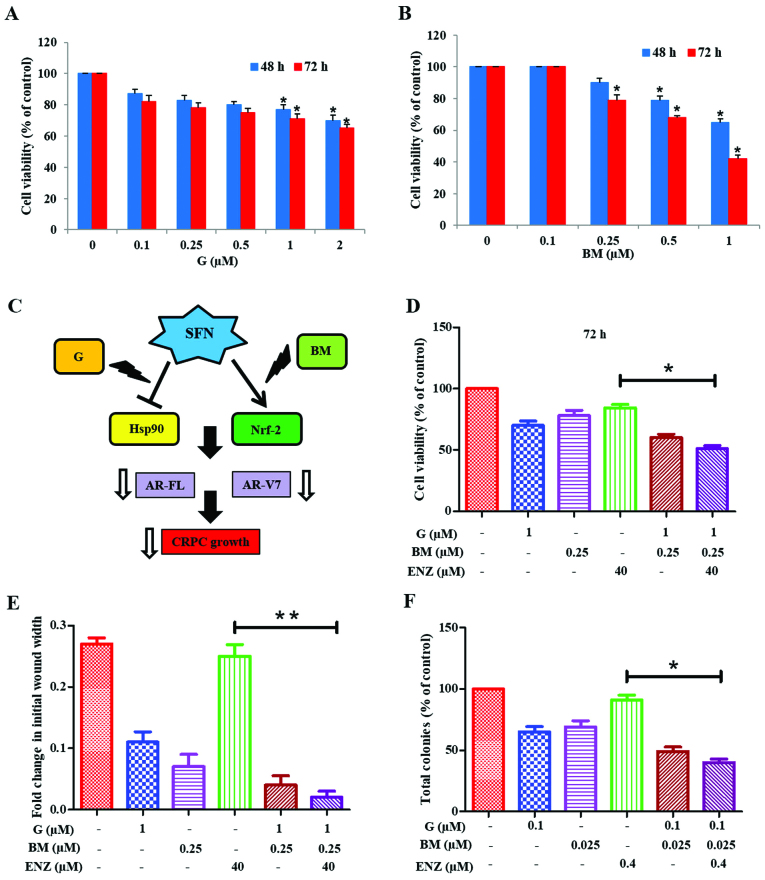
Co-targeting of Hsp90 and Nrf2 sensitizes 22Rv1 cells to the anticancer effects of ENZ
We investigated whether physiologically achievable concentrations of G and BM can be used to sensitize 22Rv1 cells to the anticancer effects of ENZ (Fig. 6). MTT cell viability assays were first carried out to document the cytotoxic effects of increasing doses of G (0.1–2.0 µM) or BM (0.1–1.0 µM) alone (Fig. 6A and B). Since the anticancer effects of SFN may involve targeting of both Hsp90 and Nrf2 pathways, we investigated whether co-exposure to G+BM would show potent anticancer and drug sensitizing effects, as well (Fig. 6C). Our investigations clearly showed that combined treatment with G (1 µM) and BM (0.25 µM) resulted in reduction of cell viability and sensitized 22Rv1 cells to ENZ (40 µM) treatment. This suppressive effect was evident at 48 h (not shown) and significantly (P<0.05) more profound at 72 h (Fig. 6D). Furthermore, combination studies using both wound-healing assays (Fig. 6E) and CFU assays (Fig. 6F) demonstrated the efficacy of G+BM combination in suppressing both the migratory behavior and clonogenic ability, respectively of 22Rv1 cells. Thus, our observations show that the knowledge of multimodal effects of SFN may be exploited to formulate a potent drug combination that works at nanomolar concentrations, in order to sensitize the AR-V7 expressing CRPC cells to ENZ.
Figure 6.
Combined efficacy of ganetespib and bardoxolone-methyl in overcoming ENZ-resistance of 22Rv1 cells. Effects of ganetespib (A) or bardoxolone-methyl (B) on 22Rv1 cell viability after 48 or 72 h post-exposure. (C) Multimodal actions of SFN, as both Hsp90 inhibitor and an Nrf2 inducer, implicate the effectiveness of combining ganetespib (G) and bardoxolone-methyl (BM). (D) Effect of G (1.0 µM) and BM (0.25 µM) alone and in combination with ENZ (40 µM) on 22Rv1 cell viability. (E) and (F) Effect of G+BM co-exposure on ENZ-mediated suppression of 22Rv1 cell migration and clonogenic ability, respectively. (E) Fold changes in wound-width at 48 h are shown in 22Rv1 cells and (F) the percentage change in CFUs is shown. In all graphs, error bars represent ± SEM and significant differences between groups are shown as P-values (*P<0.05; **P<0.01).
Discussion
The AR signaling axis is critical in both the development and progression of PCa to CRPC (1). Nuclear translocation of AR and induction of androgen response elements (ARE) regulated genes can augment tumor proliferation, invasion and therapeutic resistance (2–4). Therefore, strategies to suppress AR function have been a standard of care in PCa patients (5). Efficacy of this approach is corroborated by the survival benefits of newer and more potent AR-targeted agents, such as enzalutamide (ENZ) and abiraterone acetate (ABI) (51,52). However, despite the initial favorable response, the CRPC tumors develop typically within 1–2 years in nearly all men (1). Numerous clinical evidence suggests that AR variants may be the functional drivers of PCa progression to CRPC (13–21). The most clinically significant AR variant is reported to be AR-V7 (~80 kDa) (13–17). Hu et al showed an average of 20-fold higher expression of AR variant mRNA in CRPC tumors than in hormone-naïve PCa samples. These investigators also showed that AR-V7 mRNA, but not AR-V1 mRNA, was highly predictive of biochemical recurrence and CRPC progression (15).
There is an urgent need to suppress both AR-FL and AR-V7 for better therapeutic efficacy in patients with CRPC. Although recent studies using the anti-helminthic drug, niclosamide have documented its potent ability to suppress AR-V7 levels; no significant suppression of AR-FL levels was documented (53,54). Sarwar et al, used the phosphatidylinositol-4-phosphate 5-kinase α (PIP5Kα) inhibitor, ISA-2011B to similarly disrupt stabilization of AR-V7 protein, which circumvented resistance to the anti-androgen enzalutamide (21). However, although ISA-2011B was very potent and targeted both AR-FL and AR-V7 levels at nanomolar doses, this experimental drug is not currently in any clinical trials and translation to CRPC patients will involve significant time. Our in vitro studies using 22Rv1 cells showed that the phytochemical SFN, or the pharmaceutical agents G and BM, that are both in several clinical trials (55–61), may provide a more promising approach and can be rapidly implemented as an adjunct agent in PCa patients, especially those with therapeutic resistance.
The 22Rv1 cell line is a CRPC line that expresses AR-FL and multiple AR splice variants, out of which AR-V7 is the most abundant (62). Similar to multiple previous studies (13–17,20,21), our investigations, comparing the anticancer efficacy of hormone-deprivation and ENZ confirmed that 22Rv1 cells are more resistant than C4-2B and LNCaP cells. Overexpression of AR-V7 was found to be sufficient in driving the growth of LNCaP cells even under hormone-deprived conditions (16). In addition, studies demonstrated that the knockdown of AR-V7 inhibits growth of 22Rv1 cells under castrate conditions both in vitro and in vivo (16,63). We observed that SFN can rapidly decrease AR-FL and AR-V7 protein levels in 22Rv1 cells. The precipitous decrease in AR proteins may enable the potent reduction in cell proliferation, migration and clonogenic ability, observed with SFN in 22Rv1 cells.
Intracellular proteins are continually degraded to their constituent amino acids via the UPS pathway and cancer cells can protect important proteins from degradation via several Hsp chaperones (64). The stability of AR in CRPC cells is also maintained via chaperone binding, ubiquitination and transit to the 26S proteasomal complex (65). Thus, rapid degradation of AR-FL and AR-V7 via SFN is most likely associated with the increased proteasomal activity and ubiquitination rates observed following exposure to SFN. This is in line with earlier reports showing SFN-mediated enhancement of proteasomal activity (66). Of note, this was shown to be via the upregulation of Hsp27, another chaperone protein known to support cancer cell survival under stressful conditions by regulating both ubiquitination and proteasomal activity. Our studies using both Triton soluble and insoluble fractions (TS and TI) clearly showed that the SFN-mediated reduction in AR protein levels in the TS fraction was not reversed by MG132 co-exposure. However, AR levels increased in the TI fraction in SFN and SFN+MG132 treated groups. Indeed, significant increases in insoluble protein aggregates have been reported following exposure to proteasomal inhibitors (67–69). Protein aggregation is enhanced when the accumulation of ubiquitinated proteins exceed beyond the capacity of proteasomes to degrade them (70).
The Hsp90 protein is necessary for the stabilization and correct folding of AR in both normal and malignant prostate cells (71). However, as compared to normal prostate epithelial cells, the expression of Hsp90 is reported to significantly increase in the malignant PCa (72). Studies have shown that SFN hyper-acetylates and inactivates Hsp90 (43) by targeting the function of histone deacetylase 6 (HDAC6) (44,45) which may be associated with its ability to degrade both AR-FL and AR-V7. However, we also observed that potent inhibition of Hsp90 via high doses of G (2 µM) was unable to fully abrogate AR protein levels. Recent studies have shown that Hsp90 inhibitors alone are not very effective against CRPC tumors (73). This clearly underscores the importance of targeting parallel pathways such as oxidative stress or Nrf2 to increase the efficacy of Hsp90 inhibitors against CRPC tumors.
Cancer cells are characterized by increased reactive oxygen species (ROS) levels and oxidative stress (74). Nrf2 is a transcription factor which is known to upregulate many cellular antioxidant proteins. In its inactive state, Nrf2 is found in the cytoplasm bound by its negative regulator Kelch-like ECH-associated protein 1 (Keap-1) which prevents Nrf2 nuclear translocation and directs it for proteasomal degradation. SFN has been shown to interact with Keap-1 thereby eliminating its inhibitory effect on Nrf2 (31,40,41). In line with these earlier studies, we observed that treatment of 22Rv1 cells with a pharmacological activator of Nrf2, BM drastically reduced both AR-FL and AR-V7 protein levels. Noteworthy, co-exposure to low-dose BM enhanced the suppressive effect of G. Thus, knowledge of the multimodal actions of SFN, i.e., Hsp90 inhibition and Nrf2 activation, can be used to formulate a safe pharmaceutical combination to potently decrease AR-FL and AR-V7 levels in CRPC cells. The efficacy of this combination is further supported by our observations that similar to SFN, combined treatment with physiologic doses (nanomolar) of G and BM suppressed proliferation, migration and clonogenic ability, and most importantly, resensitized these AR-V7 expressing cells to the anticancer effects of ENZ.
Although we did not carry out in depth mechanistic studies using the G+BM combination, numerous investigators have shown the potent Hsp90 inhibitory effect of G (72,75) and the potent Nrf2 inductive effect of BM (76,77). In addition, our previous published finding documented that overexpression of Nrf2 can suppress the expression and function of AR-FL in both LNCaP and C4-2B cells (42). Since SFN functions via the above two mechanisms (31,40,41,43–45), it is likely that the G+BM combination can simultaneously target them in order to decrease both AR-FL and AR-V7 levels.
Although we have not investigated the in vivo efficacies of either SFN or our pharmaceutical drug combination, i.e. G+BM, our findings clearly implicate in vivo potential of these agents as evident from numerous past studies (55–61). Our in vitro findings on the potent effects of SFN alone (micromolar doses) or the G+BM combination (nanomolar doses) in suppressing proliferation, migration and clonogenic ability of 22Rv1 cells, parameters that dictate aggressive tumor growth in vivo (49,78,79), clearly suggest that these agents may be effective against in vivo tumors. In summary, our findings suggest that either SFN, or the combination of G+BM, may provide an effective adjunct to current treatment in CRPC patients.
Acknowledgements
This study was supported by funds from the Louisiana Cancer Research Consortium (LCRC) to D.M. and from the Laboratory Training funds to S.C.S.
References
- 1.Yap TA, Zivi A, Omlin A, de Bono JS. The changing therapeutic landscape of castration-resistant prostate cancer. Nat Rev Clin Oncol. 2011;8:597–610. doi: 10.1038/nrclinonc.2011.117. [DOI] [PubMed] [Google Scholar]
- 2.Hodgson MC, Bowden WA, Agoulnik IU. Androgen receptor footprint on the way to prostate cancer progression. World J Urol. 2012;30:279–285. doi: 10.1007/s00345-011-0743-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chang KH, Ercole CE, Sharifi N. Androgen metabolism in prostate cancer: From molecular mechanisms to clinical consequences. Br J Cancer. 2014;111:1249–1254. doi: 10.1038/bjc.2014.268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Scher HI, Buchanan G, Gerald W, Butler LM, Tilley WD. Targeting the androgen receptor: Improving outcomes for castration resistant prostate cancer. Endocr Relat Cancer. 2004;11:459–476. doi: 10.1677/erc.1.00525. [DOI] [PubMed] [Google Scholar]
- 5.Godbole AM, Njar VC. New insights into the androgen-targeted therapies and epigenetic therapies in prostate cancer. Prostate Cancer. 2011;2011:918707. doi: 10.1155/2011/918707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim W, Ryan CJ. Androgen receptor directed therapies in castration-resistant metastatic prostate cancer. Curr Treat Options Oncol. 2012;13:189–200. doi: 10.1007/s11864-012-0188-2. [DOI] [PubMed] [Google Scholar]
- 7.Harris WP, Mostaghel EA, Nelson PS, Montgomery B. Androgen deprivation therapy: Progress in understanding mechanisms of resistance and optimizing androgen depletion. Nat Clin Pract Urol. 2009;6:76–85. doi: 10.1038/ncpuro1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lamont KR, Tindall DJ. Minireview: Alternative activation pathways for the androgen receptor in prostate cancer. Mol Endocrinol. 2011;25:897–907. doi: 10.1210/me.2010-0469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brooke GN, Bevan CL. The role of androgen receptor mutations in prostate cancer progression. Curr Genomics. 2009;10:18–25. doi: 10.2174/138920209787581307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Armstrong CM, Gao AC. Drug resistance in castration resistant prostate cancer: Resistance mechanisms and emerging treatment strategies. Am J Clin Exp Urol. 2015;3:64–76. [PMC free article] [PubMed] [Google Scholar]
- 11.Antonarakis ES, Armstrong AJ, Dehm SM, Luo J. Androgen receptor variant-driven prostate cancer: Clinical implications and therapeutic targeting. Prostate Cancer Prostatic Dis. 2016;19:231–241. doi: 10.1038/pcan.2016.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lokhandwala PM, Riel SL, Haley L, Lu C, Chen Y, Silberstein J, Zhu Y, Zheng G, Lin MT, Gocke CD, et al. Analytical validation of androgen receptor splice variant 7 detection in a clinical laboratory improvement amendments (CLIA) laboratory setting. J Mol Diagn. 2017;19:115–125. doi: 10.1016/j.jmoldx.2016.08.003. [DOI] [PubMed] [Google Scholar]
- 13.Del Re M, Biasco E, Crucitta S, Derosa L, Rofi E, Orlandini C, Miccoli M, Galli L, Falcone A, Jenster GW, et al. The detection of androgen receptor splice variant 7 in plasma-derived exosomal RNA strongly predicts resistance to hormonal therapy in metastatic prostate cancer patients. Eur Urol. 2017;71:680–687. doi: 10.1016/j.eururo.2016.08.012. [DOI] [PubMed] [Google Scholar]
- 14.Dehm SM, Schmidt LJ, Heemers HV, Vessella RL, Tindall DJ. Splicing of a novel androgen receptor exon generates a constitutively active androgen receptor that mediates prostate cancer therapy resistance. Cancer Res. 2008;68:5469–5477. doi: 10.1158/0008-5472.CAN-08-0594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hu R, Dunn TA, Wei S, Isharwal S, Veltri RW, Humphreys E, Han M, Partin AW, Vessella RL, Isaacs WB, et al. Ligand-independent androgen receptor variants derived from splicing of cryptic exons signify hormone-refractory prostate cancer. Cancer Res. 2009;69:16–22. doi: 10.1158/0008-5472.CAN-08-2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guo Z, Yang X, Sun F, Jiang R, Linn DE, Chen H, Chen H, Kong X, Melamed J, Tepper CG, et al. A novel androgen receptor splice variant is up-regulated during prostate cancer progression and promotes androgen depletion-resistant growth. Cancer Res. 2009;69:2305–2313. doi: 10.1158/0008-5472.CAN-08-3795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sun S, Sprenger CC, Vessella RL, Haugk K, Soriano K, Mostaghel EA, Page ST, Coleman IM, Nguyen HM, Sun H, et al. Castration resistance in human prostate cancer is conferred by a frequently occurring androgen receptor splice variant. J Clin Invest. 2010;120:2715–2730. doi: 10.1172/JCI41824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang X, Morrissey C, Sun S, Ketchandji M, Nelson PS, True LD, Vakar-Lopez F, Vessella RL, Plymate SR. Androgen receptor variants occur frequently in castration resistant prostate cancer metastases. PLoS One. 2011;6:e27970. doi: 10.1371/journal.pone.0027970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dehm SM, Tindall DJ. Alternatively spliced androgen receptor variants. Endocr Relat Cancer. 2011;18:R183–R196. doi: 10.1530/ERC-11-0141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Antonarakis ES, Lu C, Wang H, Luber B, Nakazawa M, Roeser JC, Chen Y, Mohammad TA, Chen Y, Fedor HL, et al. AR-V7 and resistance to enzalutamide and abiraterone in prostate cancer. N Engl J Med. 2014;371:1028–1038. doi: 10.1056/NEJMoa1315815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sarwar M, Semenas J, Miftakhova R, Simoulis A, Robinson B, Gjörloff Wingren A, Mongan NP, Heery DM, Johnsson H, Abrahamsson PA, et al. Targeted suppression of AR-V7 using PIP5K1α inhibitor overcomes enzalutamide resistance in prostate cancer cells. Oncotarget. 2016;7:63065–63081. doi: 10.18632/oncotarget.11757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Watson PA, Chen YF, Balbas MD, Wongvipat J, Socci ND, Viale A, Kim K, Sawyers CL. Constitutively active androgen receptor splice variants expressed in castration-resistant prostate cancer require full-length androgen receptor; Proc Natl Acad Sci USA; 2010; pp. 16759–16765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cao B, Qi Y, Zhang G, Xu D, Zhan Y, Alvarez X, Guo Z, Fu X, Plymate SR, Sartor O, et al. Androgen receptor splice variants activating the full-length receptor in mediating resistance to androgen-directed therapy. Oncotarget. 2014;5:1646–1656. doi: 10.18632/oncotarget.1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu D, Zhan Y, Qi Y, Cao B, Bai S, Xu W, Gambhir SS, Lee P, Sartor O, Flemington EK, et al. Androgen receptor splice variants dimerize to transactivate target genes. Cancer Res. 2015;75:3663–3671. doi: 10.1158/0008-5472.CAN-15-0381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang Y, Tang L. Discovery and development of sulforaphane as a cancer chemopreventive phytochemical. Acta Pharmacol Sin. 2007;28:1343–1354. doi: 10.1111/j.1745-7254.2007.00679.x. [DOI] [PubMed] [Google Scholar]
- 26.Clarke JD, Dashwood RH, Ho E. Multi-targeted prevention of cancer by sulforaphane. Cancer Lett. 2008;269:291–304. doi: 10.1016/j.canlet.2008.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cheung KL, Kong AN. Molecular targets of dietary phenethyl isothiocyanate and sulforaphane for cancer chemoprevention. AAPS J. 2010;12:87–97. doi: 10.1208/s12248-009-9162-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fawzy Elbarbry, Nehad Elrody. Potential health benefits of sulforaphane: A review of the experimental, clinical and epidemiological evidences and underlying mechanisms. J Med Plants Res. 2011;5:473–484. [Google Scholar]
- 29.Shapiro TAI, Fahey JW, Dinkova-Kostova AT, Holtzclaw WD, Stephenson KK, Wade KL, Ye L, Talalay P. Safety, tolerance, and metabolism of broccoli sprout glucosinolates and isothiocyanates: a clinical phase I study. Nutr Cancer. 2006;55:53–62. doi: 10.1207/s15327914nc5501_7. [DOI] [PubMed] [Google Scholar]
- 30.Petri N, Tannergren C, Holst B, Mellon FA, Bao Y, Plumb GW, Bacon J, O'Leary KA, Kroon PA, Knutson L, et al. Absorption/metabolism of sulforaphane and quercetin, and regulation of phase II enzymes, in human jejunum in vivo. Drug Metab Dispos. 2003;31:805–813. doi: 10.1124/dmd.31.6.805. [DOI] [PubMed] [Google Scholar]
- 31.Keum YS, Khor TO, Lin W, Shen G, Kwon KH, Barve A, Li W, Kong AN. Pharmacokinetics and pharmacodynamics of broccoli sprouts on the suppression of prostate cancer in transgenic adenocarcinoma of mouse prostate (TRAMP) mice: Implication of induction of Nrf2, HO-1 and apoptosis and the suppression of Akt-dependent kinase pathway. Pharm Res. 2009;26:2324–2331. doi: 10.1007/s11095-009-9948-5. [DOI] [PubMed] [Google Scholar]
- 32.Traka MH, Melchini A, Mithen RF. Sulforaphane and prostate cancer interception. Drug Discov Today. 2014;19:1488–1492. doi: 10.1016/j.drudis.2014.07.007. [DOI] [PubMed] [Google Scholar]
- 33.Kim SH, Singh SV. D,L-Sulforaphane causes transcriptional repression of androgen receptor in human prostate cancer cells. Mol Cancer Ther. 2009;8:1946–1954. doi: 10.1158/1535-7163.MCT-09-0104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wiczk A, Hofman D, Konopa G, Herman-Antosiewicz A. Sulforaphane, a cruciferous vegetable-derived isothiocyanate, inhibits protein synthesis in human prostate cancer cells. Biochim Biophys Acta. 2012;1823:1295–1305. doi: 10.1016/j.bbamcr.2012.05.020. [DOI] [PubMed] [Google Scholar]
- 35.Burnett JP, Lim G, Li Y, Shah RB, Lim R, Paholak HJ, McDermott SP, Sun L, Tsume Y, Bai S, et al. Sulforaphane enhances the anticancer activity of taxanes against triple negative breast cancer by killing cancer stem cells. Cancer Lett. 2017;394:52–64. doi: 10.1016/j.canlet.2017.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li QQ, Xie YK, Wu Y, Li LL, Liu Y, Miao XB, Liu QZ, Yao KT, Xiao GH. Sulforaphane inhibits cancer stem-like cell properties and cisplatin resistance through miR-214-mediated downregulation of c-MYC in non-small cell lung cancer. Oncotarget. 2017;8:12067–12080. doi: 10.18632/oncotarget.14512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Singh SV, Srivastava SK, Choi S, Lew KL, Antosiewicz J, Xiao D, Zeng Y, Watkins SC, Johnson CS, Trump DL, et al. Sulforaphane-induced cell death in human prostate cancer cells is initiated by reactive oxygen species. J Biol Chem. 2005;280:19911–19924. doi: 10.1074/jbc.M412443200. [DOI] [PubMed] [Google Scholar]
- 38.Xiao D, Powolny AA, Antosiewicz J, Hahm ER, Bommareddy A, Zeng Y, Desai D, Amin S, Herman-Antosiewicz A, Singh SV. Cellular responses to cancer chemopreventive agent D,L-sulforaphane in human prostate cancer cells are initiated by mitochondrial reactive oxygen species. Pharm Res. 2009;26:1729–1738. doi: 10.1007/s11095-009-9883-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pei Y, Wu B, Cao Q, Wu L, Yang G. Hydrogen sulfide mediates the anti-survival effect of sulforaphane on human prostate cancer cells. Toxicol Appl Pharmacol. 2011;257:420–428. doi: 10.1016/j.taap.2011.09.026. [DOI] [PubMed] [Google Scholar]
- 40.Zhang C, Su Z-Y, Khor TO, Shu L, Kong AN. Sulforaphane enhances Nrf2 expression in prostate cancer TRAMP C1 cells through epigenetic regulation. Biochem Pharmacol. 2013;85:1398–1404. doi: 10.1016/j.bcp.2013.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kensler TW, Egner PA, Agyeman AS, Visvanathan K, Groopman JD, Chen JG, Chen TY, Fahey JW, Talalay P. Keap1-nrf2 signaling: A target for cancer prevention by sulforaphane. Top Curr Chem. 2013;329:163–177. doi: 10.1007/128_2012_339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schultz MA, Hagan SS, Datta A, Zhang Y, Freeman ML, Sikka SC, Abdel-Mageed AB, Mondal D. Nrf1 and Nrf2 transcription factors regulate androgen receptor transactivation in prostate cancer cells. PLoS One. 2014;9:e87204. doi: 10.1371/journal.pone.0087204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gibbs A, Schwartzman J, Deng V, Alumkal J. Sulforaphane destabilizes the androgen receptor in prostate cancer cells by inactivating histone deacetylase 6; Proc Natl Acad Sci USA; 2009; pp. 16663–16668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Myzak MC, Hardin K, Wang R, Dashwood RH, Ho E. Sulforaphane inhibits histone deacetylase activity in BPH-1, LnCaP and PC-3 prostate epithelial cells. Carcinogenesis. 2006;27:811–819. doi: 10.1093/carcin/bgi265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tortorella SM, Royce SG, Licciardi PV, Karagiannis TC. Dietary sulforaphane in cancer chemoprevention: The role of epigenetic regulation and HDAC inhibition. Antioxid Redox Signal. 2015;22:1382–1424. doi: 10.1089/ars.2014.6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Khurana N, Talwar S, Chandra PK, Sharma P, Abdel-Mageed AB, Mondal D, Sikka SC. Sulforaphane increases the efficacy of anti-androgens by rapidly decreasing androgen receptor levels in prostate cancer cells. Int J Oncol. 2016;49:1609–1619. doi: 10.3892/ijo.2016.3641. [DOI] [PubMed] [Google Scholar]
- 47.Wu HC, Hsieh JT, Gleave ME, Brown NM, Pathak S, Chung LW. Derivation of androgen-independent human LNCaP prostatic cancer cell sublines: Role of bone stromal cells. Int J Cancer. 1994;57:406–412. doi: 10.1002/ijc.2910570319. [DOI] [PubMed] [Google Scholar]
- 48.Li Y, Karagöz GE, Seo YH, Zhang T, Jiang Y, Yu Y, Duarte AM, Schwartz SJ, Boelens R, Carroll K, et al. Sulforaphane inhibits pancreatic cancer through disrupting Hsp90-p50(Cdc37) complex and direct interactions with amino acids residues of Hsp90. J Nutr Biochem. 2012;23:1617–1626. doi: 10.1016/j.jnutbio.2011.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Uygur B, Wu W-S. SLUG promotes prostate cancer cell migration and invasion via CXCR4/CXCL12 axis. Mol Cancer. 2011;10:139. doi: 10.1186/1476-4598-10-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chou TC. Drug combination studies and their synergy quantification using the Chou-Talalay method. Cancer Res. 2010;70:440–446. doi: 10.1158/0008-5472.CAN-09-1947. [DOI] [PubMed] [Google Scholar]
- 51.Scher HI, Fizazi K, Saad F, Taplin ME, Sternberg CN, Miller K, de Wit R, Mulders P, Chi KN, Shore ND, et al. AFFIRM Investigators: Increased survival with enzalutamide in prostate cancer after chemotherapy. N Engl J Med. 2012;367:1187–1197. doi: 10.1056/NEJMoa1207506. [DOI] [PubMed] [Google Scholar]
- 52.Ryan CJ, Cheng ML. Abiraterone acetate for the treatment of prostate cancer. Expert Opin Pharmacother. 2013;14:91–96. doi: 10.1517/14656566.2013.745852. [DOI] [PubMed] [Google Scholar]
- 53.Liu C, Lou W, Zhu Y, Nadiminty N, Schwartz CT, Evans CP, Gao AC. Niclosamide inhibits androgen receptor variants expression and overcomes enzalutamide resistance in castration-resistant prostate cancer. Clin Cancer Res. 2014;20:3198–210. doi: 10.1158/1078-0432.CCR-13-3296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu C, Armstrong C, Zhu Y, Lou W, Gao AC. Niclosamide enhances abiraterone treatment via inhibition of androgen receptor variants in castration resistant prostate cancer. Oncotarget. 2016;7:32210–32220. doi: 10.18632/oncotarget.8493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Goldman JW, Raju RN, Gordon GA, El-Hariry I, Teofilivici F, Vukovic VM, Bradley R, Karol MD, Chen Y, Guo W, et al. A first in human, safety, pharmacokinetics, and clinical activity phase I study of once weekly administration of the Hsp90 inhibitor ganetespib (STA-9090) in patients with solid malignancies. BMC Cancer. 2013;13:152. doi: 10.1186/1471-2407-13-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Thakur MK, Heilbrun LK, Sheng S, Stein M, Liu G, Antonarakis ES, Vaishampayan U, Dzinic SH, Li X, Freeman S, et al. A phase II trial of ganetespib, a heat shock protein 90 Hsp90) inhibitor, in patients with docetaxel-pretreated metastatic castrate-resistant prostate cancer (CRPC)-a prostate cancer clinical trials consortium (PCCTC) study. Invest New Drugs. 2016;34:112–118. doi: 10.1007/s10637-015-0307-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Goyal L, Wadlow RC, Blaszkowsky LS, Wolpin BM, Abrams TA, McCleary NJ, Sheehan S, Sundaram E, Karol MD, Chen J, et al. A phase I and pharmacokinetic study of ganetespib (STA-9090) in advanced hepatocellular carcinoma. Invest New Drugs. 2015;33:128–137. doi: 10.1007/s10637-014-0164-8. [DOI] [PubMed] [Google Scholar]
- 58.Hong DS, Kurzrock R, Supko JG, He X, Naing A, Wheler J, Lawrence D, Eder JP, Meyer CJ, Ferguson DA, et al. A phase I first-in-human trial of bardoxolone methyl in patients with advanced solid tumors and lymphomas. Clin Cancer Res. 2012;18:3396–3406. doi: 10.1158/1078-0432.CCR-11-2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gao X, Deeb D, Liu Y, Arbab AS, Divine GW, Dulchavsky SA, Gautam SC. Prevention of prostate cancer with oleanane synthetic triterpenoid CDDO-Me in the TRAMP mouse model of prostate cancer. Cancers (Basel) 2011;3:3353–3369. doi: 10.3390/cancers3033353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Alumkal JJ, Slottke R, Schwartzman J, Cherala G, Munar M, Graff JN, Beer TM, Ryan CW, Koop DR, Gibbs A, et al. A phase II study of sulforaphane-rich broccoli sprout extracts in men with recurrent prostate cancer. Invest New Drugs. 2015;33:480–489. doi: 10.1007/s10637-014-0189-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shapiro TA, Fahey JW, Dinkova-Kostova AT, Holtzclaw WD, Stephenson KK, Wade KL, Ye L, Talalay P. Safety, tolerance, and metabolism of broccoli sprout glucosinolates and isothiocyanates: a clinical phase I study. Nutr Cancer. 2006;55:53–62. doi: 10.1207/s15327914nc5501_7. [DOI] [PubMed] [Google Scholar]
- 62.Cunningham D, You Z. In vitro and in vivo model systems used in prostate cancer research. J Biol Methods. 2015;2:e17. doi: 10.14440/jbm.2015.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li Y, Hwang TH, Oseth LA, Hauge A, Vessella RL, Schmechel SC, Hirsch B, Beckman KB, Silverstein KA, Dehm SM. AR intragenic deletions linked to androgen receptor splice variant expression and activity in models of prostate cancer progression. Oncogene. 2012;31:4759–4767. doi: 10.1038/onc.2011.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lilienbaum A. Relationship between the proteasomal system and autophagy. Int J Biochem Mol Biol. 2013;4:1–26. [PMC free article] [PubMed] [Google Scholar]
- 65.Jaworski T. Degradation and beyond: control of androgen receptor activity by the proteasome system. Cell Mol Biol Lett. 2006;11:109–131. doi: 10.2478/s11658-006-0011-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gan N, Wu YC, Brunet M, Garrido C, Chung FL, Dai C, Mi L. Sulforaphane activates heat shock response and enhances proteasome activity through up-regulation of Hsp27. J Biol Chem. 2010;285:35528–35536. doi: 10.1074/jbc.M110.152686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Miyahara K, Kazama H, Kokuba H, Komatsu S, Hirota A, Takemura J, Hirasawa K, Moriya S, Abe A, Hiramoto M, et al. Targeting bortezomib-induced aggresome formation using vinorelbine enhances the cytotoxic effect along with ER stress loading in breast cancer cell lines. Int J Oncol. 2016;49:1848–1858. doi: 10.3892/ijo.2016.3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Guthrie CR, Kraemer BC. Proteasome inhibition drives HDAC6-dependent recruitment of tau to aggresomes. J Mol Neurosci. 2011;45:32–41. doi: 10.1007/s12031-011-9502-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zaarur N, Meriin AB, Bejarano E, Xu X, Gabai VL, Cuervo AM, Sherman MY. Proteasome failure promotes positioning of lysosomes around the aggresome via local block of microtubule-dependent transport. Mol Cell Biol. 2014;34:1336–1348. doi: 10.1128/MCB.00103-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Moriya S, Komatsu S, Yamasaki K, Kawai Y, Kokuba H, Hirota A, Che XF, Inazu M, Gotoh A, Hiramoto M, et al. Targeting the integrated networks of aggresome formation, proteasome, and autophagy potentiates ER stress-mediated cell death in multiple myeloma cells. Int J Oncol. 2015;46:474–486. doi: 10.3892/ijo.2014.2773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Azad AA, Zoubeidi A, Gleave ME, Chi KN. Targeting heat shock proteins in metastatic castration-resistant prostate cancer. Nat Rev Urol. 2015;12:26–36. doi: 10.1038/nrurol.2014.320. [DOI] [PubMed] [Google Scholar]
- 72.He S, Zhang C, Shafi AA, Sequeira M, Acquaviva J, Friedland JC, Sang J, Smith DL, Weigel NL, Wada Y, et al. Potent activity of the Hsp90 inhibitor ganetespib in prostate cancer cells irrespective of androgen receptor status or variant receptor expression. Int J Oncol. 2013;42:35–43. doi: 10.3892/ijo.2012.1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Shafi AA, Cox MB, Weigel NL. Androgen receptor splice variants are resistant to inhibitors of Hsp90 and FKBP52, which alter androgen receptor activity and expression. Steroids. 2013;78:548–554. doi: 10.1016/j.steroids.2012.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Shiota M, Yokomizo A, Naito S. Oxidative stress and androgen receptor signaling in the development and progression of castration-resistant prostate cancer. Free Radic Biol Med. 2011;51:1320–1328. doi: 10.1016/j.freeradbiomed.2011.07.011. [DOI] [PubMed] [Google Scholar]
- 75.Gillis JL, Selth LA, Centenera MM, Townley SL, Sun S, Plymate SR, Tilley WD, Butler LM. Constitutively-active androgen receptor variants function independently of the HSP90 chaperone but do not confer resistance to HSP90 inhibitors. Oncotarget. 2013;4:691–704. doi: 10.18632/oncotarget.975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wang YY, Yang YX, Zhe H, He ZX, Zhou SF. Bardoxolone methyl (CDDO-Me) as a therapeutic agent: An update on its pharmacokinetic and pharmacodynamic properties. Drug Des Devel Ther. 2014;8:2075–2088. doi: 10.2147/DDDT.S68872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Probst BL, McCauley L, Trevino I, Wigley WC, Ferguson DA. Cancer cell growth is differentially affected by constitutive activation of NRF2 by KEAP1 deletion and pharmacological activation of NRF2 by the synthetic triterpenoid, RTA 405. PLoS One. 2015;10:e0135257. doi: 10.1371/journal.pone.0135257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Huo C, Kao Y-H, Chuu C-P. Androgen receptor inhibits epithelial-mesenchymal transition, migration, and invasion of PC-3 prostate cancer cells. Cancer Lett. 2015;369:103–111. doi: 10.1016/j.canlet.2015.08.001. [DOI] [PubMed] [Google Scholar]
- 79.Aapro MS, Eliason JF, Krauer F, Alberto P. Colony formation in vitro as a prognostic indicator for primary breast cancer. J Clin Oncol. 1987;5:890–896. doi: 10.1200/JCO.1987.5.6.890. [DOI] [PubMed] [Google Scholar]