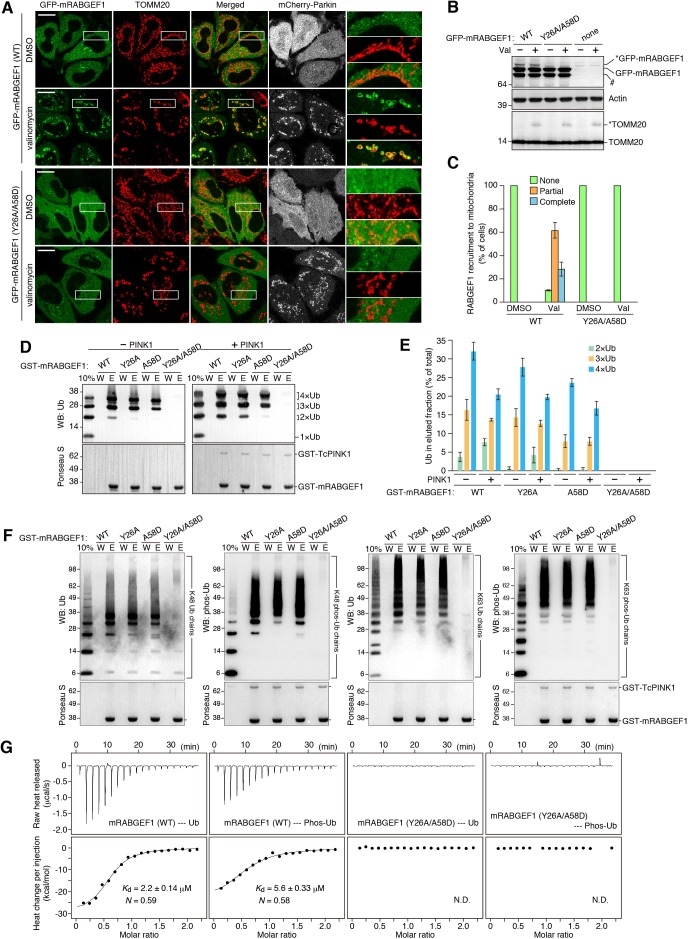
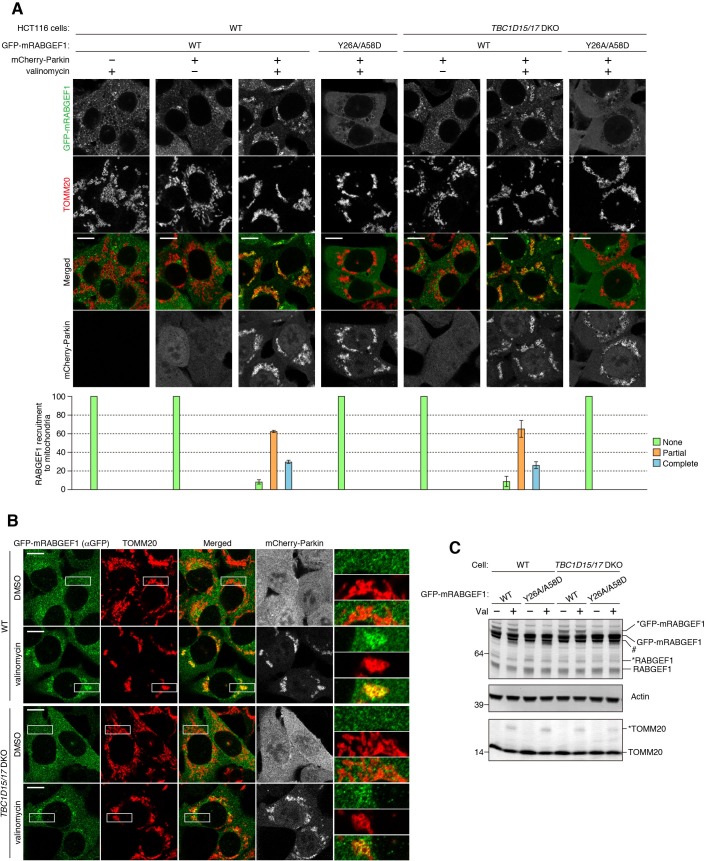
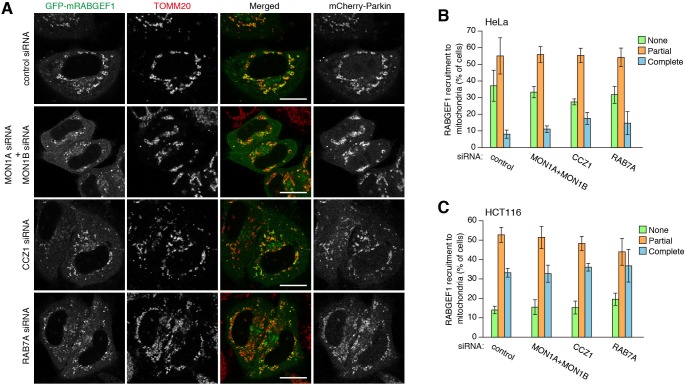
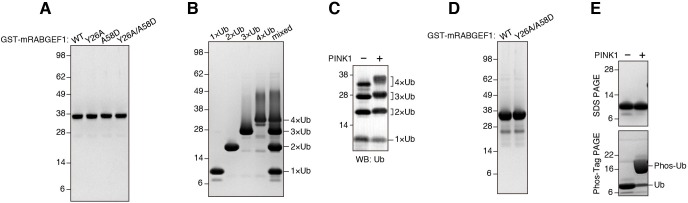
Figure 8. RABGEF1 is recruited to the damaged mitochondria in a ubiquitin-binding dependent manner.
(A) HeLa cells transiently expressing mChery-Parkin and GFP-mRABGEF1 were treated with DMSO or valinomycin for 3 hr followed by immunostaining. The magnified pictures were shown in the right. Bars, 10 μm. (B) Total cell lysates of (A) were analyzed by immunoblotting. Anti-GFP antibody was used for the GFP-mRABGEF1 detection. * and # denote ubiquitinated forms and truncated forms, respectively. (C) Quantification of RABGEF1 recruitment to damaged mitochondria in (A). None, partial and complete denote that GFP-mRABGEF1 signals were overlapped with no, some of, and all mitochondria, respectively. (D) Recombinant ubiquitin (Ub) pre-treated with or without GST-TcPINK1 was subjected to pull-down assay with GST-mRABGEF1. W and E indicate wash and eluted fractions, respectively. 10%, 10% of input. (E) Percentages of the amount of ubiquitin in the eluted fraction in (D) were shown. The error bars represent mean ±SE from three independent experiments. (F) K48-linked and K63-linked Ub chains pre-treated with or without GST-TcPINK1 were subjected to pull-down assay with GST-mRABGEF1. (G) Interactions between GST-mRABGEF1 (WT or Y26A/A58D) and ubiquitin or phosphorylated ubiquitin were measured by ITC. N, stoichiometry of binding.