Abstract
OBJECTIVE
The phenotypic diversity of type 1 diabetes suggests heterogeneous etiopathogenesis. We investigated the relationship of type 2 diabetes–associated transcription factor 7 like 2 (TCF7L2) single nucleotide polymorphisms (SNPs) with immunologic and metabolic characteristics at type 1 diabetes diagnosis.
RESEARCH DESIGN AND METHODS
We studied TrialNet participants with newly diagnosed autoimmune type 1 diabetes with available TCF7L2 rs4506565 and rs7901695 SNP data (n = 810; median age 13.6 years; range 3.3–58.6). We modeled the influence of carrying a TCF7L2 variant (i.e., having 1 or 2 minor alleles) on the number of islet autoantibodies and oral glucose tolerance test (OGTT)–stimulated C-peptide and glucose measures at diabetes diagnosis. All analyses were adjusted for known confounders.
RESULTS
The rs4506565 variant was a significant independent factor of expressing a single autoantibody, instead of multiple autoantibodies, at diagnosis (odds ratio [OR] 1.66 [95% CI 1.07, 2.57], P = 0.024). Interaction analysis demonstrated that this association was only significant in participants ≥12 years old (n = 504; OR 2.12 [1.29, 3.47], P = 0.003) but not younger ones (n = 306, P = 0.73). The rs4506565 variant was independently associated with higher C-peptide area under the curve (AUC) (P = 0.008) and lower mean glucose AUC (P = 0.0127). The results were similar for the rs7901695 SNP.
CONCLUSIONS
In this cohort of individuals with new-onset type 1 diabetes, type 2 diabetes–linked TCF7L2 variants were associated with single autoantibody (among those ≥12 years old), higher C-peptide AUC, and lower glucose AUC levels during an OGTT. Thus, carriers of the TCF7L2 variant had a milder immunologic and metabolic phenotype at type 1 diabetes diagnosis, which could be partly driven by type 2 diabetes–like pathogenic mechanisms.
Introduction
Although the autoimmune destruction of β-cells has a major role in the development of type 1 diabetes, there is growing evidence that the differences in clinical, metabolic, immunologic, and genetic characteristics among patients (1) likely reflect diverse etiology and pathogenesis (2). Factors that govern this heterogeneity are poorly understood, yet these may have important implications for prognosis, therapy, and prevention.
The transcription factor 7 like 2 (TCF7L2) locus contains the single nucleotide polymorphism (SNP) most strongly associated with type 2 diabetes risk, with an ∼30% increase per risk allele (3). In a U.S. cohort, heterozygous and homozygous carriers of the at-risk alleles comprised 40.6% and 7.9%, respectively, of the control subjects and 44.3% and 18.3%, respectively, of the individuals with type 2 diabetes (3). The locus has no known association with type 1 diabetes overall (4–8), with conflicting reports in latent autoimmune diabetes in adults (8–16). Patients with type 2 diabetes and healthy subjects carrying the T allele or the TT genotype at the rs7903146 locus exhibit impaired insulin secretion and defects in glucagon suppression, particularly in response to incretins and oral glucose, as well as abnormal insulin processing (17–19). In the hepatocyte, TCF7L2 risk alleles are associated with gain of function and increased hepatic glucose release during fasting (17). Recent studies suggest a possible regulatory role of a TCF7L2 variant on acyl-CoA synthetase long-chain family member 5 (ACSL5) expression and, thus, potentially on insulin sensitivity (20,21).
Our studies in two separate cohorts have shown that the type 2 diabetes–associated TCF7L2 genetic variant is more frequent among specific subsets of individuals with autoimmune type 1 diabetes, specifically those with fewer markers of islet autoimmunity (22,23). These observations support a role of this genetic variant in the pathogenesis of diabetes at least in a subset of individuals with autoimmune diabetes. However, whether individuals with type 1 diabetes and this genetic variant have distinct metabolic abnormalities has not been investigated. We aimed to study the immunologic and metabolic characteristics of individuals with type 1 diabetes who carry a type 2 diabetes–associated allele of the TCF7L2 locus. If nonautoimmune diabetogenic factors play a role in the pathogenesis of type 1 diabetes, preventative and therapeutic strategies addressing the underlying mechanisms could be tried.
Research Design and Methods
Participants
TrialNet is a National Institutes of Health (NIH)–funded, international network of centers with the mission to prevent type 1 diabetes and stop disease progression (TN01; clinical trial reg. no. NCT00097292, clinicaltrials.gov) (24). The observational arm of TrialNet, Pathway to Prevention (PTP), prospectively monitors at-risk individuals without diabetes (first- or second-degree relatives of patients with type 1 diabetes) for progression of islet autoimmunity and development of type 1 diabetes (25). Here, we focused on those participants who did develop type 1 diabetes during the course of their follow-up in the PTP cohort. In addition, we studied individuals with newly diagnosed type 1 diabetes from the general population who participated in TrialNet New Onset clinical trials to preserve insulin production. Type 1 diabetes was diagnosed according to American Diabetes Association criteria (26). Subjects were included in the study if TCF7L2 SNP data were available and islet autoantibody testing was performed within 180 days of diagnosis, with positivity for at least one autoantibody. Our analysis involved a cohort of 810 newly diagnosed patients, including 249 autoantibody-positive TrialNet PTP participants and 561 subjects enrolled in TrialNet New Onset clinical trials. No subjects were in both groups. All study participants gave informed consent, and the study was approved by the responsible ethics committee at each study site.
Procedures
All subjects were screened for autoantibodies to GAD65, insulin (microinsulin antibody assay), and insulinoma-associated antigen 2 (IA2). If any of these were positive, autoantibodies to zinc transporter 8 (ZnT8) and islet cell antibodies (ICA) were also tested. Oral glucose tolerance tests (OGTT) were conducted with an oral glucose dose of 1.75 g/kg (maximum 75 g). C-peptide (ng/mL) and glucose (mg/dL) were measured fasting and at 30, 60, 90, and 120 min.
Islet autoantibody (25) and C-peptide (27) assays have been previously described. HLA genotyping was performed at the Type 1 Diabetes Genetics Consortium Laboratories. The Illumina Immunochip was used to genotype TCF7L2 rs7901695 and rs4506565 at the Center for Public Health Genomics at the University of Virginia. The Immunochip is a custom array of 186,000 SNPs selected from regions of the genome robustly associated with autoimmune diseases.
Statistical Analyses
Islet autoimmunity was defined as confirmed positive for at least one islet autoantibody, including insulin, GAD65, IA2, ZnT8, and ICA. The trapezoid method was used to calculate the area under the curve (AUC) for C-peptide and glucose during the OGTT. HLA DR3 was defined as DRB1*03:01 – DQA1*05:01 – DQB1*02:01, and HLA DR4-DQ8 was defined as DQA1*03:01 – DQB1*03:02 with DRB1*04:01, *04:02, or *04:05. Because TCF7L2 rs7903146 was not included in the Immunochip, we analyzed frequencies of TCF7L2 rs4506565 and rs7901695, which are in nearly complete linkage disequilibrium (r2 = 0.913 and 0.909, respectively, in a population of European ancestry) with rs7903146 (Genome Variation Server; http://gvs.gs.washington.edu/GVS). We examined ordered outcomes for numbers of minor alleles (zero, one, or two) and found high concordance (99.0%) between the two TCF7L2 SNPs, rs4506565 and rs7901695 (95% CI 0.98, 1.00). Minor allele frequencies were evaluated as an ordered variable as well as dichotomized by carrier (one or two minor alleles) versus noncarrier (zero minor alleles) and also homozygous for the minor allele (two minor alleles) versus not (zero or one minor alleles). All analyses were conducted for each of these SNPs, although the results presented focus on one (rs4506565, minor allele: T) given the concordance of the results. Hardy-Weinberg equilibrium was verified for the full sample.
All clinical and metabolic factors were summarized and evaluated using descriptive statistics. Univariate and multivariable logistic regression models were used to evaluate the influence of various factors on the dichotomous outcome of interest (e.g., single autoantibody–positive at diagnosis vs. positive for multiple autoantibodies). When the influence of carrier status on outcome was evaluated, models were adjusted for age at diagnosis, sex, BMI (z-score or overweight/obese vs. not), mean AUC for C-peptide at diagnosis, presence of HLA DR3 and DR4 (vs. absence), and subgroup (PTP progressors vs. New Onset trial participants). Mantel-Haenszel-Cochran tests were also used to evaluate the influence of carrier status on whether subjects were single autoantibody–positive at diagnosis versus not while stratifying by subgroup, thus further accounting for the differences between the two subgroups (Supplementary Table 2). Race and ethnicity were evaluated by univariate analyses. Because race and ethnicity were not significantly associated with the outcomes of interest and the percentage of nonwhite and/or Hispanic subjects in this cohort was very small, this factor was not included in the multivariable analysis.
Generalized linear regression models were used to evaluate influential factors on the mean AUC for C-peptide and AUC for glucose at diagnosis. Given the skewed distribution of these variables, these metabolic markers were log-transformed (e.g., log[AUC C-peptide]) in the models. Cut point analyses were conducted using recursive partitioning algorithms (http://CRAN.R-project.org/package=rpart). Statistical significance was determined if P < 0.05, except for tests of interactions, where those with P < 0.10 were explored further. All analyses were conducted using the R statistical program (https://CRAN.R-project.org/package=rpart).
Results
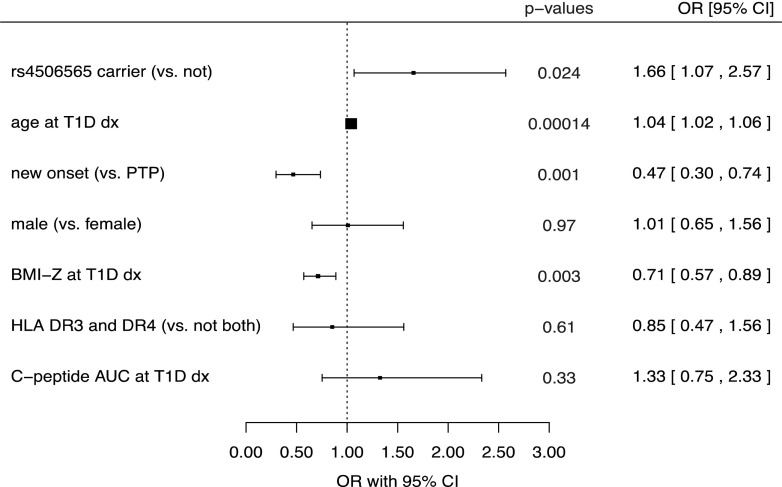
Characteristics of the cohort under study are presented in Table 1. The median age at diagnosis was 13.6 years (range 3.3–58.6), and 72% of the participants were younger than 18 years. Among the 119 participants who had a single positive autoantibody at diagnosis, 70 (59%) carried a type 2 diabetes–associated TCF7L2 allele (including 11 homozygotes), compared with 332 carriers (48%; including 7 homozygotes) among 691 participants with multiple autoantibodies (P = 0.038). After adjusting for possible confounders, we found that rs4506565 T allele carriers (i.e., participants who had one or two alleles of the variant associated with type 2 diabetes) were more likely to express a single positive autoantibody at type 1 diabetes diagnosis compared with noncarriers (i.e., those who had no minor alleles). Specifically, single autoantibody positivity was 66% more likely in participants carrying a TCF7L2 rs4506565T allele than in noncarriers (odds ratio [OR] 1.66 [95% CI 1.07, 2.57], P = 0.024) after adjustment for age, sex, BMI z-score, presence of highest-risk HLA genotype (HLA DR3/DR4-DQ8), C-peptide AUC, and subgroup (Fig. 1). Additional multivariable models demonstrated that HLA DR3 alone or combined with DR4-DQ8 was not an independent factor for single autoantibody positivity or for carrying the TCF7L2 variant. The protective HLA DRB1*15:01-DQB1*0602 haplotype was found in four noncarriers and in three carriers of the type 2 diabetes–associated TCF7L2 allele.
Table 1.
Characteristics of study subjects (n = 810)
| Age at T1D diagnosis | |
| Median (range), years | 13.6 (3.3–58.6) |
| <18 years | 584 (72) |
| ≥18 years | 226 (28) |
| Sex | |
| Female | 363 (45) |
| Male | 444 (55) |
| Not reported (n) | 3 |
| Race | |
| Nonwhite | 56 (7) |
| White | 744 (93) |
| Missing/unknown (n) | 10 |
| Ethnicity | |
| Hispanic or Latino | 76 (9.5) |
| Not Hispanic or Latino | 724 (90.5) |
| Missing/unknown (n) | 10 |
| BMI z-score at diagnosis | |
| Median (range) | 0.49 (−2.8 to 3.1) |
| Normal/underweight | 644 (81.1) |
| Overweight/obese | 150 (18.9) |
| Missing (n) | 16 |
| Positive islet autoantibodies at diagnosis (n) | |
| 1 | 119 (14.7) |
| 2 | 192 (23.7) |
| 3 | 276 (34.1) |
| 4 | 202 (24.9) |
| 5 | 21 (2.6) |
| Single | 119 (14.7) |
| Multiple (≥2) | 691 (85.3) |
| HLA DR3/DR4-DQ8† | |
| No | 648 (82.4) |
| Yes | 138 (17.6) |
| Missing (n) | 24 |
| HLA DR3 and/or DR4-DQ8† | |
| No | 200 (25.4) |
| Yes | 586 (74.6) |
| Missing (n) | 24 |
| Fasting glucose | |
| Median (range), mmol/L | 5.89 (3–16.06) |
| Missing (n) | 25 |
| Mean AUC glucose | |
| Median (range), mmol/L | 9.29 (4.76–18.71) |
| Missing (n) | 23 |
| Fasting C-peptide | |
| Median (range), nmol/L | 0.36 (0.02–3.53) |
| Missing (n) | 27 |
| Mean AUC C-peptide | |
| Median (range), nmol/L | 0.69 (0.01–2.70) |
| Missing (n) | 51 |
| rs4506565_T: minor allele distribution | |
| 0 | 408 (50.4) |
| 1 | 341 (42.1) |
| 2 | 61 (7.5) |
| Carrier of minor allele | 402 (49.6) |
| Homozygous for minor allele | 61 (7.5) |
Data are presented as n (%) except where noted otherwise. T1D, type 1 diabetes.
†Note that DR3 was defined as HLA DRB1*0301, DQA1*0501, DQB1*0201, and DR4-DQ8 was defined as HLA DRB1*0401, *0402, or *0405, DQA1*0301, DQB1*0302.
Figure 1.
Forest plot representing the influence of the shown measures on the likelihood of having single (vs. multiple) autoantibody positivity at diagnosis of type 1 diabetes (T1D dx) from a multivariable logistic regression model. Results shown are the corresponding ORs and 95% CIs. ORs >1 reflect higher relative likelihood of being single autoantibody–positive at diagnosis, and ORs <1 reflect a lower relative likelihood (lower odds) of having a single autoantibody (i.e., higher relative odds of being multiple autoantibody–positive at diagnosis).
There was a significant interaction effect of age at diagnosis with being a carrier of a TCF7L2 rs4506565 T allele (P < 0.03) in relation to expression of single autoantibody positivity. To study the influence of age, we conducted cut point analyses, which indicated that 12 years was an optimal cut point in differentiating those who presented with single versus multiple positive autoantibodies at type 1 diabetes diagnosis. Among participants 12 years or older at diagnosis (n = 504), 250 (50%), 215 (43%), and 39 (8%) carried, respectively, zero, one, or two minor alleles, and those who carried one or two minor alleles were more than twice as likely to have a single positive autoantibody at diagnosis than those who did not carry the allele (OR 2.17 [95% CI 1.32, 3.57], P = 0.002) after controlling for age, BMI z-score, and subgroup. On the other hand, when we evaluated this same model in children younger than 12 years at diagnosis (n = 306), 158 (52%), 126 (41%), and 22 (7%) carried, respectively, zero, one, or two minor alleles, and the model was not statistically significant (P = 0.73). Additional adjustment for HLA, sex, and fasting C-peptide did not affect either of the multivariable models, and these were not significant factors for having single or multiple autoantibody positivity at diagnosis overall or in either age category. Of note, the age of onset distributions were very similar in carriers (median 13.8 years, interquartile range 10.3–18.1, range 3.4–48.6) and noncarriers (median 13.5 years, interquartile range 9.9–18.9, range 3.3–58.6) (Wilcoxon rank sum test, P = 0.82).
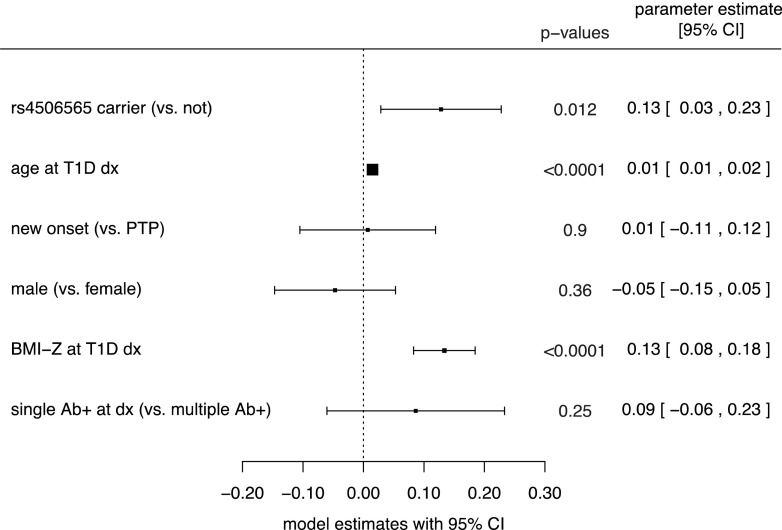
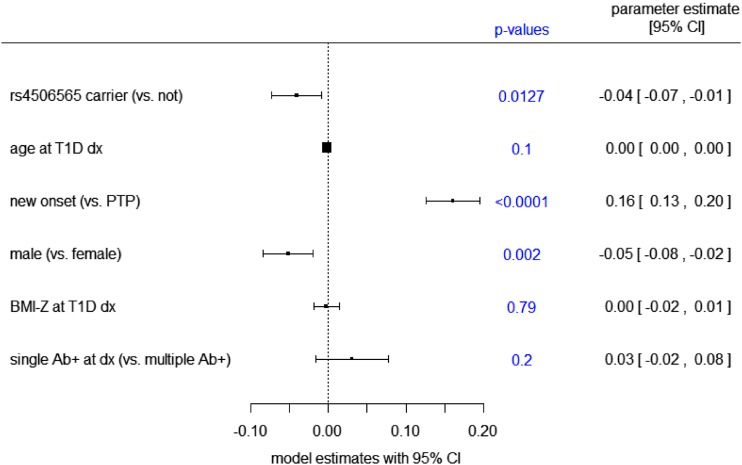
Next, we modeled the influence of carrying a TCF7L2 rs450656 T allele on the OGTT-stimulated mean C-peptide AUC. Race and cohort were not significant factors and were excluded from the model. Participants who carried a TCF7L2 minor allele had a higher C-peptide AUC than noncarriers (P = 0.012) after adjustment for age, sex, BMI z-score, number of islet autoantibodies (single or multiple), and subgroup (Fig. 2). Modeling of the relationship between carrying a TCF7L2 minor allele and the mean glucose AUC indicated that the SNP was an independent, significant factor of a lower glucose AUC (P = 0.013) after adjustment for age, sex, BMI z-score, number of islet autoantibodies (single or multiple), and cohort (Fig. 3).
Figure 2.
Forest plot representing the influence of the shown measures on the mean log-transformed AUC for C-peptide at type 1 diabetes diagnosis (T1D dx) based on a multivariable generalized linear regression model. Parameter estimates are the estimated regression coefficients from the model. Estimates <0 indicate a negative relationship with the log-transformed mean AUC for C-peptide levels, and estimates >0 indicate a positive relationship with the log-transformed mean AUC for C-peptide levels. Ab+, autoantibody-positive.
Figure 3.
Forest plot representing the influence of the shown measures on mean log-transformed AUC glucose at type 1 diabetes (T1D) diagnosis (dx) based on a multivariable generalized linear regression model. Parameter estimates are the estimated regression coefficients from the model. Estimates <0 indicate a negative relationship with the log-transformed mean AUC for glucose levels, and estimates >0 indicate a positive relationship with the log-transformed mean AUC for glucose levels. Ab+, autoantibody-positive.
Restricting the analysis to non-Hispanic white participants (n = 689), we found that the association between single autoantibody positivity and carrying a type 2 diabetes–associated TCF7L2 allele was a significant factor in those 12 years old and older at diagnosis (OR 1.92, P = 0.016), even adjusting for continuous age at diagnosis, BMI z-score, subgroup, and HLA DR3/DR4. These results were consistent with those of the overall cohort regardless of race and ethnicity. Similarly, the results remained consistent for models of glucose and C-peptide measures when the analyses were restricted to the non-Hispanic white participants. Furthermore, the results held up for the end points of single autoantibody positivity as well as glucose and C-peptide measures when analyzing white participants only, regardless of ethnicity. Supplementary Table 3 illustrates TCF7L2 allele frequencies by race/ethnicity.
Conclusions
We studied 810 TrialNet participants with newly diagnosed type 1 diabetes and found that among individuals 12 years and older, the type 2 diabetes–associated TCF7L2 genetic variant is more frequent in those presenting with a single autoantibody than in participants who had multiple autoantibodies. These TCF7L2 variants were also associated with higher mean C-peptide AUC and lower mean glucose AUC levels at the onset of type 1 diabetes. To our knowledge, this is the first study to report that a type 2 diabetes–associated TCF7L2 SNP is associated with a metabolic phenotype among individuals with type 1 diabetes.
These findings suggest that, besides the well-known link with type 2 diabetes, the TCF7L2 locus may play a role in the development of type 1 diabetes. The type 2 diabetes–associated TCF7L2 genetic variant identifies a subset of individuals with autoimmune type 1 diabetes and fewer markers of islet autoimmunity, lower glucose, and higher C-peptide at diagnosis. Our data provide a genetic explanation for some of the heterogeneity of the disease manifestations and support the concept of differential diabetes phenotypes, which has implications for disease prevention and treatment. For example, selected individuals with or at risk for type 1 diabetes may benefit from therapies such as lifestyle modification, metformin, or incretin enhancers.
A possible interpretation of these data is that TCF7L2-encoded diabetogenic mechanisms may contribute to diabetes development in individuals with limited autoimmunity, as exemplified by a single autoantibody (28). Because the risk of progression to type 1 diabetes is lower in individuals with single compared with multiple autoantibodies, it is possible that in the absence of this type 2 diabetes–associated TCF7L2 variant, these individuals may have not manifested diabetes. If that is the case, we would postulate that disease development in these patients may have a type 2 diabetes–like pathogenesis in which islet autoimmunity is a significant component but not necessarily the primary driver. The mechanisms that underlie the metabolic abnormalities observed in carriers of the TCF7L2 genetic variants in the current study are unknown. However, TCF7L2 has been reported to play a role in various aspects of glucose metabolism (17–19), including a possible regulatory role of a TCF7L2 variant on ACSL5 expression and thus, potentially, on insulin sensitivity (20,21). Further testing of this hypothesis may be achieved by treating patients with therapies that are relevant to the disease phenotype and genetic background. If a therapeutic benefit is shown, this would provide credence for the presence of a type 2 diabetes–like pathway and that the phenotypic characterization of diabetes subtypes may be useful to inform individualized treatment decisions in clinical practice (2,29).
Age was a significant modifier of the relationship of the TCF7L2 genetic variant with the number of islet autoantibodies at diagnosis of type 1 diabetes, despite similar age distributions between carriers and noncarriers of the type 2 diabetes–associated allele. The association between this genetic variant and single autoantibody positivity was present in individuals 12 years or older but not in children younger than 12 years. This may explain why the difference in the proportion of carriers between single and multiple autoantibody–positive children and young adults was not statistically significant in a previous analysis by Yu et al. (30), which did not stratify by age, as well as findings in previously negative studies (5–7). The results in the current study suggest that the type 2 diabetes–associated TCF7L2 genetic variant plays a larger role in older individuals. There is mounting evidence that the pathogenesis of type 1 diabetes varies by age (31). Younger individuals appear to have a more aggressive form of disease, with faster decline of β-cell function before and after onset of disease, higher frequency and severity of diabetic ketoacidosis, which is a clinical correlate of severe insulin deficiency, and lower C-peptide at presentation (31–35). Furthermore, older patients are less likely to have type 1 diabetes–associated HLA alleles and islet autoantibodies (28). Individuals with less aggressive, slowly progressive islet autoimmunity, who would otherwise never or only later in life progress to clinical type 1 diabetes (28), may present earlier (but still not younger than 12 years) because of the concurrence of a second diabetogenic factor, namely, a TCF7L2 variant. It is possible that, in individuals with aggressive islet autoimmunity, the abnormalities associated with TCF7L2 are not obvious as a result of the stronger effect on glucose metabolism of profound insulin deficiency. The heterogeneity of autoimmune diabetes is underscored by the influence of age and obesity on the effects of the TCF7L2 variants observed by us and others (8,10,11).
Limitations of this study include its cross-sectional design in capturing autoantibody-positive at-risk subjects and lack of prediagnosis data on those in the New Onset trials, which does not allow causal inference. Studies are underway to understand the role of the TCF7L2 genetic variant before the diagnosis of type 1 diabetes. Although some of the positive islet autoantibodies could possibly correspond to assay false positives, the probability of this is low in both subgroups included in this study: TrialNet New Onset clinical trial participants had a prior clinical diagnosis of type 1 diabetes and were subsequently tested for autoantibody positivity before inclusion in clinical trials, and for the TrialNet PTP participants, single autoantibody positivity must be confirmed on a second blood sample according to the study protocol. A strength of the study is the extensive immunologic and metabolic characterization of this large sample of individuals with newly diagnosed type 1 diabetes.
Taken together, we have demonstrated that individuals with autoimmune type 1 diabetes who carry the type 2 diabetes–associated TCF7L2 genetic variant have a distinct phenotype characterized by milder immunologic and metabolic characteristics than noncarriers, closer to those of type 2 diabetes, with an important effect of age. These results provide a genetic basis for the phenotypic heterogeneity of type 1 diabetes that may provide new insights into therapy and prevention.
Supplementary Material
Article Information
Funding. The sponsor of the trial was the Type 1 Diabetes TrialNet Study Group. The Type 1 Diabetes TrialNet Study Group is a clinical trials network funded by the National Institutes of Health (NIH) through the National Institute of Diabetes and Digestive and Kidney Diseases, the National Institute of Allergy and Infectious Diseases, and the Eunice Kennedy Shriver National Institute of Child Health and Human Development, through the cooperative agreements U01-DK-061010, U01-DK-061034, U01-DK-061042, U01-DK-061058, U01-DK-085465, U01-DK-085453, U01-DK-085461, U01-DK-085466, U01-DK-085499, U01-DK-085504, U01-DK-085509, U01-DK-103180, U01-DK-103153, U01-DK-085476, U01-DK-103266, U01-DK-103282, U01-DK-106984, U01-DK-106994, U01-DK-107013, U01-DK-107014, and UC4-DK-106993; National Institute of Diabetes and Digestive and Kidney Diseases grant U01-DK-103180-01 to M.J.R. (primary investigator); and JDRF.
The contents of this article are solely the responsibility of the authors and do not necessarily represent the official views of the NIH or the JDRF.
Duality of Interest. No potential conflicts of interest relevant to this article were reported.
Author Contributions. M.J.R. designed the study, interpreted the data, and wrote the manuscript. S.G. contributed to the study design, analyzed the data, contributed to data interpretation, and reviewed and edited the manuscript. A.K.S., J.S., M.A., P.A., A.M., and J.W. contributed to data interpretation and manuscript review and edits. P.X. contributed to data analysis. A.P. contributed to study design, reviewed data, contributed to data interpretation, and reviewed and edited the manuscript. All authors are members of the Type 1 Diabetes TrialNet Study Group (Supplementary Table 1). M.J.R. is the guarantor of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Prior Presentation. Parts of this study were presented in abstract form at the 76th Scientific Sessions of the American Diabetes Association, New Orleans, LA, 10–14 June 2016.
Footnotes
This article contains Supplementary Data online at http://care.diabetesjournals.org/lookup/suppl/doi:10.2337/dc17-0961/-/DC1.
A list of the Type 1 Diabetes TrialNet Study Group members can be found in Supplementary Table 1 online.
See accompanying article, p. 224.
References
- 1.Tuomi T, Santoro N, Caprio S, Cai M, Weng J, Groop L. The many faces of diabetes: a disease with increasing heterogeneity. Lancet 2014;383:1084–1094 [DOI] [PubMed] [Google Scholar]
- 2.Skyler JS, Bakris GL, Bonifacio E, et al. . Differentiation of diabetes by pathophysiology, natural history, and prognosis. Diabetes 2017;66:241–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grant SF, Thorleifsson G, Reynisdottir I, et al. . Variant of transcription factor 7-like 2 (TCF7L2) gene confers risk of type 2 diabetes. Nat Genet 2006;38:320–323 [DOI] [PubMed] [Google Scholar]
- 4.Bonetti S, Trombetta M, Malerba G, et al. . Variants and haplotypes of TCF7L2 are associated with β-cell function in patients with newly diagnosed type 2 diabetes: the Verona Newly Diagnosed Type 2 Diabetes Study (VNDS) 1. J Clin Endocrinol Metab 2011;96:E389–E393 [DOI] [PubMed] [Google Scholar]
- 5.Qu HQ, Polychronakos C. The TCF7L2 locus and type 1 diabetes. BMC Med Genet 2007;8:51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Raj SM, Howson JM, Walker NM, et al. . No association of multiple type 2 diabetes loci with type 1 diabetes. Diabetologia 2009;52:2109–2116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Field SF, Howson JM, Smyth DJ, Walker NM, Dunger DB, Todd JA. Analysis of the type 2 diabetes gene, TCF7L2, in 13,795 type 1 diabetes cases and control subjects. Diabetologia 2007;50:212–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bakhtadze E, Cervin C, Lindholm E, et al. . Common variants in the TCF7L2 gene help to differentiate autoimmune from non-autoimmune diabetes in young (15-34 years) but not in middle-aged (40-59 years) diabetic patients. Diabetologia 2008;51:2224–2232 [DOI] [PubMed] [Google Scholar]
- 9.Andersen MK, Sterner M, Forsén T, et al. . Type 2 diabetes susceptibility gene variants predispose to adult-onset autoimmune diabetes. Diabetologia 2014;57:1859–1868 [DOI] [PubMed] [Google Scholar]
- 10.Lukacs K, Hosszufalusi N, Dinya E, Bakacs M, Madacsy L, Panczel P. The type 2 diabetes-associated variant in TCF7L2 is associated with latent autoimmune diabetes in adult Europeans and the gene effect is modified by obesity: a meta-analysis and an individual study. Diabetologia 2012;55:689–693 [DOI] [PubMed] [Google Scholar]
- 11.Mishra R, Chesi A, Cousminer DL, et al.; Bone Mineral Density in Childhood Study . Relative contribution of type 1 and type 2 diabetes loci to the genetic etiology of adult-onset, non-insulin-requiring autoimmune diabetes. BMC Med 2017;15:88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Basile KJ, Guy VC, Schwartz S, Grant SF. Overlap of genetic susceptibility to type 1 diabetes, type 2 diabetes, and latent autoimmune diabetes in adults. Curr Diab Rep 2014;14:550. [DOI] [PubMed] [Google Scholar]
- 13.Zampetti S, Spoletini M, Petrone A, et al.; NIRAD Study Group. Association of TCF7L2 gene variants with low GAD autoantibody titre in LADA subjects (NIRAD Study 5). Diabet Med 2010;27:701–704 [DOI] [PubMed] [Google Scholar]
- 14.Grant SF, Hakonarson H, Schwartz S. Can the genetics of type 1 and type 2 diabetes shed light on the genetics of latent autoimmune diabetes in adults? Endocr Rev 2010;31:183–193 [DOI] [PubMed] [Google Scholar]
- 15.Szepietowska B, Moczulski D, Wawrusiewicz-Kurylonek N, Grzeszczak W, Gorska M, Szelachowska M. Transcription factor 7-like 2-gene polymorphism is related to fasting C peptide in latent autoimmune diabetes in adults (LADA). Acta Diabetol 2010;47:83–86 [DOI] [PubMed] [Google Scholar]
- 16.Cervin C, Lyssenko V, Bakhtadze E, et al. . Genetic similarities between latent autoimmune diabetes in adults, type 1 diabetes, and type 2 diabetes. Diabetes 2008;57:1433–1437 [DOI] [PubMed] [Google Scholar]
- 17.Lyssenko V, Lupi R, Marchetti P, et al. . Mechanisms by which common variants in the TCF7L2 gene increase risk of type 2 diabetes. J Clin Invest 2007;117:2155–2163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shu L, Matveyenko AV, Kerr-Conte J, Cho JH, McIntosh CH, Maedler K. Decreased TCF7L2 protein levels in type 2 diabetes mellitus correlate with downregulation of GIP- and GLP-1 receptors and impaired beta-cell function. Hum Mol Genet 2009;18:2388–2399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shah M, Varghese RT, Miles JM, et al. . TCF7L2 genotype and α-cell function in humans without diabetes. Diabetes 2016;65:371–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xia Q, Chesi A, Manduchi E, et al. . The type 2 diabetes presumed causal variant within TCF7L2 resides in an element that controls the expression of ACSL5. Diabetologia 2016;59:2360–2368 [DOI] [PubMed] [Google Scholar]
- 21.van de Bunt M. An alternative effector gene at the type 2 diabetes-associated TCF7L2 locus? Diabetologia 2016;59:2292–2294 [DOI] [PubMed] [Google Scholar]
- 22.Redondo MJ, Muniz J, Rodriguez LM, et al. . Association of TCF7L2 variation with single islet autoantibody expression in children with type 1 diabetes. BMJ Open Diabetes Res Care 2014;2:e000008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Redondo MJ, Grant SF, Davis A, Greenbaum C; T1D Exchange Biobank. Dissecting heterogeneity in paediatric type 1 diabetes: association of TCF7L2 rs7903146 TT and low-risk human leukocyte antigen (HLA) genotypes. Diabet Med 2017;34:286–290 [DOI] [PubMed] [Google Scholar]
- 24.Skyler JS, Greenbaum CJ, Lachin JM, et al.; Type 1 Diabetes TrialNet Study Group . Type 1 Diabetes TrialNet–an international collaborative clinical trials network. Ann N Y Acad Sci 2008;1150:14–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mahon JL, Sosenko JM, Rafkin-Mervis L, et al.; TrialNet Natural History Committee; Type 1 Diabetes TrialNet Study Group . The TrialNet natural history study of the development of type 1 diabetes: objectives, design, and initial results. Pediatr Diabetes 2009;10:97–104 [DOI] [PubMed] [Google Scholar]
- 26.American Diabetes Association Classification and diagnosis of diabetes. Sec. 2. In Standards of Medical Care in Diabetes—2017. Diabetes Care 2017;40(Suppl. 1):S11–S2427979889 [Google Scholar]
- 27.Little RR, Rohlfing CL, Tennill AL, et al. . Standardization of C-peptide measurements. Clin Chem 2008;54:1023–1026 [DOI] [PubMed] [Google Scholar]
- 28.Frohnert BI, Ide L, Dong F, et al. . Late-onset islet autoimmunity in childhood: the Diabetes Autoimmunity Study in the Young (DAISY). Diabetologia 2017;60:998–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Florez JC. Precision medicine in diabetes: is it time? Diabetes Care 2016;39:1085–1088 [DOI] [PubMed] [Google Scholar]
- 30.Yu J, Steck AK, Babu S, et al. . Single nucleotide transcription factor 7-like 2 (TCF7L2) gene polymorphisms in antiislet autoantibody-negative patients at onset of diabetes. J Clin Endocrinol Metab 2009;94:504–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wherrett DK, Chiang JL, Delamater AM, et al.; Type 1 Diabetes TrialNet Study Group . Defining pathways for development of disease-modifying therapies in children with type 1 diabetes: a consensus report. Diabetes Care 2015;38:1975–1985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miller KM, Foster NC, Beck RW, et al.; T1D Exchange Clinic Network . Current state of type 1 diabetes treatment in the U.S.: updated data from the T1D Exchange Clinic Registry. Diabetes Care 2015;38:971–978 [DOI] [PubMed] [Google Scholar]
- 33.Parikka V, Näntö-Salonen K, Saarinen M, et al. . Early seroconversion and rapidly increasing autoantibody concentrations predict prepubertal manifestation of type 1 diabetes in children at genetic risk. Diabetologia 2012;55:1926–1936 [DOI] [PubMed] [Google Scholar]
- 34.Greenbaum CJ, Beam CA, Boulware D, et al.; Type 1 Diabetes TrialNet Study Group . Fall in C-peptide during first 2 years from diagnosis: evidence of at least two distinct phases from composite Type 1 Diabetes TrialNet data. Diabetes 2012;61:2066–2073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Redondo MJ, Rodriguez LM, Escalante M, O’Brian Smith E, Balasubramanyam A, Haymond MW. Beta cell function and BMI in ethnically diverse children with newly diagnosed autoimmune type 1 diabetes. Pediatr Diabetes 2012;13:564–571 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.