Abstract
Mitophagy is a cellular quality-control pathway, which is essential for elimination of unhealthy mitochondria. While mitophagy is critical to pancreatic β-cell function, the posttranslational signals governing β-cell mitochondrial turnover are unknown. Here, we report that ubiquitination is essential for the assembly of a mitophagy regulatory complex, comprised of the E3 ligase Nrdp1, the deubiquitinase enzyme USP8, and Clec16a, a mediator of β-cell mitophagy with unclear function. We discover that the diabetes gene Clec16a encodes an E3 ligase, which promotes nondegradative ubiquitin conjugates to direct its mitophagy effectors and stabilize the Clec16a-Nrdp1-USP8 complex. Inhibition of the Clec16a pathway by the chemotherapeutic lenalidomide, a selective ubiquitin ligase inhibitor associated with new-onset diabetes, impairs β-cell mitophagy, oxygen consumption, and insulin secretion. Indeed, patients treated with lenalidomide develop compromised β-cell function. Moreover, the β-cell Clec16a-Nrdp1-USP8 mitophagy complex is destabilized and dysfunctional after lenalidomide treatment as well as after glucolipotoxic stress. Thus, the Clec16a-Nrdp1-USP8 complex relies on ubiquitin signals to promote mitophagy and maintain mitochondrial quality control necessary for optimal β-cell function.
Introduction
Pancreatic β-cells rely on mitochondrial bioenergetics to fuel glucose-stimulated insulin secretion (GSIS), which is essential for normal glucose homeostasis and is impaired in all forms of diabetes (1). Beyond respiratory function, the mass of healthy mitochondria, regulated by a balance of mitochondrial biogenesis and turnover, is vital to maintain energy demands for proper stimulus-secretion coupling. Mitochondrial autophagy (mitophagy) is a quality-control mechanism necessary for the selective elimination of dysfunctional mitochondria to maintain optimal mitochondrial respiration in many cell types, including β-cells (2,3). Despite the importance of mitophagy to β-cell function (4–7), the molecular signals dictating mitophagy and their significance to mitochondrial health remain largely undefined.
Ubiquitination of protein substrates, catalyzed by the linkage of activated ubiquitin by three distinct enzymes (E1, E2, and E3), leads to diverse responses, including substrate degradation by the ubiquitin-proteasome system, protein trafficking, complex assembly, and cellular signaling (8). Posttranslational modification of downstream mitophagy effectors, including the E3 ligase Parkin, by ubiquitination is necessary for efficient mitochondrial turnover (9); however, evidence of posttranslational control of proximal mitophagy regulators has not been identified. The E3 ubiquitin ligase neuregulin receptor degradation protein 1 (Nrdp1) and the deubiquitination enzyme ubiquitin-specific protease 8 (USP8) counterbalance ubiquitin dynamics to direct Parkin action or its proteasomal degradation (10,11). Nrdp1 controls Parkin-mediated mitophagy in β-cells through its interaction with, and regulation by, the diabetes gene C-type lectin domain family 16, member A (Clec16a) (4,5); however, the precise action of Clec16a in mitophagy remains poorly understood. While these factors (Nrdp1, USP8, and Clec16a) act upstream as sentinels to ensure that mitophagy is utilized appropriately, the role of ubiquitination as a control mechanism for their actions is unknown.
Here, we discover that formation of a ubiquitin-dependent Clec16a-Nrdp1-USP8 complex is necessary for control of mitophagy in β-cells. We identify that inhibition of Clec16a-mediated Nrdp1 ubiquitination by the chemotherapeutic agent lenalidomide impairs Clec16a-mediated mitophagy and, ultimately, reduces β-cell function. We demonstrate that Clec16a is an E3 ubiquitin ligase, which ubiquitinates Nrdp1 to promote assembly of the Clec16a-Nrdp1-USP8 mitophagy complex. Lastly, we determine that mobilization and function of the β-cell Clec16a-Nrdp1-USP8 mitophagy complex is disrupted by glucolipotoxicity.
Research Design and Methods
Patient Samples
Samples were provided from de-identified multiple myeloma patients without diabetes by the University of Michigan Hematologic Malignancies Tissue Bank and in compliance with the University of Michigan Institutional Review Board. Fasting samples were collected prior to chemotherapy and again 12 weeks later. Patients received a combination of perifosine, bortezomib, liposomal doxorubicin, Bexxar, and/or carfilzomib in the presence/absence of lenalidomide (25 mg PO daily for 28 days × 4 cycles). All patients received corticosteroids (dexamethasone 40 mg PO daily × 12 doses/cycle). C-peptide and glucose concentrations were measured by chemiluminescence and the glucose oxidase method, respectively, by the Michigan Diabetes Research Center Clinical Core at the University of Michigan. HOMA of β-cell function (HOMA-β) and insulin resistance were calculated by HOMA2 (12).
Animals
Animal studies were approved by the University of Michigan Institutional Animal Care and Use Committee. Clec16aloxP mice (5) were mated to MIP-CreERT2 mice (13) to generate experimental groups. Recombination was induced with three i.p. injections of 150 μg/g body wt tamoxifen (Cayman Chemical) every other day over 5 days. 10-week old C57BL/6 mice were injected daily with 50 mg/kg of lenalidomide i.p. or vehicle control for 3 weeks. Mice were left to recover for 1 week before undergoing glucose tolerance testing. Animals were housed on a standard 12-h light/12-h dark cycle with ad libitum access to food and water unless fasted prior to testing.
Antibodies
A full listing of antibodies is provided in Supplementary Table 1.
Proximity Ligation
Studies were performed with a commercially available kit (Duolink; Sigma-Aldrich) per the manufacturers’ protocols utilizing Nrdp1-specific (Bethyl Laboratories) and USP8-specific (Sigma-Aldrich) antisera. Counterstaining to identify β-cells was performed in dissociated human islets utilizing PDX-1 antisera (Santa Cruz Biotechnology). Proximity ligation events were quantified via ImageJ similar to published approaches (14). Imaging was performed with an IX81 microscope (Olympus). Z-stack images were captured using an ORCA-Flash.4 CMOS digital camera (Hamamatsu Photonics) and then subjected to deconvolution (cellSens; Olympus). Co- localization analysis was performed as previously described (5).
Islet Isolation and Tissue Culture
Primary dermal fibroblasts from wild-type (WT) and Ubc-CreERT;Clec16alox/lox mice were treated ex vivo with tamoxifen as previously described (5). Stably expressing Flag-Clec16a NIH3T3 cells, and Min6 cells (a gift from Dr. Doris Stoffers), were maintained as previously described (5). Mouse islets were isolated and cultured as previously described (15). Cell treatments included DMSO (Fisher BioReagents), lenalidomide (Ark Pharm), chloroquine (Santa Cruz Biotechnology), carbonyl cyanide 4-(trifluoromethoxy)phenylhydrazone (FCCP) (Sigma-Aldrich), bafilomycin A1, MG132, and cycloheximide (all from EMD Millipore). Palmitate/BSA couplings were generated as previously described (16).
Human Islets
Studies performed on human islets procured from the Integrated Islet Distribution Program approved by the University of Michigan Institutional Review Board. Donor information is provided in Supplementary Table 2. Human islets were cultured in PIM(S) (Prodo Laboratories) supplemented with 10% FBS, 1% antibiotic/antimycotic (Life Technologies), and 1% PIM(G) (Prodo Laboratories). After culture and treatments, islets were dissociated into single cells using trypsin, cytocentrifuged to glass slides, and prepared for immunostaining.
Transfections
Transfection of human embryonic kidney (HEK)293T, HeLa, and NIH3T3 cells (Lipofectamine 2000; Life Technologies) and Min6 cells (Amaxa Nucleofector) was performed as previously described (5). A detailed list of plasmids utilized or generated for this study can be found in Supplementary Data.
Protein Expression/Purification
Recombinant Clec16a and Nrdp1 were expressed in NiCo21 (DE3) strain Escherichia coli (New England Biolabs) after culture in auto-induction media as previously described (17). Glutathione S-transferase (GST)-tagged and 6xHis-tagged proteins were purified utilizing glutathione matrix or nickel-charged resin (HiCap and Ni-NTA agarose; Qiagen) per the manufacturers’ protocols. In vitro transcribed/translated Clec16a and Nrdp1 were expressed in T7-driven mammalian expression systems (Promega) per the manufacturers’ protocols. Clec16a and Nrdp1 protein expression/purification was confirmed by silver staining (Bio-Rad) and/or Western blotting.
Ubiquitination Assays
Detection of protein ubiquitination in vivo was performed in cell lines similar to previously described approaches (18). Detection of protein ubiquitination in vitro was performed after incubation of 1 µmol/L GST-Clec16a-Flag or Clec16a-6xHis-Flag with a commercially available kit (E2 Select; Bio-Techne). Ubiquitin conjugation assays were performed with 1 µmol/L GST-Clec16a-Flag/Clec16a-6xHis-Flag, 2.5 µmol/L GST–Nrdp1 CSHQ, 50 nmol/L E1, 5 µmol/L Ube2d3/Ube2c, 50 µmol/L HA-tagged (HA-)ubiquitin, Mg-ATP, and conjugation reaction buffers (Bio-Techne). (A description of Nrdp1 CSHQ appears below.) Conjugation assays were performed in solution for 60 min at 37°C. Ubiquitination and protein interaction assays from in vitro transcribed/translated Flag-tagged (Flag)-Clec16a and HA-Nrdp1 were incubated in 40 mmol/L Tris-HCl (pH 7.6), 5 mmol/L MgCl2, and 2 mmol/L dithiothreitol with or without the addition of 75 µmol/L 4[4-(5-nitro-furan-2-ylmethylene)-3,5-dioxo-pyrazolidin-1-yl]-benzoic acid ethyl ester (PYR-41) (EMD Millipore) for 3 h at 37°C, similar to previously published approaches (19).
Glucose/Insulin Measurements, Respirometry, Gene Expression, Immunoprecipitation, Immunofluorescence, and Western Blotting Assays
All assays were performed as previously described (5).
Statistics
Data are presented as mean ± SEM, and error bars denote SEM. Statistical comparisons were performed using one- or two-tailed Student t test or two-way ANOVA as appropriate (Prism GraphPad). A P value <0.05 was considered significant.
Results
Inhibition of Nrdp1 Ubiquitination Impairs Human β-Cell Function and Mitophagy
We hypothesized that ubiquitination of upstream mitophagy regulators regulates β-cell mitochondrial quality control to preserve the well-functioning mitochondria necessary for proper stimulus-secretion coupling and GSIS. To this end, we assayed β-cell function in multiple-myeloma patients who received lenalidomide, a chemotherapeutic agent and selective ubiquitin ligase inhibitor (20), which, along with its parent compound thalidomide, is associated with new-onset glucose intolerance (21,22). Lenalidomide has also been shown to impair ubiquitination of the E3 ligase Nrdp1 (23), an essential regulator of β-cell mitophagy (4,5). To evaluate the effect of lenalidomide in vivo, we estimated β-cell function by HOMA-β in patients treated with or without lenalidomide for 12 weeks (Fig. 1A and Supplementary Table 3). Although all patients received glucocorticoids as part of their treatment regimen (see methods described above), none had prior documented diabetes or glucose intolerance. Interestingly, we observed decreased HOMA-β (Fig. 1A) in lenalidomide-treated patients. Impaired β-cell function was not secondary to advanced age or increased insulin resistance, as lenalidomide-treated patients were younger and had no differences in HOMA of insulin resistance compared with control subjects (Supplementary Table 3 and data not shown). We also observed that short-term i.p. lenalidomide treatment (3 weeks) induced modest glucose intolerance in mice (Supplementary Fig. 1A and B); however, the milder effects of lenalidomide in mice could be attributed to the rapid clearance of i.p. lenalidomide (versus oral) as well as a shorter duration of exposure (24). We also confirmed that lenalidomide impaired ubiquitination of Nrdp1 in Min6 β-cells (Supplementary Fig. 1C). Further, lenalidomide significantly reduced both GSIS and oxygen consumption in isolated islets (Fig. 1B and C).
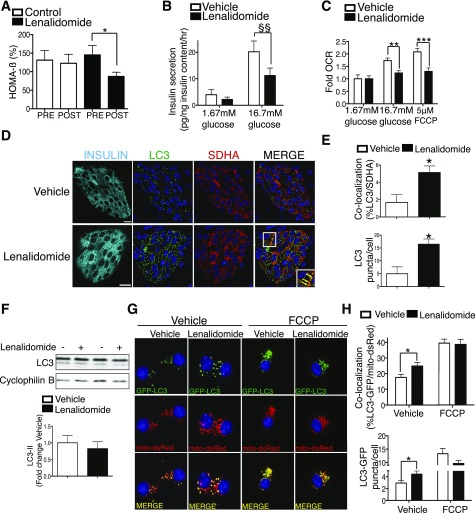
Figure 1.
The selective ubiquitin ligase inhibitor lenalidomide impairs β-cell function, Nrdp1 ubiquitination, and mitophagy. A: Calculated HOMA-β measured in multiple myeloma patients preinitiation (pre) and 12 weeks postinitiation (post) of chemotherapy (n = 11 control and 14 lenalidomide). B: GSIS after static incubations in 1.67 mmol/L and 16.7 mmol/L glucose performed in isolated CD1 islets pretreated with vehicle (DMSO) or 10 μmol/L lenalidomide for 24 h (hr) (n = 4/group). C: Fold oxygen consumption rates (OCR) in isolated islets pretreated with vehicle or 10 μmol/L lenalidomide for 24 h prior to dispersion to single cells for analysis as adherent monolayers on Seahorse flux analyzer (n = 10/group). D: Representative images of pancreas sections from C57BL/6 mice treated for 3 weeks in vivo with 50 mg/kg lenalidomide or vehicle alone, stained for insulin (cyan), LC3 (green), and SHDA (red). Nuclei are demarcated with DAPI (blue). Inset (lower right): Focused area of LC3/SDHA colocalized structures demarcated by yellow arrows. Scale bars = 10 µm. n = 4/group. E: Quantification of LC3/SDHA colocalization and total LC3+ puncta in islets from C57BL/6 mice treated in vivo with or without 50 mg/kg lenalidomide (n = 4/group and ∼900 β-cells and ∼25,000 total LC3/SDHA structures quantified/group). F: Representative Western blot of LC3 levels in isolated islets treated with vehicle or 10 μmol/L lenalidomide for 24 h with quantification (by densitometry) of LC3-II levels normalized to cyclophilin B. n = 4/group. G: Representative deconvolution images of Min6 β-cells (transfected with LC3-GFP and mito-DsRed plasmids) then treated with vehicle or 10 μmol/L lenalidomide for 24 h with addition of 5 µmol/L FCCP for the final 4 h. n = 4/group. H: Quantification of LC3-GFP/mito-DsRed and mito-DsRed colocalization and total LC3-GFP puncta in Min6 β-cells treated with vehicle or 10 μmol/L lenalidomide for 24 h with addition of 5 µmol/L FCCP for the final 4 h. (n = 4/group and ∼1,500 total GFP and DsRed puncta quantified per experiment.) *P < 0.05, **P < 0.01, ***P < 0.001 by nonpaired Student t test; $$P < 0.01 by paired Student t test.
Given the importance of Nrdp1 to mitophagy and maintenance of insulin secretion, we hypothesized that lenalidomide would lead to dysfunctional mitophagy in β-cells. Indeed, the β-cells of lenalidomide-treated mice developed an accumulation of autophagosomes containing mitochondria, as shown by increased colocalization of the autophagosome marker LC3 with the mitochondrial protein succinate dehydrogenase A (SDHA) Fig. 1D and E). Additionally, lenalidomide-treated islets developed an increase in the total number of LC3+ puncta (Fig. 1E) without overt changes in LC3-I/II protein levels (Fig. 1F), suggesting that lenalidomide disrupts selective rather than bulk autophagy. As a complementary approach, we investigated the effects of lenalidomide on mitophagic flux by assessing the colocalization of mitochondria with autophagosomes using fluorescent-labeled markers (mitochondria, mito-DsRed; autophagosomes, LC3-GFP) in Min6 β-cells at baseline and after mitochondrial damage by the uncoupler FCCP (5). Concordantly, lenalidomide treatment led to increased colocalization of LC3-GFP and mito-DsRed (Fig. 1G and H). It is unlikely that lenalidomide enhances mitophagic flux, however, as the increased colocalization of mito-DsRed and LC3-GFP at baseline is not maintained after FCCP treatment (Fig. 1G and H), suggesting that dysfunctional mitochondria are not efficiently removed. Taken together, we observe that lenalidomide impairs glucose homeostasis, GSIS, mitochondrial respiration, and Nrdp1 ubiquitination, and our results implicate the importance of Nrdp1 ubiquitination in maintenance of mitophagy and β-cell function.
Clec16a Promotes Ubiquitination of Nrdp1
Protein ubiquitination leads to several posttranslational effects, including targeting for proteasomal degradation, trafficking, or the activation of downstream signaling cascades. Our observation of a lenalidomide-sensitive effect on Nrdp1 ubiquitination and β-cell mitophagy led us to assess the role of Nrdp1 interaction partners that have downstream consequences on mitophagy. Nrdp1 interacts with two upstream regulators of mitophagy: Clec16a and the deubiquitinase (DUB) enzyme USP8 (5,25). Specifically, Clec16a prevents the proteasomal degradation of Nrdp1 to control downstream events in β-cell mitophagy (5); therefore, we investigated the role of Clec16a on Nrdp1 stability and ubiquitination. Clec16a deficiency decreased Nrdp1 levels and protein stability (Supplementary Fig. 2A and C), while Clec16a overexpression increased Nrdp1 protein accumulation, consistent with previous reports in β-cells (4). Surprisingly, we observed that Clec16a overexpression resulted in enhanced Nrdp1 ubiquitination, while Clec16a knockout primary fibroblasts exhibited reduced Nrdp1 ubiquitination (Fig. 2A–C and Supplementary Fig. 2B).
Figure 2.
Clec16a promotes the ubiquitination of Nrdp1. A: Representative Western blot (WB) of in vivo ubiquitination assay of overexpressed HA-Nrdp1 performed in HEK293T cells transfected with Myc-tagged ubiquitin (Myc-Ub), Flag–empty vector, Flag-Clec16a, or Flag-USP8 (as well as whole-cell lysate HA-Nrdp1, Flag-Clec16a, and Flag-USP8 levels) treated with DMSO (-) or 20 μmol/L MG132 (+) overnight. n = 3/group. B: Representative Western blot of in vivo ubiquitination assay of endogenous Nrdp1 performed in 293T cells transfected with Myc-ubiquitin and Flag–empty vector or Flag-Clec16a (as well as whole-cell lysate Nrdp1 and Flag-Clec16a levels). n = 3/group. C: Representative WB of in vivo endogenous ubiquitination assay of endogenous Nrdp1 ubiquitination performed in primary dermal fibroblasts derived from WT and Clec16a-deficient mice (as well as whole-cell lysate Nrdp1 levels). n = 3/group. D: Representative Western blot of in vivo ubiquitination assay of overexpressed HA-WT or C34S H36Q mutant (CSHQ) Nrdp1 performed in HEK293T cells transfected with Myc-ubiquitin and Flag–empty vector or Flag-Clec16a (as well as whole-cell lysate HA-Nrdp1 and Flag-Clec16a levels). n = 3/group. E: Representative Western blot of in vivo ubiquitination assay of overexpressed HA-Nrdp1 performed in HEK293T cells transfected with Myc-ubiquitin and Flag–empty vector or Flag-Clec16a (as well as whole-cell lysate HA-Nrdp1 and Flag-Clec16a levels) treated with DMSO (−) or 10 μmol/L lenalidomide (+) for 24 h. n = 3/group. F: GSIS after static incubations in 1.67 mmol/L and 16.7 mmol/L glucose performed in isolated MIP-CreERT2 control or β-Clec16aKO islets pretreated with vehicle (DMSO) or 10 μmol/L lenalidomide for 24 h (hr) (n = 4/group). *P < 0.05 nonpaired Student t test. IP, immunoprecipitation.
Clec16a-mediated enhancement of Nrdp1 ubiquitination and stability is in contrast to the effects of USP8, a DUB enzyme that stabilizes Nrdp1 by opposing its auto-ubiquitination and preventing proteasomal degradation (Fig. 2A and ref. 25). We tested the importance of USP8 to Clec16a-mediated Nrdp1 stability by both gain- and loss-of-function approaches. We reproduced published findings of enhanced Nrdp1 levels after USP8 overexpression (Supplementary Fig. 2D) (25). We observed no changes in basal Nrdp1 levels with USP8 short hairpin RNA (shRNA) (Supplementary Fig. 2E), consistent with previous studies (26), possibly suggesting that other factors stabilize Nrdp1 after USP8 loss of function. Clec16a increased Nrdp1 levels independent of USP8 levels (Supplementary Fig. 2D and E), suggesting that USP8 and Clec16a act via distinct, yet complementary, means to promote Nrdp1 protein accumulation. While Clec16a is an endolysosomal protein, it also appears to promote Nrdp1 levels independent of lysosomal degradation (Supplementary Fig. 2F). To investigate whether Clec16a promotes Nrdp1 ubiquitination by enhancing the auto-ubiquitination activity of Nrdp1, we generated an Nrdp1 RING-domain mutant (C34S H36Q, also known as Nrdp1 CSHQ) lacking E3 ligase activity (27). In the presence of Clec16a, both Nrdp1 WT and Nrdp1 CSHQ exhibited enhanced ubiquitination (Fig. 2D), suggesting that Clec16a promotes Nrdp1 ubiquitination independent of Nrdp1 self-ubiquitination.
The similar effects of Clec16a loss of function and lenalidomide treatment on Nrdp1 ubiquitination led us to hypothesize that lenalidomide may act through Clec16a to impact β-cell function. Indeed, lenalidomide abrogated Clec16a-mediated Nrdp1 ubiquitination (Fig. 2E). To investigate how Clec16a-mediated Nrdp1 ubiquitination affects β-cell function, we studied tamoxifen-inducible and β-cell–specific Clec16a knockout mice (MIP-CreERT2; Clec16aloxP/loxP, hereafter referred to as β-Clec16aKO). β-Clec16aKO mice exhibited reduced Clec16a mRNA levels and impaired glucose tolerance similar to what has been previously described (5) (Supplementary Fig. 3A–C). As expected, lenalidomide treatment or Clec16a deficiency resulted in decreased GSIS compared with vehicle-treated MIP-CreERT2 controls (Fig. 2F). Importantly, we did not observe any additive impairment of lenalidomide on β-Clec16aKO islets (Fig. 2F), suggesting that lenalidomide acts through the Clec16a pathway to regulate Nrdp1 ubiquitination and β-cell function. Notably, lenalidomide treatment did not affect Clec16a mRNA and protein levels (Fig. 2E and Supplementary Fig. 3D). These results suggest that lenalidomide inhibits Clec16a ubiquitination of Nrdp1 and that this activity is essential for downstream maintenance of maximal glucose-stimulated insulin release from β-cells.
Clec16a Is an E3 Ubiquitin Ligase
Despite the connection between Clec16a and mitophagy/autophagy (5,28,29), the direct molecular/enzymatic function for Clec16a has remained elusive. We questioned whether Clec16a possesses intrinsic ubiquitin ligase activity and investigated its capacity to induce ubiquitination in vitro. A vital characteristic of E3 ubiquitin ligases is their ability to auto-ubiquitinate (30), and thus we screened 26 independent E2 conjugation enzymes for their ability to participate in Clec16a auto-ubiquitination. Recombinant Clec16a protein was incubated with each E2 enzyme and evaluated for the presence of an increased molecular weight shift by SDS-PAGE, which would be suggestive of ubiquitinated Clec16a.
Interestingly, we observed that both UbE2C and UbE2D3 were able to promote high–molecular weight Clec16a protein (Fig. 3A). Concordantly, we observed that recombinant Clec16a (Fig. 3B and C) was sufficient to promote auto-ubiquitination in an ATP-, UbE2C/UbE2D3-, and ubiquitin-dependent manner. Moreover, addition of Clec16a to an auto-ubiquitination–deficient Nrdp1 mutant (Nrdp1 CSHQ) led to enhanced Nrdp1 ubiquitination, suggesting that Clec16a directly ubiquitinates Nrdp1 (Fig. 3D).
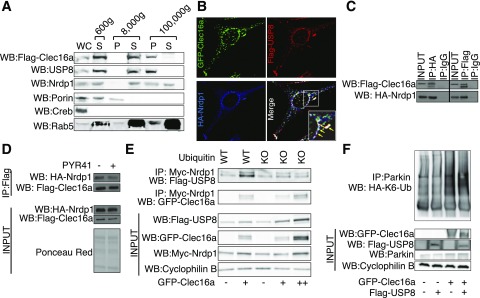
Figure 3.
Clec16a possesses E3 ligase activity and directly ubiquitinates Nrdp1 via a non-K48 linkage. A: Representative α-Flag Western blot (WB) from purified Clec16a-6xHis-Flag protein after incubation in vitro with ATP, ubiquitin, E1, and 26 unique E2-conjugating enzymes at 37°C for 1 h. Arrows demarcate both the expected and novel high–molecular weight Clec16a proteins identified by Western blot. n = 3/group. B and C: Representative Western blot of in vitro ubiquitination assay of recombinant GST-Clec16a-Flag (B) or Clec16a-6xHis-Flag (C) protein after incubation with ATP, HA-ubiquitin (HA-Ub), and E1 in the presence or absence of E2 conjugation enzymes UbE2D3 and/or UbE2C at 37°C for 1 h. n = 3/group. D: Representative Western blot of in vitro ubiquitination assay of recombinant GST-Nrdp1-CSHQ after incubation with ATP, HA-ubiquitin, and E1 in the presence or absence of UbE2D3 and recombinant Clec16a-6xHis-Flag at 37°C for 1 h. n = 3/group. E: Representative WB of in vivo ubiquitination assay of overexpressed Myc-tagged Nrdp1 (Myc-Nrdp1) performed in HEK293T cells transfected with Flag–empty vector or Flag-Clec16a in the presence of HA-WT ubiquitin as well as ubiquitin mutants with all lysines converted to arginines (KO), ubiquitin only capable of K48-linked ubiquitination (K48), ubiquitin incapable of K48-linked ubiquitination (K48R), or ubiquitin only capable of K63-linked ubiquitination (K63) (as well as whole-cell lysate Myc-Nrdp1 and Flag-Clec16a levels). n = 3/group. IP, immunoprecipitation.
Ubiquitination leads to diverse cellular effects, dependent in part upon the linkages in the ubiquitin chain. Linkages can occur on one of seven distinct ubiquitin lysine residues, and proteasomal degradation generally only follows K48-linked ubiquitination (8). Furthermore, K48-independent ubiquitination plays crucial signaling roles in mitophagy (10,31,32). Given that Clec16a promotes Nrdp1 protein accumulation, we postulated that Clec16a might link nondegradative ubiquitin moieties to Nrdp1. Clec16a potentiated WT ubiquitin linkages on Nrdp1, which was lost after the addition of a ubiquitin mutant rendered incapable of ubiquitin laddering by mutagenesis of all seven lysine residues to arginine (KO) (Fig. 3E). Furthermore, Clec16a potentiated Nrdp1 ubiquitination in the presence of a ubiquitin mutant incapable of K48-degradative linkages (K48R) but not in the presence of mutants capable of K48 and K63 linkages only (K48 and K63) (Fig. 3E). This suggests that Clec16a relies upon K48- and K63-independent nondegradative ubiquitin linkages to posttranslationally regulate Nrdp1. Taken altogether, Clec16a is an E3 ligase, which directly ubiquitinates its mitophagy partner and substrate Nrdp1 with nondegradative conjugates.
Clec16a, USP8, and Nrdp1 Interact in a Ubiquitin-Dependent Complex
The convergence of Clec16a and USP8 in the regulation of Nrdp1 and control of mitophagy led us to question whether Clec16a, Nrdp1, and USP8 occupy the same subcellular compartment to participate in a tripartite complex. Clec16a is an endolysosomal protein and may colocalize with Nrdp1 and USP8, which have previously been observed within the endosomal compartment (33). Indeed, we observed that Clec16a as well as Nrdp1 and USP8 were isolated within the membrane protein fraction (which contains endolysosomal proteins) after differential centrifugation (Fig. 4A). Furthermore, we observed Clec16a, Nrdp1, and USP8 colocalization by immunostaining, (Fig. 4B), indicating they do indeed occupy a common compartment.
Figure 4.
Ubiquitination is necessary to maintain the Clec16a-Nrdp1-USP8 protein complex. A: Representative Western blot (WB) after cell fractionation and centrifugation of stably expressing Flag-Clec16a NIH3T3 cells probed for Flag (Clec16a), endogenous Nrdp1, and endogenous USP8 as well as porin (mitochondria), Creb (nuclei), and Rab5 (endosomal/soluble small GTPase) as markers of cell compartments. n = 3/group. B: Representative deconvolution image of NIH3T3 fibroblasts after transfection with GFP-Clec16a, Flag-USP8, and HA-Nrdp1 plasmids with indirect immunofluorescence staining for GFP-Clec16a (green), Flag-USP8 (red), and HA-Nrdp1 (blue). Inset: triple colocalized structures (white) highlighted with yellow arrows. n = 3/group. C: Representative Western blot of reticulocyte lysates after in vitro transcription/translation of Flag-Clec16a and HA-Nrdp1 with anti-HA (left) or anti-Flag (right) immunoprecipitation (IP). n = 3/group. D: Representative Western blot of reticulocyte lysates after in vitro transcription/translation of Flag-Clec16a and HA-Nrdp1 protein treated in solution with 75 μmol/L PYR41 at 37°C for 3 h, after anti-Flag immunoprecipitation. n = 3/group. E: Representative Western blot after anti-Myc immunoprecipitation in HEK293T cells transfected with Myc-tagged Nrdp1 (Myc-Nrdp1) and Flag-USP8 and either GFP–empty vector or GFP-Clec16a, along with WT or KO ubiquitin vectors. n = 3/group. F: Representative WB of in vivo K6-linkage–specific ubiquitination assay of overexpressed Parkin performed in HEK293T cells transfected with HA-K6-ubiquitin, Flag–empty vector or Flag-USP8, and GFP–empty vector or GFP-Clec16a (as well as whole-cell lysate GFP-Clec16a, Flag-USP8, and Parkin levels). n = 3/group. P, pellet; S, supernatant; WC, whole cell.
To determine the role of ubiquitination on complex assembly, we studied the interaction of in vitro transcribed/translated Clec16a and Nrdp1 expressed in reticulocyte lysates and verified their interaction in this system (Fig. 4C). We next coincubated in vitro–derived Clec16a and Nrdp1 with the E1 initiation inhibitor PYR-41 to test the role of ubiquitination independent of its cytotoxic effects observed in vivo (19). PYR-41 reduced the interaction between Clec16a and Nrdp1 (Fig. 4D), implicating ubiquitination in maintenance of the Clec16a-Nrdp1 complex. We next determined the role of ubiquitination and Clec16a in stabilization of Nrdp1-USP8 binding utilizing WT and mutant ubiquitin. While Clec16a enhanced Nrdp1-USP8 complex assembly, addition of the ubiquitin KO mutant reduced Nrdp1 levels as well as Nrdp1 interaction with both Clec16a and USP8 (Fig. 4E). To clarify whether impaired ubiquitination affected the Clec16a-Nrdp1-USP8 complex as a result of reduced levels of constituents or by impaired complex assembly, we also doubled the amount of Flag-Clec16a plasmid in the presence of KO ubiquitin, resulting in increased levels of Clec16a, USP8, and Nrdp1 (Fig. 4E, lane ++). While levels of constituent proteins were rescued, interactions with Nrdp1 were only partially restored with Clec16a—yet not with USP8. Collectively, these findings suggest that functional ubiquitination is necessary for maintaining levels of the constituents of the Clec16a-Nrdp1-USP8 complex as well as providing vital signals for complex assembly.
USP8 shares functional overlap with Clec16a/Nrdp1 in the regulation of mitophagic flux, coinciding in their opposing regulation of the E3 ligase Parkin, a critical effector in mitophagy (34). While Clec16a and Nrdp1 limit excessive Parkin activity to ensure healthy mitochondria are not degraded (5), USP8 promotes Parkin translocation to the mitochondrial outer membrane by removal of K6-linked ubiquitin conjugates (10). Therefore, we investigated whether Clec16a influences the K6-linked ubiquitination state of Parkin. Utilizing a mutant ubiquitin only capable of K6-linked conjugates, we observed the expected reduction of K6-linked Parkin ubiquitination by USP8 (Fig. 4F). Conversely, Clec16a promotes K6-linked Parkin ubiquitination independent of USP8 (Fig. 4F) indicating that Clec16a opposes USP8-mediated K6 deubiquitination of Parkin. Consequently, we conclude that Clec16a, Nrdp1, and USP8 assemble in a tripartite ubiquitin-dependent mitophagy complex to promote maintenance of Parkin.
Nrdp1 Ubiquitination Is Pivotal for Complex Formation and β-Cell Function
Given the importance of ubiquitination to Clec16a-Nrdp1-USP8 complex assembly, we hypothesized that specific ubiquitination of Nrdp1 may influence formation of the Clec16a-Nrdp1-USP8 complex. To investigate how Nrdp1 ubiquitination could influence Clec16a-mediated processes, we generated an Nrdp1 mutant (Nrdp1 K-R), in which all 11 lysine residues were mutated to arginine, thereby rendering it incapable of ubiquitination. As expected, ubiquitin laddering was not observed on the Nrdp1 K-R mutant despite Clec16a overexpression (Fig. 5A). Not surprisingly, overall levels of the Nrdp1 K-R mutant protein were higher than those of WT Nrdp1, potentially due to a loss of degradative Nrdp1 auto-ubiquitination (25) (Fig. 5A). Furthermore, Clec16a was incapable of increasing Nrdp1 levels in the presence of the K-R mutant, reinforcing the importance of Clec16a-mediated Nrdp1 ubiquitination to enhance Nrdp1 stability (Fig. 5A). We next assessed the role of Nrdp1 ubiquitination on formation of the Clec16a-Nrdp1-USP8 complex. Interestingly, Clec16a interacts with endogenous USP8, and this interaction is maintained in the presence or absence of Nrdp1 ubiquitin ligase activity (Fig. 5B). However, Clec16a-USP8 binding as well as USP8 levels are reduced after overexpression of the Nrdp1 K-R mutant (Fig. 5B). These results suggest, again, that Clec16a drives assembly of the Clec16a-Nrdp1-USP8 complex by providing essential ubiquitin signals and stabilizing its component proteins.
Figure 5.
Specific Nrdp1 ubiquitination is vital for β-cell function. A: Representative Western blot (WB) of in vivo ubiquitination assay of overexpressed HA-WT or K-R mutant Nrdp1 performed in HEK293T cells transfected with Myc-tagged ubiquitin (Myc-Ub) and Flag–empty vector or Flag-Clec16a (as well as whole-cell lysate HA-Nrdp1 and Flag-Clec16a levels). n = 3/group. B: Representative Western blot of endogenous USP8 after anti–Flag-Clec16a immunoprecipitation (IP) in HeLa cells transfected with Flag–empty vector or Flag-Clec16a and HA–empty vector or HA-Nrdp1 expression vectors (WT, CSHQ, or K-R). n = 3/group. C: Representative Western blot of HA-Nrdp1 levels in Min6 β-cells 72 h after transfection with nontargeting (NT) or Clec16a-specific shRNA plasmids, as well as HA–empty vector, HA-Nrdp1 WT, or HA-Nrdp1 K-R. (n = 5/group.) D: GSIS after static incubations in 2 mmol/L and 20 mmol/L glucose performed in Min6 β-cells 72 h after transfection with nontargeting or Clec16a-specific shRNA plasmids, as well as HA–empty vector, HA-Nrdp1 WT, or HA-Nrdp1 K-R. (n = 5/group.) E: Top, representative deconvolution image of Nrdp1:USP8 Proximity Ligation Assay (PLA, red) and DNA (DAPI, blue) in Min6 β-cells 24 h after treatment with vehicle (DMSO) or 10 μmol/L lenalidomide; bottom, quantification of relative Nrdp1:USP8 PLA events in Min6 β-cells (fold change vs. vehicle controls) after treatment conditions as noted. (n = 3/group and ∼2,900 PLA events quantified/group per experiment.) #P < 0.05, paired Student t test, **P < 0.01, nonpaired Student t test.
To determine whether the Clec16a-Nrdp1-USP8 axis regulates β-cell function, we assessed the role of Clec16a-mediated ubiquitination on GSIS in Min6 β-cells. We found that shRNA-mediated reduction of Clec16a decreased GSIS (Supplementary Fig. 3E and F). Clec16a shRNA treatment also led to reduced WT-Nrdp1 levels, whereas Clec16a does not modulate levels of the ubiquitin-deficient Nrdp1 K-R mutant (Fig. 5C). Overexpression of WT-Nrdp1, but not Nrdp1 K-R, restored GSIS in shClec16a-treated Min6 β-cells (Fig. 5D), suggesting that Clec16a-mediated ubiquitination of Nrdp1 is necessary for optimal β-cell function. To assess the role of Clec16a-mediated ubiquitination on formation of the β-cell mitophagy complex, we utilized a proximity ligation assay (PLA) to visualize Nrdp1-USP8 association in situ after lenalidomide treatment (35). Lenalidomide impaired Nrdp1-USP8 interaction in Min6 β-cells (Fig. 5E), suggesting that Clec16a-mediated ubiquitination maintains the Clec16a-Nrdp1-USP8 complex as well as β-cell function.
The Clec16a-Nrdp1-USP8 Complex Is Perturbed by Glucolipotoxicity
Defects in β-cell mitochondrial structure and function have been observed under conditions of diabetogenic stimuli, notably, glucolipotoxicity (36,37). To determine whether the combination of reduced Clec16a expression and/or glucolipotoxicity impairs the upstream mitophagy complex, we assessed Nrdp1-USP8 interaction by PLA. As expected, Clec16a- specific shRNA reduced Nrdp1-USP8 binding in situ (Fig. 6A). Nrdp1-USP8 binding was also decreased after palmitate treatment, indicating that glucolipotoxicity destabilizes upstream mitophagy regulators (Fig. 6A). However, palmitate treatment did not additively reduce Nrdp1-USP8 binding in shClec16a-treated Min6 β-cells (Fig. 6A), suggesting that palmitate acts to destabilize the Nrdp1-USP8 complex through effects on the Clec16a pathway. We next investigated the impact of glucolipotoxicity on the Clec16-Nrdp-USP8 complex in human islets. Similarly, glucolipotoxicity decreased Nrdp1-USP8 interaction in primary human β-cells (Fig. 6B). Given our findings suggesting that both Nrdp1 stability and ubiquitination regulate formation of the Clec16a-Nrdp1-USP8 complex, we next measured Nrdp1 levels after palmitate treatment in primary islets. Indeed, we observed a significant reduction in Nrdp1 levels during glucolipotoxic conditions (Fig. 6C). Furthermore, we observed that both Nrdp1 stability and ubiquitination were reduced in Min6 β-cells after palmitate treatment (Fig. 6D), thus demonstrating effects consistent with impact on the Clec16a pathway to maintain the upstream mitophagy complex.
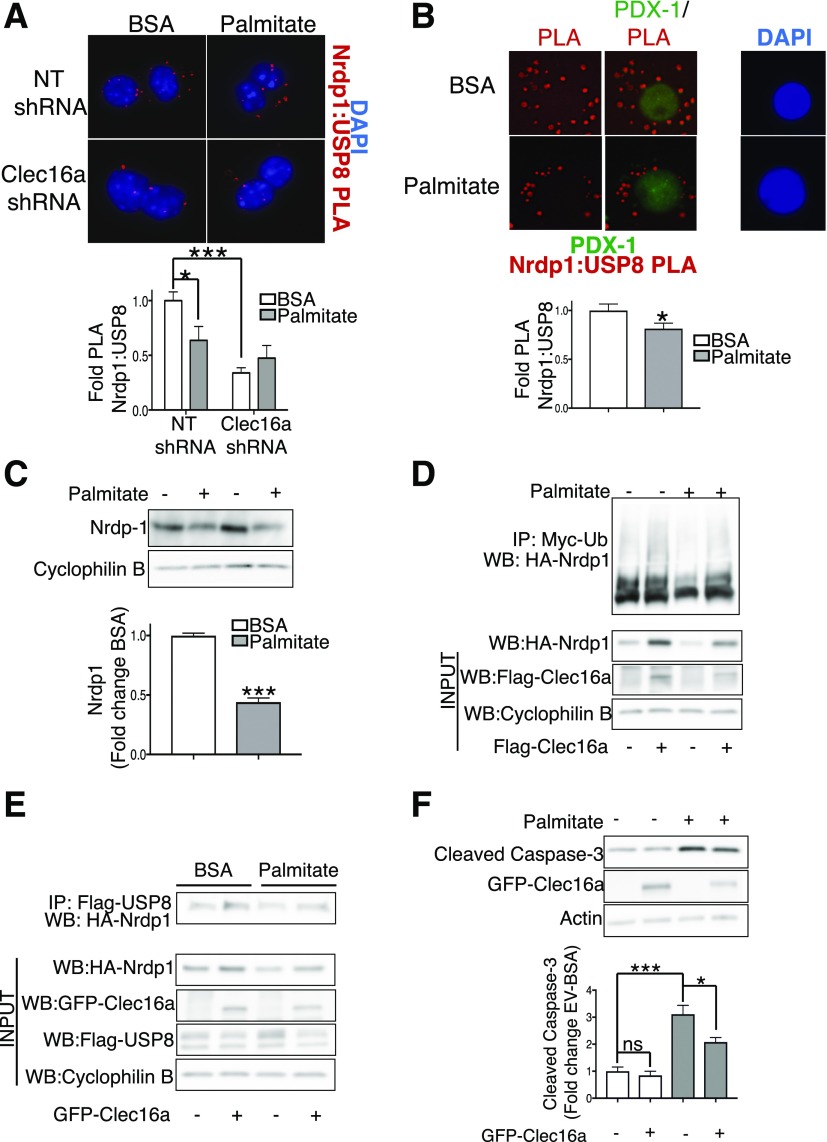
Figure 6.
The Clec16a-Nrdp1-USP8 complex is perturbed by glucolipotoxicity. A: Top, representative deconvolution image of Nrdp1:USP8 Proximity Ligation Assay (PLA, red; DAPI, blue) in Min6 β-cells 72 h after transfection with nontargeting (NT) or Clec16a-specific shRNA plasmids after treatment with BSA control or 0.4 mmol/L palmitate for 48 h; bottom, quantification of relative Nrdp1:USP8 PLA events in Min6 β-cells (fold change vs. NT shRNA BSA controls). (n = 4/group and ∼1,200 PLA events quantified/group per experiment.) B: Top, representative deconvolution image of Nrdp1:USP8 PLA (PLA, red; PDX1, green; and DAPI, blue) in β-cells of dissociated human islets after 48 h treatment with BSA control (BSA) or 0.4 mmol/L palmitate and 0.92% BSA + 20 mmol/L glucose (palmitate); bottom, quantification of relative Nrdp1:USP8 PLA events in β-cells of dissociated human islets after control BSA (white bar) or 0.4 mmol/L palmitate and 0.92% BSA + 20 mmol/L glucose (gray bar) treatment for 48 h (n = 3 donors and ∼900 β-cell PLA events quantified/group per donor). C: Top, representative Western blot (WB) of Nrdp1 levels in isolated mouse islets treated for 48 h with or without 0.4 mmol/L palmitate + 20 mmol/L glucose; bottom, relative quantification (by densitometry) of Nrdp1 levels by Western blot in isolated islets from mice treated for 48 h with (gray bars) or without (white bars) 0.4 mmol/L palmitate and 0.92% BSA + 20 mmol/L glucose. n = 3/group. D: Representative Western blot of in vivo ubiquitination assay of HA-Nrdp1 in Min6 cells transfected with HA-Nrdp1, Myc-tagged ubiquitin (Myc-Ub), and Flag–empty vector or Flag-Clec16a plasmids and treated for 48 h with or without 0.4 mmol/L palmitate/BSA coupling. n = 3/group. E: Representative Western blot after Flag-immunoprecipitation (IP) in Min6 cells transfected with HA-Nrdp1, Flag-USP8, and either GFP–empty vector or GFP-Clec16a plasmids and treated for 48 h with or without 0.4 mmol/L palmitate. n = 4/group. F: Top, representative WB of cleaved caspase-3 levels in Min6 cells transfected with or without GFP-Clec16a and treated for 48 h with BSA or palmitate; bottom, relative quantification (by densitometry) of cleaved caspase-3 levels (normalized to actin loading control) by Western blot in Min6 cells transfected with or without GFP-Clec16a and treated for 48 h with BSA (white bars) or palmitate (gray bars) couplings. n = 5/group. *P < 0.05, ***P < 0.001, nonpaired Student t test. ns, not significant.
To determine whether glucolipotoxicity affects Clec16a to impact formation of the Clec16a-Nrdp1-USP8 complex, we overexpressed Clec16a in palmitate-treated Min6 β-cells. Importantly, Clec16a overexpression in palmitate-treated cells raised Nrdp1 levels, Nrdp1 ubiquitination, and Nrdp1-USP8 binding back to baseline levels (BSA alone) (Fig. 6D and E). While palmitate-treated Clec16a-overexpressing cells did not rescue the above parameters completely to the same degree as Clec16a overexpression alone, it is noteworthy that palmitate treatment itself reduced levels of Clec16a (Fig. 6D and E), suggestive of an effect of palmitate on Clec16a.
Beyond mitochondrial functional defects, chronic glucolipotoxicity can ultimately promote β-cell apoptosis (1,36). To determine the importance of maintenance of the Clec16a-Nrdp1-USP8 complex to β-cell survival during glucolipotoxicity, we assessed β-cell apoptosis by cleaved caspase-3 levels after Clec16a overexpression. As expected, palmitate significantly induced β-cell apoptosis in Min6 β-cells (Fig. 6F). Interestingly, Clec16a overexpression reduced cleaved caspase-3 levels after palmitate treatment (Fig. 6F), suggesting that restoration of the Clec16a pathway partially ameliorates the negative effects of glucolipotoxicity on β-cell survival. Taken altogether, these results indicate that glucolipotoxic insults destabilize the ubiquitin-dependent Clec16-Nrdp1-USP8 mitophagy complex, which could ultimately impact the ability of β-cells to withstand noxious stimuli.
Discussion
Here, we describe that ubiquitination is essential for the function and assembly of upstream mitophagy regulators in their maintenance of β-cell function. Pharmacologic impairment of Clec16a-mediated ubiquitination by the chemotherapeutic agent lenalidomide impairs mitophagy and insulin release. We identify that the diabetes gene Clec16a encodes an E3 ligase that promotes Nrdp1 ubiquitination. Furthermore, Clec16a-mediated Nrdp1 ubiquitination is essential for assembly of the Clec16a-Nrdp1-USP8 complex and preservation of β-cell function (Fig. 7). We observe that glucolipotoxicity perturbs the assembly and function of this complex in β-cells. Thus, Clec16a, Nrdp1, and USP8 function in a ubiquitin-dependent protein complex to regulate β-cell mitophagy.
Figure 7.
Schematic model of the Clec16a-Nrdp1-USP8 mitophagy complex. A: Under physiologic conditions, Clec16a promotes nondegradative ubiquitination (Ub) of the E3 ligase Nrdp1 to facilitate complex formation between Clec16a, USP8, and Nrdp1. This complex functions to promote normal mitophagy initiation and balance mitochondrial quality control, which in turn leads to normal β-cell function and regulated insulin release. B: However, under conditions of impaired Clec16a function, such as pharmacologic inhibition by the chemotherapeutic lenalidomide or glucolipotoxicity, Clec16a-mediated Nrdp1 ubiquitination is inhibited and the Clec16a-USP8-Nrdp1 complex is destabilized. This leads to dysfunctional mitophagy, loss of mitophagic quality control, and dysregulated mitochondrial respiration, ultimately leading to compromised β-cell insulin release.
Posttranslational control of Parkin by ubiquitination and phosphorylation is necessary for the execution of mitophagy (38). However, the direct convergence of Clec16a, Nrdp1, and USP8, and the dependence on ubiquitination for upstream control of mitophagy, has not previously been appreciated. The tripartite Clec16a-Nrdp1-USP8 complex that we identify coordinates mitophagy in part through their integrated actions on Parkin and cohesively harmonizes the effects of these mitophagy regulators (5,10,25). Several DUB enzymes, including USP8, regulate Parkin action during mitophagy (39–41). While Parkin auto-inhibition and K6 ubiquitination are relieved by USP8 to promote its mitochondrial localization, it is unclear how Clec16a opposes USP8-mediated deubiquitination of Parkin. Nrdp1 binds and opposes USP8 action on cytokine receptor signaling (42), and Clec16a could function through Nrdp1 to counter USP8 effects on Parkin. Clec16a may also directly (or indirectly through other E3 ligases) promote K6-linked conjugates on Parkin. While the dynamics of the Clec16a-Nrdp1-USP8 complex are still a mystery, it is clear that Clec16a-mediated ubiquitination orchestrates mitophagic flux in β-cells.
Our discovery of Clec16a as an E3 ligase, whose function is inhibited by lenalidomide, expands our insight into its functional role in mitophagy. Initial prediction of Clec16a function was challenging owing to a lack of recognizable conserved motifs, including the misidentified predicted C-lectin domain, suggesting that Clec16a is a misnomer (5,43). While it is clear that Clec16a has E3 ligase activity, we could not identify a consensus domain consistent with known ubiquitin ligases. Indeed, future studies of Clec16a domains coupled to structural prediction of lenalidomide-Clec16a binding sites may reveal a potential ubiquitin ligase domain. Additionally, lenalidomide inhibits other E3 ligases, notably cereblon (20). Cereblon function in β-cells is unknown; therefore, additional study is needed to determine the contribution of cereblon and/or Clec16a inhibition to β-cell dysfunction caused by lenalidomide in humans.
Clec16a ubiquitinates its principal partner Nrdp1, leading to ubiquitin laddering of unknown composition, which could occur at one or several lysine residues. Clec16a-mediated Nrdp1 ubiquitin conjugates may also be modified by the cooperative actions of USP8 to favor nondegradative linkages while removing K48 linkages. Alternatively, there may be Clec16a-independent effects to Nrdp1 activity that regulate β-cell function. Notably, the endoplasmic reticulum structural protein reticulon 4A (Rtn4a or Nogo-A) and retinoic acid derivatives modulate Nrdp1 activity (44,45) in non-β-cells, and they do not intersect with the Clec16a pathway in our previous interactome studies (5). Rtn4a has also been implicated in the regulation of GSIS (46). Therefore, elucidating Clec16a/USP8-dependent and -independent effects on Nrdp1 function will provide insight into shared and unique pathways that could be evaluated for their contribution to β-cell function.
Single nucleotide polymorphisms (SNPs) within the CLEC16A locus on chromosome 16 are associated with diabetes as well as several human diseases of both autoimmune and nonautoimmune origin (47,48). We previously described that a diabetogenic SNP within the CLEC16A locus correlated with impaired β-cell function, diminished glycemic control, and functioned as an expression quantitative trait locus for human islet CLEC16A levels, while not affecting levels of other nearby genes (ref. 5 and data not shown). While a diabetogenic CLEC16A SNP does correlate with abnormal β-cell function, we can only speculate that these SNPs directly lead to abnormalities within the CLEC16A-mitophagy pathway in human β- cells. Clec16a is also a ubiquitously expressed gene with functions in other cell types, including immune cells and neurons (5,28,49). An effect of disease-related CLEC16A SNPs to influence CLEC16A expression or the expression of nearby related genes in immune cells remains controversial (48). While Clec16a and Nrdp1 both play vital roles within immune cells (28,50), the function of CLEC16A SNPs (on mitophagy or other processes) within other human cell types requires future investigation.
These studies clarify a role for ubiquitin-dependent mitophagy complex assembly in the maintenance of β-cell function. Mitochondrial competence is essential for β-cell function, and deficient mitochondrial quality control could exacerbate β-cell dysfunction by diabetogenic insults. Previous observations of defective mitochondrial integrity after glucolipotoxicity (7,36), coupled with our findings here of glucolipotoxicity-mediated destabilization of the Clec16a-Nrdp1-USP8 complex, could inspire future in vivo studies to evaluate the importance of dysfunctional mitophagy to β-cell failure in diabetes. Additional examination will be required to determine whether nutrient excess, insulin resistance, and/or inflammation (among other diabetogenic stressors) interfere with the mitophagy machinery but may prove to illuminate a more profound understanding of diabetes pathogenesis.
Supplementary Material
Article Information
Acknowledgments. The authors acknowledge D. Giacherio and J. Whalen of the Chemistry Laboratory Core of the Michigan Diabetes Research Center (University of Michigan) (P30-DK-020572) for assistance with human hormone studies. The authors thank K. Claiborn, J. Spaeth, P. Arvan, L. Satin, L. Qi, M. Myers, and R. Stein and members of the Soleimanpour laboratory for helpful advice.
Funding. The authors acknowledge funding support from JDRF (CDA-2016-189 and SRA-2018-539), the National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health (K08-DK-089117, R03-DK-106304, and R01-DK-108921), the Central Society for Clinical and Translational Research, the Brehm family, and the Anthony family. The JDRF Career Development Award to S.A.S. is partly supported by the Danish Diabetes Academy, which is supported by the Novo Nordisk Foundation.
Duality of Interest. No potential conflicts of interest relevant to this article were reported.
Author Contributions. G.P. conceived of, designed, and performed experiments; interpreted results, and drafted and reviewed the manuscript. B.C., T.V., and X.L. designed and performed experiments and interpreted results. M.K. provided human samples from the University of Michigan Hematologic Malignancies Tissue Bank. R.C.P. designed ubiquitination studies, interpreted results, and reviewed the manuscript. S.A.S. conceived of and designed the studies, interpreted results, and edited and reviewed the manuscript. S.A.S. is the guarantor of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Prior Presentation. Parts of this study were presented in abstract form at the 77th Scientific Sessions of the American Diabetes Association, San Diego, CA, 9–13 June 2017.
Footnotes
This article contains Supplementary Data online at http://diabetes.diabetesjournals.org/lookup/suppl/doi:10.2337/db17-0321/-/DC1.
References
- 1.Supale S, Li N, Brun T, Maechler P. Mitochondrial dysfunction in pancreatic β cells. Trends Endocrinol Metab 2012;23:477–487 [DOI] [PubMed] [Google Scholar]
- 2.Jin SM, Youle RJ. PINK1- and Parkin-mediated mitophagy at a glance. J Cell Sci 2012;125:795–799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kaufman BA, Li C, Soleimanpour SA. Mitochondrial regulation of β-cell function: maintaining the momentum for insulin release. Mol Aspects Med 2015;42:91–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Soleimanpour SA, Ferrari AM, Raum JC, et al. . Diabetes susceptibility genes Pdx1 and Clec16a function in a pathway regulating mitophagy in β-cells. Diabetes 2015;64:3475–3484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Soleimanpour SA, Gupta A, Bakay M, et al. . The diabetes susceptibility gene Clec16a regulates mitophagy. Cell 2014;157:1577–1590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hoshino A, Ariyoshi M, Okawa Y, et al. . Inhibition of p53 preserves Parkin-mediated mitophagy and pancreatic β-cell function in diabetes. Proc Natl Acad Sci USA 2014;111:3116–3121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Twig G, Elorza A, Molina AJA, et al. . Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. EMBO J 2008;27:433–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wong E, Cuervo AM. Integration of clearance mechanisms: the proteasome and autophagy. Cold Spring Harb Perspect Biol 2010;2:a006734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Durcan TM, Fon EA. The three 'P's of mitophagy: PARKIN, PINK1, and post-translational modifications. Genes Dev 2015;29:989–999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Durcan TM, Tang MY, Pérusse JR, et al. . USP8 regulates mitophagy by removing K6-linked ubiquitin conjugates from parkin. EMBO J 2014;33:2473–2491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhong L, Tan Y, Zhou A, Yu Q, Zhou J. RING finger ubiquitin-protein isopeptide ligase Nrdp1/FLRF regulates parkin stability and activity. J Biol Chem 2005;280:9425–9430 [DOI] [PubMed] [Google Scholar]
- 12.Wallace TM, Levy JC, Matthews DR. Use and abuse of HOMA modeling. Diabetes Care 2004;27:1487–1495 [DOI] [PubMed] [Google Scholar]
- 13.Tamarina NA, Roe MW, Philipson L. Characterization of mice expressing Ins1 gene promoter driven CreERT recombinase for conditional gene deletion in pancreatic β-cells. Islets 2014;6:e27685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McKenna B, Guo M, Reynolds A, Hara M, Stein R. Dynamic recruitment of functionally distinct Swi/Snf chromatin remodeling complexes modulates Pdx1 activity in islet β cells. Cell Reports 2015;10:2032–2042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pearson GL, Mellett N, Chu KY, et al. . Lysosomal acid lipase and lipophagy are constitutive negative regulators of glucose-stimulated insulin secretion from pancreatic beta cells. Diabetologia 2014;57:129–139 [DOI] [PubMed] [Google Scholar]
- 16.Busch AK, Cordery D, Denyer GS, Biden TJ. Expression profiling of palmitate- and oleate-regulated genes provides novel insights into the effects of chronic lipid exposure on pancreatic beta-cell function. Diabetes 2002;51:977–987 [DOI] [PubMed] [Google Scholar]
- 17.Studier FW. Stable expression clones and auto-induction for protein production in E. coli. In Structural Genomics: General Applications. Chen YW, Ed. Totowa, NJ, Humana Press, 2014, p. 17–32 [DOI] [PubMed] [Google Scholar]
- 18.Choo YS, Zhang Z. Detection of protein ubiquitination. J Vis Exp 2009;30:1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang Y, Kitagaki J, Dai R-M, et al. . Inhibitors of ubiquitin-activating enzyme (E1), a new class of potential cancer therapeutics. Cancer Res 2007;67:9472–9481 [DOI] [PubMed] [Google Scholar]
- 20.Krönke J, Udeshi ND, Narla A, et al. . Lenalidomide causes selective degradation of IKZF1 and IKZF3 in multiple myeloma cells. Science 2014;343:301–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Issa ZA, Zantout MS, Azar ST. Multiple myeloma and diabetes. ISRN Endocrinol 2011;2011:815013 [DOI] [PMC free article] [PubMed]
- 22.Iqbal N, Zayed M, Boden G. Thalidomide impairs insulin action on glucose uptake and glycogen synthesis in patients with type 2 diabetes. Diabetes Care 2000;23:1172–1176 [DOI] [PubMed] [Google Scholar]
- 23.Basiorka AA, McGraw KL, De Ceuninck L, et al. . Lenalidomide stabilizes the erythropoietin receptor by inhibiting the E3 ubiquitin ligase RNF41. Cancer Res 2016;76:3531–3540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rozewski DM, Herman SEM, Towns WH 2nd, et al. . Pharmacokinetics and tissue disposition of lenalidomide in mice. AAPS J 2012;14:872–882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu X, Yen L, Irwin L, Sweeney C, Carraway KL 3rd. Stabilization of the E3 ubiquitin ligase Nrdp1 by the deubiquitinating enzyme USP8. Mol Cell Biol 2004;24:7748–7757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cao Z, Wu X, Yen L, Sweeney C, Carraway KL 3rd. Neuregulin-induced ErbB3 downregulation is mediated by a protein stability cascade involving the E3 ubiquitin ligase Nrdp1. Mol Cell Biol 2007;27:2180–2188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Qiu X-B, Goldberg AL. Nrdp1/FLRF is a ubiquitin ligase promoting ubiquitination and degradation of the epidermal growth factor receptor family member, ErbB3. Proc Natl Acad Sci U S A 2002;99:14843–14848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schuster C, Gerold KD, Schober K, et al. . The autoimmunity-associated gene CLEC16A modulates thymic epithelial cell autophagy and alters T cell selection. Immunity 2015;42:942–952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim S, Naylor SA, DiAntonio A. Drosophila Golgi membrane protein Ema promotes autophagosomal growth and function. Proc Natl Acad Sci U S A 2012;109:E1072–E1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ardley HC, Robinson PA. E3 ubiquitin ligases. Essays Biochem 2005;41:15–30 [DOI] [PubMed] [Google Scholar]
- 31.Birsa N, Norkett R, Wauer T, et al. . Lysine 27 ubiquitination of the mitochondrial transport protein Miro is dependent on serine 65 of the Parkin ubiquitin ligase. J Biol Chem 2014;289:14569–14582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fu M, St-Pierre P, Shankar J, Wang PT, Joshi B, Nabi IR. Regulation of mitophagy by the Gp78 E3 ubiquitin ligase. Mol Biol Cell 2013;24:1153–1162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mizuno E, Iura T, Mukai A, Yoshimori T, Kitamura N, Komada M. Regulation of epidermal growth factor receptor down-regulation by UBPY-mediated deubiquitination at endosomes. Mol Biol Cell 2005;16:5163–5174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Narendra D, Tanaka A, Suen D-F, Youle RJ. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J Cell Biol 2008;183:795–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Söderberg O, Gullberg M, Jarvius M, et al. . Direct observation of individual endogenous protein complexes in situ by proximity ligation. Nat Methods 2006;3:995–1000 [DOI] [PubMed] [Google Scholar]
- 36.Molina AJA, Wikstrom JD, Stiles L, et al. . Mitochondrial networking protects β-cells from nutrient-induced apoptosis. Diabetes 2009;58:2303–2315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Poitout V, Robertson RP. Glucolipotoxicity: fuel excess and β-cell dysfunction. Endocr Rev 2008;29:351–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shiba-Fukushima K, Imai Y, Yoshida S, et al. . PINK1-mediated phosphorylation of the Parkin ubiquitin-like domain primes mitochondrial translocation of Parkin and regulates mitophagy. Sci Rep 2012;2:1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cornelissen T, Haddad D, Wauters F, et al. . The deubiquitinase USP15 antagonizes Parkin-mediated mitochondrial ubiquitination and mitophagy. Hum Mol Genet 2014;23:5227–5242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cunningham CN, Baughman JM, Phu L, et al. . USP30 and parkin homeostatically regulate atypical ubiquitin chains on mitochondria. Nat Cell Biol 2015;17:160–169 [DOI] [PubMed] [Google Scholar]
- 41.Bingol B, Tea JS, Phu L, et al. . The mitochondrial deubiquitinase USP30 opposes parkin-mediated mitophagy. Nature 2014;510:370–375 [DOI] [PubMed] [Google Scholar]
- 42.De Ceuninck L, Wauman J, Masschaele D, Peelman F, Tavernier J. Reciprocal cross-regulation between RNF41 and USP8 controls cytokine receptor sorting and processing. J Cell Sci 2013;126:3770–3781 [DOI] [PubMed] [Google Scholar]
- 43.Finn RD, Bateman A, Clements J, et al. . Pfam: the protein families database. Nucleic Acids Res 2014;42:D222–D230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hatakeyama J, Wald JH, Rafidi H, Cuevas A, Sweeney C, Carraway KL 3rd. The ER structural protein Rtn4A stabilizes and enhances signaling through the receptor tyrosine kinase ErbB3. Sci Signal 2016;9:ra65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Byun S, Shin SH, Lee E, et al. . The retinoic acid derivative, ABPN, inhibits pancreatic cancer through induction of Nrdp1. Carcinogenesis 2015;36:1580–1589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bonal CB, Baronnier DE, Pot C, et al. . Nogo-A downregulation improves insulin secretion in mice. Diabetes 2013;62:1443–1452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fujimaki T, Kato K, Yokoi K, et al. . Association of genetic variants in SEMA3F, CLEC16A, LAMA3, and PCSK2 with myocardial infarction in Japanese individuals. Atherosclerosis 2010;210:468–473 [DOI] [PubMed] [Google Scholar]
- 48.Berge T, Leikfoss IS, Harbo HF. From identification to characterization of the multiple sclerosis susceptibility gene CLEC16A. Int J Mol Sci 2013;14:4476–4497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Redmann V, Lamb CA, Hwang S, et al. . Clec16a is critical for autolysosome function and Purkinje cell survival. Sci Rep 2016;6:23326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang M, Chen T, Li X, et al. K33-linked polyubiquitination of Zap70 by Nrdp1 controls CD8+ T cell activation. 2015;16:1253 [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.