Abstract
The present study aimed to examine the effect of RNA interference targeting cell division cycle-associated protein 4 (CDCA4) on the proliferation and apoptosis of the MCF-7/ADR' human breast cancer cell line. CDCA4 has been shown to have a unique role in regulating cell cycle. In the present study, the expression of CDCA4 was suppressed by CDCA4-specific short hairpin (sh)RNA transfection of the human breast cancer cells, following which changes in the proliferation and apoptosis of the CDCA4-knockdown cells were compared with those of control shRNA-transfected cells. The results confirmed that CDCA4 RNA interference reduced the percentage of human breast cancer cells to <50%. In addition, RNA interference of CDCA4 resulted in a significant increase in the apoptotic rate of cells. Taken together, these results suggested that CDCA4 enhanced proliferation and reduced apoptosis in the MCF-7/ADM human breast cancer cells in vitro.
Keywords: apoptosis, breast cancer, cell division cycle-associated protein 4, proliferation, cell cycle
Introduction
Cell division cycle-associated protein 4 (CDCA4), also known as HEPP/SEI-3/TRIP-Br3, was originally identified by Abdullah et al in 2001, as a novel gene involved in the intrinsic regulation of hematopoietic stem cell and progenitor cell lineage commitment and differentiation (1). Gene-based analysis suggested that genes of the chenodeoxycholic acid (CDCA) family were closely co-expressed with known cell cycle genes, including CDC2, CDC7 and cyclins (2). CDCA4 was categorized as SEI3 of the selected with INK4A (SEI) gene family, based on its characteristic SEI-1, RBT1 and TARA domain and as a transcriptional regulator interacting with the PHD bromodomain (TRIP-Br)3 of the TRIP-Br gene family, according to its unique PHD-bromo-binding domain (3,4). The SEI gene family consists of four genes: SEI-1, SEI-2, RBT1 and CDCA4. The close association between the overexpression of SEI family genes and tumorigenesis has been confirmed in various studies (5,6). Previous studies have indicated that SEI-1 and SEI-2 interact with dimerization partner 1 (DP1) and regulate the transcriptional activity of E2F-1 (7). SEI genes also interact with bromodomain-containing transcriptional cofactors, including lysine acetyltransferase 2B (PCAF), SPT7-like STAGA complex γ subunit and kinesin-II-associated protein 1 (3). The overexpression of SEI family genes, including CDCA4, can enhance the transactivation function of p53, leading to p53-independent growth inhibition in HeLa and U2OS cells (4). CDCA4 and SEI-1 can inhibit breast cancer cell apoptosis by interacting with an X-linked inhibitor of apoptosis protein, a potent apoptosis inhibitor (8,9). Human CDCA4 mRNA is expressed at higher levels in actively proliferating tissues, for example the pancreas, thymus, testis, spleen, liver and placenta. Of note, the mRNA expression of CDCA4 is only marginal or is undetected in tissues consisting of permanent cells, including the brain, skeletal muscle and heart (10). CDCA4 affects the mRNA expression of the Jun proto-oncogene and determines cell fate (11). In addition, CDCA4 has a dual role in cell cycle regulation by modulating the transcriptional activity of activator E2F transcription factors and p53 (10). Evidence suggests that the SEI-1 protein can inhibit the MCF-7 cell senescence induced by doxorubicin (12). SEI-2 is frequently overexpressed in several types of human tumor. Clinicopathological evidence and Kaplan-Meier survival analysis indicates that the overexpression of SEI-2 is directly linked to poor clinical outcomes in hepatocellular carcinoma (5). MicroRNA-15a downregulates the expression of CDCA4 through targeting the 3′-untranslated region of CDCA4, and the downregulation of CDCA4 can inhibit proliferation, causing cell cycle arrest and reducing the invasiveness of melanoma cells (13). In addition, The Cancer Genome Atlas (TCGA) public databases show that CDCA4 has been found in numerous samples of breast cancer tissue; its mRNA expression being significantly higher compared with its expression in adjacent tissues. In addition, GeneChip analysis has suggested that CDCA4 is a downstream gene of the nuclear factor erthyroid 2-related factor 2 (Nrf2) signaling pathway. Nrf2 has been shown to regulate the resistance of cancer cells to chemotherapeutic drugs (14). Based on the aforementioned information, the present study hypothesized that CDCA4 may be closely associated with breast cancer cells. The present study focused on the effect of RNA interference targeting CDCA4 on cell proliferation, cycle progression and apoptosis in a human breast cancer cell line. The results suggested that CDCA4 RNA interference reduced the proliferation of human breast cancer cells to <50%, and that CDCA4 regulated cell proliferation, at least in part, through cell cycle progression. In addition, RNA interference of CDCA4 resulted in a significant increase in the apoptotic rate of cells.
Materials and methods
GeneChip analysis
Total RNA was extracted from three different batches of Nrf2 shRNA-transfected MCF-7/ADR cells and are referred to as knockdown (KD) group samples and three different batches of control shRNA-transfected MCF-7/ADR cells that are referred to as nonsilencing control (NC) group samples using TRIzol reagent (Shanghai Pufei Biotechnology, Co., Ltd., Shanghai, China), and then quantified using a Thermo Nanodrop 2000 spectrophotometer (Thermo Fisher Scientific, Inc., Waltham, MA, USA) and Agilent 2100 Bioanalyzer (Agilent Technologies, Inc., Santa Clara, CA, USA). The samples for microarray hybridization were prepared based on the manufacturer's protocols. The cDNA was hybridized to the arrays at 2 × g for 18 h at 45°C. The chips were processed in the GeneChip Fluidics Station 450 (Affymetrix, Inc., Santa Clara, CA, USA). Microarray images were captured using the Scanner 3000 (Affymetrix, Inc.), and data were extracted using Affymetrix Power Tools software v1.8 (Affymetrix; Thermo Fisher Scientific, Inc., Waltham, MA, USA).
Cell culture
The MCF-7/AD-M human breast cancer cell line was obtained from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). The MCF-7/ADM cells were cultured in high-glucose Dulbecco's modified Eagle's medium (DMEM; (GE Healthcare Life Sciences, Logan, UT, USA), supplemented with 10% fetal bovine serum (FBS, HyClone; GE Healthcare Life Sciences), penicillin (100 U/ml) and streptomycin (10 µg/ml). They were incubated at 37°C and 5% CO2 in a humidified chamber atmosphere.
RNA interference
Different regions of CDCA4 mRNA and nonsilencing control were designed, chemically synthesized, and ligated into the GV115 vector according to the manufacturer's protocol (GeneChem Co., Ltd., Shanghai, China). The target sequence of human CDCA4 with the significant interference effect was 5′-TAGACCTAAGAGTAAATTA-3′. The nonsilencing short hairpin (sh)RNA was designed as a nonspecific sequence, with no matching to the human genome, using the Basic Local Alignment Search Tool (www.ncbi.nlm.nih.gov). For each interference experiment, equal numbers of MCF-7/ADM cells were transfected with shRNAs of CDCA4 and control shRNA by Lentivirus-mediated method. The antibiotic-resistant transfected cells were selected by applying puromycin (Clontech Laboratories, Inc., Mountainview, CA, USA) in DMEM, and the stably transfected MCF-7/ADM cells were checked under phase contrast and fluorescence microscopes. In a similar manner, MCF-7/ADM cells were transfected with shRNAs of Nrf2 and control shRNA. The target sequence of Nrf2 was 5′-GGCATTTCACTAAACACAA-3′.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR) analysis
Total RNA was isolated from the cultured cells using TRIzol reagent (Shanghai Pufei Biotechnology, Co., Ltd., Shanghai, China). cDNA was synthesized from the DNaseI-treated total RNA (1 µg) using oligo-(dT) primer and reverse transcriptase of Moloney murine leukemia virus (Promega Corporation, Madison, WI, USA) in a 10-µl reaction mixture. The RT-qPCR analysis was performed using SYBR Premix Ex Taq (Takara Bio, Inc., Otsu, Japan). In brief, 1 µl cDNA was diluted with 10 µl SYBR premix ex taq, 0.5 µl sense primers, 0.5 µl antisense primers and 8 µl RNase-Free H2O to 20 µl of reaction mixture, and the mixture was subjected to PCR in an MX3000pq-PCR system (Agilent Technologies, Inc., Santa Clara, CA, USA). The following primer sets were used: Forward 5′-ATTTGAAACGCTGGAGACT-3′ and reverse 5′-CCCATCATGCCTGTCAGTA-3′ for human CDCA4 and Forward 5′-TGACTTCAACAGCGACACCCA-3′ and reverse 5′- CAC CCT GTT GCT GTA GCC AAA-3′ for GAPDH. RT-qPCR was performed as followed: One cycle of pre denaturation (95°C for 30 sec), 45 cycles of amplification (95°C for 5 sec and 60°C for 30 sec), and a cooling period. The expression of GAPDH was used as the internal control for detecting mRNAs and results were analyzed by the 2−ΔΔCq method (15). In a similar manner, silencing effect of MCF-7/ADM cells was examined.
Western blot analysis
The cells were washed in an ice-cold phosphate-buffered saline buffer and then lysed in radioimmunoprecipitation assay lysis buffer. Following protein quantification using a bicinchoninic acid assay, equal quantities of 20 µg protein sample were separated using 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred onto a polyvinylidene difluoride membrane. The membranes were blocked with 5% nonfat dry milk and then incubated with primary antibody (anti-CDCA4, cat. no. QC17060, 1:500 dilution, Sigma-Aldrich; Merck MGaA, Darmstadt, Germany) for 12 h at 4°C, followed by horseradish peroxidase-conjugated secondary antibody (Anti-rabbit IgG, cat.no. sc-2004, 1:2,000 dilution, Santa Cruz Biotechnology, Inc., Dallas, TX, USA) for 1 h at 26°C. Protein bands were visualized using a Pierce™ ECL Western Blotting Substrate (Thermo Fisher Scientific, Inc.) and the levels of expression were quantified by Image J software v1.46r.
Flow cytometric analysis of cell cycle
Cells were harvested by trypsinization, centrifuged at 1,200 × g for 5 min at 26°C, and washed twice with D-Hanks (pH 7.2–7.4) at 4°C. The cell pellets were fixed by slowly adding 70% ethanol at 20°C and then stored at 4°C. To determine the DNA content, the cell pellets were centrifuged at 1,200 × g for 5 min at 26°C and washed with cold D-Hanks and resuspended in a solution of propidium iodide (Sigma-Aldrich; Merck KGaA) and RNase A (Fermentas; Thermo Fisher Scientific, Inc.) in D-Hanks for 30–60 min at 37°C. The cell samples were analyzed using a Guava easyCyte 5HT flow cytometer (EMD Millipore, Billerica, MA, USA). The data were analyzed to determine the proportion of cells in each stage of the cell cycle. The experiments were performed in triplicate.
3-(4,5-dimethylthia-zol-2-yl)-2, 5-diphenyltetrazolium bromide (MTT) assay
An MTT assay was performed to measure cell growth. The cells were seeded at 2,000 per well in 96-well plates and incubated at 37°C for a designed time period. For each test, 20 µl of MTT solution (5 mg/ml; Genview Tallahasses, FL, USA) was added to each of the wells, followed by incubation at 37°C for 4 h. Subsequently, 100 µl of dimethyl sulfoxide solution was added to resolve the formazan crystals overnight at 37°C. The optical density at 490 nm was measured the following day to determine the quantities of formazan formed by cleaving MTT in living cells. For each treatment, the cells were cultured for 5 days and the samples were prepared for the quantitative MTT assay once per day following plating. Three independent assays were performed, and two samples were measured at each time point.
Colony formation assay
About 400 cells suspended in DMEM containing 10% FBS were plated on each well of six-well plates. The plates were incubated at 37°C in a 5% CO2 incubator for 14 days and colonies of >50 cells were counted. Each dose was examined in triplicate, and the experiments were performed three times. The control shRNA (shCtrl)-transfected MCF-7/ADM cells were detected using the same methods.
Apoptosis assay using flow cytometric analysis
The cells were cultured for 5 days, washed twice in D-Hanks (pH 7.2–7.4), and then resuspended in binding buffer. Annexin V-APC (10 µl) was added to a final concentration of 1 µg/ml Annexin V-APC. The mixture was incubated for 15 min in the dark at room temperature and then analyzed using a flow cytometer (Guava easyCyte HT; EMD Millipore). The reproducibility of the assay was examined by two independent experiments.
Statistical analysis
Data are presented as the mean ± standard deviation and were analyzed using SPSS 23. 0 (IBM SPSS, Armonk, NY, USA). Student's t-test was used to measure statistical significance between the experimental and control samples. P<0.05 was considered to indicate a statistically significant difference.
Results
Expression of CDCA4 is significantly upregulated following Nrf2 knockdown in breast cancer cells
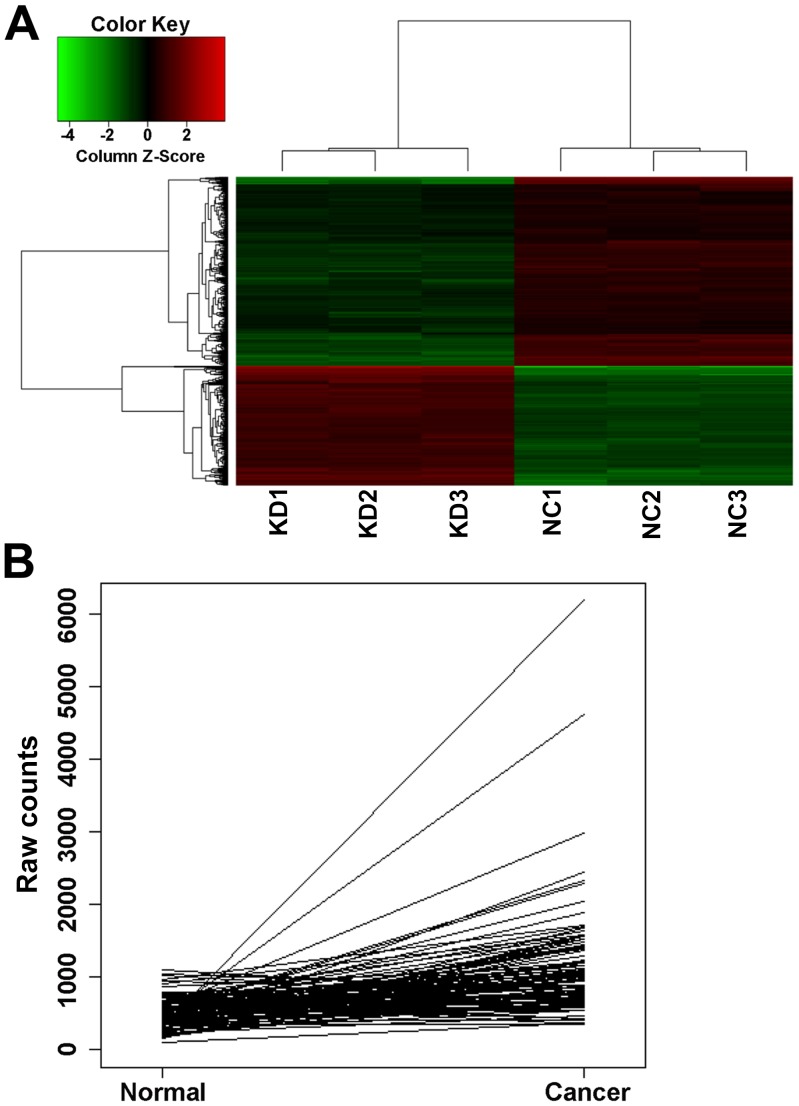
To identify the genes exhibiting change in expression when Nrf2 was knocked down, GeneChip analysis was performed. The resulting heat map showed the differences in the expression of genes between the nonsilencing control group (NC1, NC2 and NC3) and silencing/knockdown group (KD1, KD2 and KD3) samples (Fig. 1A). The expression profiles of the MCF-7/ADM breast cancer cells were analyzed prior to and following Nrf2 shRNA transfection by GeneChip. A total of 878 significantly different genes were screened. Among the identified genes, the present study focused on CDCA4, which was upregulated in the Nrf2 shRNA-transfected cells, compared with the control shRNA-transfected cells (fold-change=1.6128796). CDCA4 was found in several samples of breast cancer tissue, with The Cancer Gene Atlas (TCGA) public databases showing that the mRNA expression of CDCA4 was significantly higher, compared with its expression in adjacent tissues (Table I and Fig. 1B). These findings suggested that CDCA4 may be closely associated with breast cancer cells.
Figure 1.
(A) Heatmap of the GeneChip assay. knockdown group (KD) 1, 2 and 3 were there independent samples of experimental group which were transfected with shRNA of Nrf2. Nonsilencing control group (NC) 1, 2 and 3 were there independent samples of control group. Black represents no change in gene expression; red represents upregulated and green represents downregulated gene expression. (B) mRNA expression levels of cell division cycle-associated protein 4 were are markedly different between breast cancer tissues and normal tissues. Data are expressed as the mean ± standard deviation. *P>0.05, **P<0.05.
Table I.
Comparison of breast cancer tissues with normal tissues for mRNA levels of CDCA4 in 106 samples from The Cancer Gene Atlas public databases.
| Gene | FC | P-value | Total samples (n) | Expression unchanged (n) | Increased expression (n) | Decreased expression (n) |
|---|---|---|---|---|---|---|
| CDCA4 | 2.03 | 3.05E-42 | 106 | 51 | 55 | 0 |
CDCA4, cell division cycle-associated protein 4; FC, fold-change.
Silencing effect of shRNA on the expression of CDCA4 in the MCF-7/ADM human breast cancer cell line
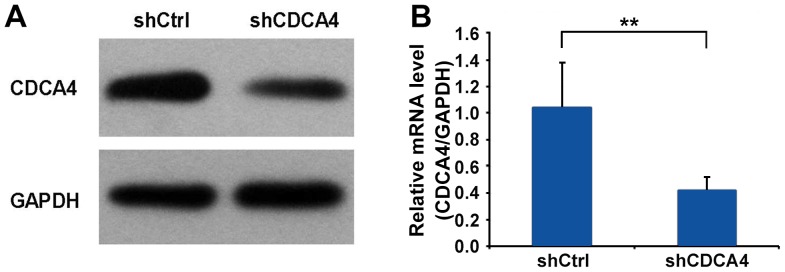
To examine the function of CDCA4 in human breast cancer, the CDCA4 shRNA-mediated suppression of expression in the MCF-7/ADM human breast cancer cell line was examined using western blot and RT-qPCR analyses. The expression of CDCA4 was significantly decreased in the MCF-7/ADM cells (Fig. 2A and B). Therefore, MCF-7/ADM cells were selected as the model for subsequent experiments.
Figure 2.
(A) Western blot analysis of the protein expression of CDCA4 following transfection. (B) Reverse transcription-quantitative polymerase chain reaction analysis was used to examine the mRNA expression levels of CDCA4 in MCF-7/ADM cells. Data are expressed as the mean ± standard deviation. *P>0.05, **P<0.05. CDCA4, cell division cycle-associated protein 4; sh, short hairpin RNA; Ctrl, control.
Knockdown of the expression of CDCA4 inhibits the growth of breast cancer cells
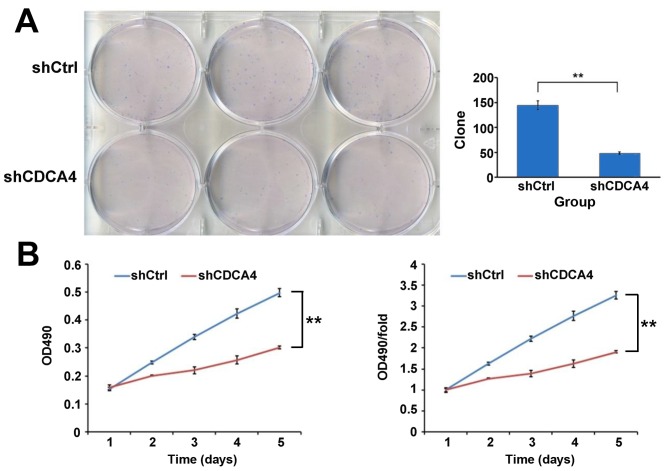
A colony formation assay was used to evaluate the growth of the breast cancer cells in which CDCA4 was silenced. As shown in Fig. 3A, CDCA4 knockdown in the MCF-7/ADM cells led to the formation of significantly fewer colonies on soft agar, compared with the shCtrl group of cells To further examine the negative effect of CDCA4 knockdown on breast cancer cell growth, an MTT assay was performed and growth curves were generated (Fig. 3B). As shown by the curves, the proliferation of shCDCA4-transfected group cells was slower, compared with that of the shCtrl-transfected group of cells during the first 5 days following plating of the cells. The marked reduction in colony formation and growth of the CDCA4-silenced cells suggested that the suppression of CDCA4 may negatively regulate breast cancer cell growth. Together, these data suggested that CDCA4 enhanced the growth and proliferation of the human breast cancer cells.
Figure 3.
(A) Colony formation assay of the MCF-7/ADM cells. (B) Growth curves of the cell line as assessed by an MTT assay. Data are expressed as the mean ± standard deviation. *P>0.05, **P<0.05. CDCA4, cell division cycle-associated protein 4; sh, short hairpin RNA; Ctrl, control; OD, optical density.
Effect of suppressing the expression of CDCA4 on cell cycle control in MCF-7/ADM breast cancer cells
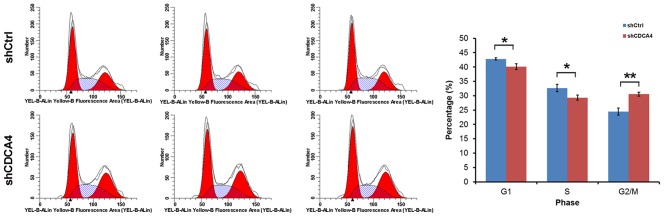
Flow cytometry was used to examine whether the promoting effect of CDCA4 on MCF-7/ADM cell growth and proliferation was mediated through cell cycle progression. In the shCDCA4-transfected group of cells, 40.10% of cells were in the G0/G1phase, 29.34% were in the S phase and 30.56% were in the G2/M phase of the cell cycle, which were significantly lower, compared with the percentages in the shCtrl group of cells (Fig. 4). These data indicated that CDCA4 was significantly associated with the cell cycle of the MCF-7/ADM cells. Therefore, CDCA4 regulated cell proliferation, at least in part, through cell cycle progression.
Figure 4.
Cell cycle distribution was measured using flow cytometry. Data are expressed as the mean ± standard deviation. *P>0.05, **P<0.05. CDCA4, cell division cycle-associated protein 4; sh, short hairpin RNA; Ctrl, control.
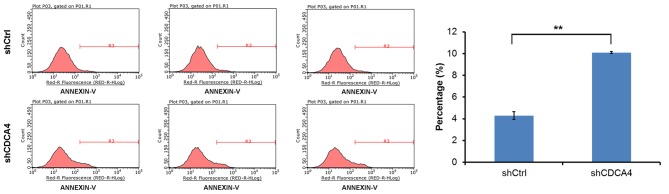
CDCA4 knockdown increases the apoptotic rate of MCF-7/ADM cells
To investigate whether CDCA4 shRNA-transfected cells and shCtrl-transfected cells showed differences in apoptotic rate, an apoptosis assay was performed on the experimental and control groups of cells using flow cytometry. Apoptosis occurred in ~4.29±0.36% of the control group cells in the 5 days following plating of the cells. The apoptotic rate of the CDCA4-knockdown MCF-7/ADM cells increased to 10.1±0.09%. The apoptotic rate of the shCDCA4 group was significantly higher, compared with that of the shCtrl group (Fig. 5). These results indicated that CDCA4 knockdown increased the apoptotic rate of the MCF-7/ADM cells. Therefore, the present study provided direct evidence that CDCA4 affected the survival of MCF-7/ADM cells.
Figure 5.
Apoptotic rates were measured using flow cytometry. Data are expressed as the mean ± standard deviation. *P>0.05, **P<0.05. CDCA4, cell division cycle-associated protein 4; sh, short hairpin RNA; Ctrl, control.
Discussion
Breast cancer is a global health challenge. Patients with early-stage breast cancer following treatment with surgery followed by postoperative radiotherapy have a satisfactory survival rate. However, a substantial number of patients with terminal breast cancer are unable to undergo surgery due to symptomatic metastasis. The tolerance of these patients is often too weak to accept chemotherapy or radiotherapy. Gene-targeted therapy has been shown to be an effective supplement for conventional therapy for several types of cancer. The results of the National Surgical Adjuvant Breast and Bowel Project trial B-31 and the North Central Cancer Treatment Group trial N9831 suggested that treatment with trastuzumab following chemotherapy substantially reduced the rate of recurrence and significantly improved outcomes for patients with HER2-positive breast cancer (16). Gene-targeted therapy can be considered as a novel cancer therapy although it requires further investigation.
In a preliminary study (data not shown), which aimed to identify potential downstream transcriptional target genes regulated by Nrf2 in breast cancer cells, GeneChip data were analyzed, and CDCA4 was shown to be upregulated in Nrf2 shRNA-transfected cells. Considering relevant findings on the SEI family and analysis of TCGA public databases, it was suggested that CDCA4 may be closely associated with breast cancer cells.
Few studies (8,11,13) have demonstrated the importance of CDCA4 in the development of cancer and cancer cell survival. However, the exact role of CDCA4 in the progression of cancer remains to be fully elucidated, and the function of CDCA4 in the cell cycle of breast cancer has not been investigated previously. Whether CDCA4 is a cell fate regulator important in human breast cancer has not been elucidated. To address these issues, the present study evaluated the expression of CDCA4 in terms of its possible direct correlations with the proliferation and apoptosis of human breast cancer cells.
The present study aimed to examine the effect of RNA interference targeting CDCA4 on cell proliferation, cell cycle progression and apoptosis in the MCF-7/ADM human breast cancer cell line. A couple of studies (10,17) have revealed that CDCA4 regulates cell proliferation in a variety of cancer cells. However, the effect of the expression of CDCA4 on the growth of breast cancer cells remains to be fully elucidated. The results of the MTT and colony formation assays in the present study indicated that the suppression of CDCA4 led to the inhibition of cell proliferation and growth. RNA interference of CDCA4 resulted in a significant increase in the apoptotic rate. These findings provided direct evidence that the expression of CDCA4 can affect the survival of human breast cells in vitro, at least in part. This may be associated with the function of CDCA4 in cell cycle control. The results of the present study suggested that CDCA4 is important in regulating the fate of human breast cancer cells through enhancing proliferation and reducing apoptosis.
In conclusion, the expression of CDCA4 was found to be associated with the fate of breast cancer cells. Using RNA interference technology, the results of in vitro experiments indicated that a downregulation in the expression of CDCA4 inhibited proliferation and induced apoptosis in the MCF-7/ADM human breast cancer cell line. These data provide a scientific rationale for potential gene-targeted therapy of breast cancer. Other potential functions of CDCA4 in breast cancer require further elucidation through additional relevant investigations.
Acknowledgements
The present study was supported by the National Natural Science Foundation of China (grant no. 81260341).
References
- 1.Abdullah JM, Jing X, Spassov DS, Nachtman RG, Jurecic R. Cloning and characterization of Hepp, a novel gene expressed preferentially in hematopoietic progenitors and mature blood cells. Blood Cells Mol Dis. 2001;27:667–676. doi: 10.1006/bcmd.2001.0434. [DOI] [PubMed] [Google Scholar]
- 2.Walker MG. Drug target discovery by gene expression analysis: Cell cycle genes. Curr Cancer Drug Targets. 2001;1:73–83. doi: 10.2174/1568009013334241. [DOI] [PubMed] [Google Scholar]
- 3.Lai IL, Wang SY, Yao YL, Yang WM. Transcriptional and subcellular regulation of the TRIP-Br family. Gene. 2007;388:102–109. doi: 10.1016/j.gene.2006.10.008. [DOI] [PubMed] [Google Scholar]
- 4.Watanabe-Fukunaga R, Iida S, Shimizu Y, Nagata S, Fukunaga R. SEI family of nuclear factors regulates p53-dependent transcriptional activation. Genes Cells. 2005;10:851–860. doi: 10.1111/j.1365-2443.2005.00881.x. [DOI] [PubMed] [Google Scholar]
- 5.Cheong JK, Gunaratnam L, Zang ZJ, Yang CM, Sun X, Nasr SL, Sim KG, Peh BK, Rashid SB, Bonventre JV, et al. TRIP-Br2 promotes oncogenesis in nude mice and is frequently overexpressed in multiple human tumors. J Transl Med. 2009;7:8. doi: 10.1186/1479-5876-7-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tang DJ, Hu L, Xie D, Wu QL, Fang Y, Zeng Y, Sham JS, Guan XY. Oncogenic transformation by SEI-1 is associated with chromosomal instability. Cancer Res. 2005;65:6504–6508. doi: 10.1158/0008-5472.CAN-05-0351. [DOI] [PubMed] [Google Scholar]
- 7.Hsu SI, Yang CM, Sim KG, Hentschel DM, O'Leary E, Bonventre JV. TRIP-Br: A novel family of PHD zinc finger- and bromodomain-interacting proteins that regulate the transcriptional activity of E2F-1/DP-1. EMBO J. 2001;20:2273–2285. doi: 10.1093/emboj/20.9.2273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li C, Jung S, Lee S, Jeong D, Yang Y, Kim KI, Lim JS, Cheon CI, Kim C, Kang YS, Lee MS. Nutrient/serum starvation derived TRIP-Br3 down-regulation accelerates apoptosis by destabilizing XIAP. Oncotarget. 2015;6:7522–7535. doi: 10.18632/oncotarget.3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hong SW, Kim CJ, Park WS, Shin JS, Lee SD, Ko SG, Jung SI, Park IC, An SK, Lee WK, et al. p34SEI-1 inhibits apoptosis through the stabilization of the X-linked inhibitor of apoptosis protein: p34SEI-1 as a novel target for anti-breast cancer strategies. Cancer Res. 2009;69:741–746. doi: 10.1158/0008-5472.CAN-08-1189. [DOI] [PubMed] [Google Scholar]
- 10.Hayashi R, Goto Y, Ikeda R, Yokoyama KK, Yoshida K. CDCA4 is an E2F transcription factor family-induced nuclear factor that regulates E2F-dependent transcriptional activation and cell proliferation. J Biol Chem. 2006;281:35633–35648. doi: 10.1074/jbc.M603800200. [DOI] [PubMed] [Google Scholar]
- 11.Tategu M, Nakagawa H, Hayashi R, Yoshida K. Transcriptional co-factor CDCA4 participates in the regulation of JUN oncogene expression. Biochimie. 2008;90:1515–1522. doi: 10.1016/j.biochi.2008.05.014. [DOI] [PubMed] [Google Scholar]
- 12.Lee SL, Hong SW, Shin JS, Kim JS, Ko SG, Hong NJ, Kim DJ, Lee WJ, Jin DH, Lee MS. p34SEI-1 inhibits doxorubicin-induced senescence through a pathway mediated by protein kinase C-delta and c-Jun-NH2-kinase 1 activation in human breast cancer MCF7 cells. Mol Cancer Res. 2009;7:1845–1853. doi: 10.1158/1541-7786.MCR-09-0086. [DOI] [PubMed] [Google Scholar]
- 13.Alderman C, Sehlaoui A, Xiao Z, Yang Y. MicroRNA-15a inhibits the growth and invasiveness of malignant melanoma and directly targets on CDCA4 gene. Tumour Biol. 2016;37:13941–13950. doi: 10.1007/s13277-016-5271-z. [DOI] [PubMed] [Google Scholar]
- 14.Wang XJ, Sun Z, Villeneuve NF, Zhang S, Zhao F, Li Y, Chen W, Yi X, Zheng W, Wondrak GT, et al. Nrf2 enhances resistance of cancer cells to chemotherapeutic drugs, the dark side of Nrf2. Carcinogenesis. 2008;29:1235–1243. doi: 10.1093/carcin/bgn095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Livak KJ, Scmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 16.Romond EH, Perez EA, Bryant J, Suman VJ, Geyer CE, Jr, Davidson NE, Tan-Chiu E, Martino S, Paik S, Kaufman PA, et al. Trastuzumab plus adjuvant chemotherapy for operable HER2-positive breast cancer. N Engl J Med. 2005;353:1673–1684. doi: 10.1056/NEJMoa052122. [DOI] [PubMed] [Google Scholar]
- 17.Wang L, Zhu G, Li Q, Li Y, Xu X, He D, Zeng C. The spindle function of CDCA4. Cell Motil Cytoskel. 2008;65:581–593. doi: 10.1002/cm.20286. [DOI] [PubMed] [Google Scholar]