Abstract
The aim of the present study was to identify risk genes in myocardial infarction. Microarray data GSE34198, containing data from the peripheral blood of 49 myocardial infarction samples and 48 corresponding control samples, were downloaded from the Gene Expression Omnibus database to screen the differentially expressed genes (DEGs). The DEGs were used to construct a protein-protein interaction (PPI) network of patient samples, from which the feature genes were identified using the neighboring score method. The recursive feature elimination (RFE) algorithm was employed to select the risk genes among feature genes, which were subsequently applied to perform a support vector machine (SVM) classifier to identify the specific signature in myocardial infarction samples. Another dataset, GSE61144, was also downloaded to verify the efficacy of the classifier. A total of 724 downregulated and 483 upregulated DEGs were screened in patient samples compared with control samples in the GSE34198 dataset. The PPI network of myocardial infarction was comprised of 1,083 nodes (genes) and 46,363 lines (connections). Using the neighborhood scoring method, the top 100 feature genes in myocardial infarction samples were identified as the disease feature genes, which distinguish the myocardial infarction samples from the control samples. The RFE algorithm screened 15 risk genes, which were employed to construct a SVM classifier with an average precision of 88% to the patient sample following visualization by a confusion matrix. The predictive precision of the classifier on another microarray dataset, GSE61144, was 0.92, with an average true positive of 0.9278 and an average false positive of 0.2361. A-kinase-anchoring protein 12 (AKAP12) and glycine receptor α2 (GLRA2) were two risk genes in the SVM classifier. Therefore, AKAP12 and GLRA2 exert potential roles in the development of myocardial infarction, potentially by influencing cardiac contractility and protecting against ischemia-reperfusion injury, which may provide clues in developing potential diagnostic biomarkers or therapeutic targets for myocardial infarction.
Keywords: myocardial infarction, microarray, protein-protein interaction network, recursive feature elimination algorithm, support vector machine classifier
Introduction
Myocardial infarction is a result of interrupted blood flow to a certain area of the heart, which subsequently damages heart muscle. Among the various symptoms, chest pain or discomfort that may travel to the shoulder, arm, neck, back or jaw is the most common (1). Shortness of breath, feeling faint, nausea and cold sweats may also be experienced by patients suffering a myocardial infarction. Myocardial infarction may trigger heart failure, cardiac arrest, an irregular heartbeat or cardiogenic shock (2), and, as a life-threatening disease that may lead to severe hemodynamic instability or sudden death, is one of the major causes of mortality worldwide (3). According to an estimation by the World Bank, the number of individuals experiencing myocardial infarction may reach >23 million by 2030 in China (4). Globally, the mortality associated with acute myocardial infarction has reduced in the past few decades, however, as a result, the incidence of heart failure has increased (5). Heart failure following myocardial infarction is associated with cardiac remodeling, which leads to ventricular dysfunction and chamber dilation (6).
Clinically, the occurrence of myocardial infarction is often unexpected and sudden, which makes it difficult to prevent and diagnose. Cardiovascular risk factors for heart disease include circulating blood lipid levels (7), smoking (8), heavy drinking (9), oral contraceptives (10), high intake of anthocyanins (11), human immunodeficiency virus infection (12) and a family history or genetic alterations. A positive family history is among the strongest cardiovascular risk factors for heart disease, therefore, numerous studies have aimed to determine the associated genetic factors of myocardial infarction. For example, Helgadottir et al (13) reported that arachidonate 5-lipoxygenase-activating protein variants are involved in the pathogenesis of myocardial infarction by increasing the inflammation in the arterial wall and the production of leukotrienes. In addition, Do et al (14) identified that multiple rare alleles of the low-density lipoprotein receptor and apolipoprotein A5 confer risk for early-onset myocardial infarction, and a meta-analysis demonstrated that the rs671 aldehyde dehydrogenase 2 family (mitochondrial) polymorphism increases the risk of myocardial infarction (15).
Despites the current findings, reliable molecular prediction in the diagnosis and prevention of myocardial infarction remains to be discovered. In the present study, using the feature genes selected from differentially expressed genes (DEGs) in patients with myocardial infarction compared with controls, a support vector machine (SVM) classifier and certain risk genes were screened. These risk genes allow patient samples to be distinguished from normal controls.
Materials and methods
Microarray data
The GSE34198 microarray dataset (16) was downloaded from the Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo) and included 49 samples from the peripheral blood of patients with myocardial infarction and 48 control samples. The platform for GSE34198 was Illumina human-6 v2.0 expression BeadChip. Affy package RMA in R version 3.3.1 (17) (http://bioconductor.org/packages/release/bioc/html/affy.html) was utilized to transfer the array data in GSE34198 into expression data, which was subsequently normalized by the Z-score method (18).
DEG identification
The DEGs between patients with myocardial infarction and control subjects were identified using the Limma version 3.32.8 (http://bioconductor.org/packages/release/bioc/html/limma.html) (19) with a threshold of P<0.05 and log|fold change (FC)|>1.
Protein-protein interaction (PPI) network construction. All screened DEGs were subjected to the human protein interaction network Human Protein Reference Database (20) (http://www.hprd.org/) for the identification of their interactions. Subsequently, the interactions were visualized using Cytoscape 3.4 software (http://www.cytoscape.org/) as the PPI network of DEGs in myocardial infarction.
Feature gene selection
Usually, significant expression connections exist between disease feature genes and their connected genes. To identify the feature genes in myocardial infarction, the neighborhood score (21) was employed to identify the feature genes in the PPI network. The formula for calculating the score was as follows:
Where i represents the node in the network, FC represents the fold change value for the expression level of the node, N(i) represents the number of the connection nodes to the selected node and score(i) represents the correlations between the node(i) and the disease.
By the neighborhood scoring algorithm, the changing degrees of the nodes under disease will be inferred, along with their influence on the connecting genes. If the score is >0, the node and its connected nodes are all highly expressed, and if the score is <0, the expression of the nodes are low. The nodes (DEGs) in the PPI network with the top 100 |score| values were considered to be the feature genes in myocardial infarction.
Tomography cluster analysis
Tomography cluster analysis was conducted to determine whether the feature genes were differentially expressed between patient and control samples using Pearson's correlation coefficient (22) and average linkage (23). The clustering results were visualized using heatmaps in R version 3.2.1 (24).
Risk gene identification
To further identify the most significant feature genes that distinguish patients with myocardial infarction from controls, the recursive feature elimination (RFE) algorithm was utilized (25). In this algorithm, the optional feature gene combinations were selected as the risk genes in myocardial infarction.
SVM classifier construction
SVM is a supervised classification algorithm that estimates the attribution of a class by distinguishing and predicting the samples by the eigenvalues of the features in each sample (26). A SVM classifier was performed using the selected risk genes by using 4 samples as the training dataset and 1 sample as the testing dataset. The receiver operating characteristic (ROC) curve was drawn to evaluate the precision and robustness of the SVM classifier. A confusion matrix in R version 3.2.1 (https://cran.r-project.org/web/packages/ROCR/index.html) was also employed to visualize the classification results of the classifier.
Verification of the SVM classifier
An additional dataset, GSE61144 (27), was downloaded from the GEO database, which is based on the GPL6106Sentrix Human-6 v2 Expression BeadChip platform. This dataset consists of 7 samples from patients prior to percutaneous coronary intervention (PCI), 7 from patients following PCI and 10 normal controls. These 24 samples were used to verify the classification effect of the SVM classifier on myocardial infarction patient samples by R version 3.2.1 e1071 1.6–8 package (https://cran.r-project.org/web/packages/e1071/index.html).
Results
Identification of DEGs
A total of 1,207 DEGs were screened from myocardial samples compared with normal controls, including 724 downregulated ones and 483 upregulated ones.
PPI network in myocardial infarction samples
The PPI network was comprised of 1,083 nodes (genes) and 46,363 lines (connections). The degrees of the nodes in the network were calculated, and their distributions are presented in Fig. 1. The degrees are referring indexes of the interaction of genes in influencing the development and process of myocardial infarction. A-kinase-anchoring protein (AKAP)12 and glycine receptor α (GLRA)2 were two DEGs with a degree of 1, which means the number of interaction genes is 1 in the PPI network.
Figure 1.
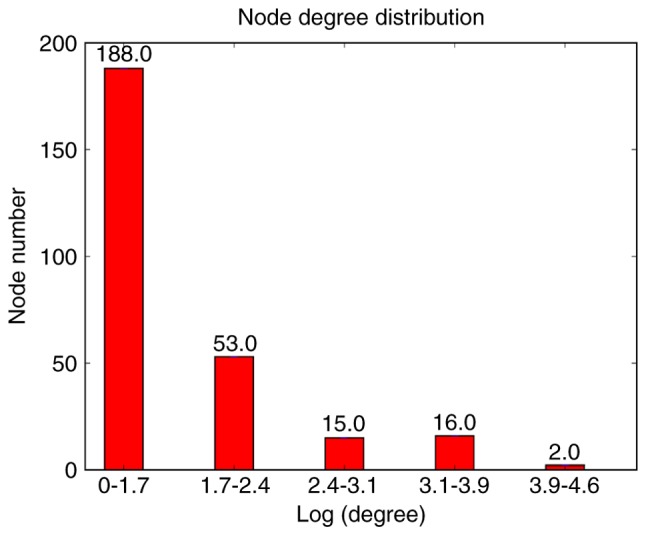
Distributions of node degrees in the protein-protein interaction network. The x-axis represents the log (degree) value; the y-axis indicates the number of responding nodes in each of the log (degree) ranges.
Feature genes and clustering analysis
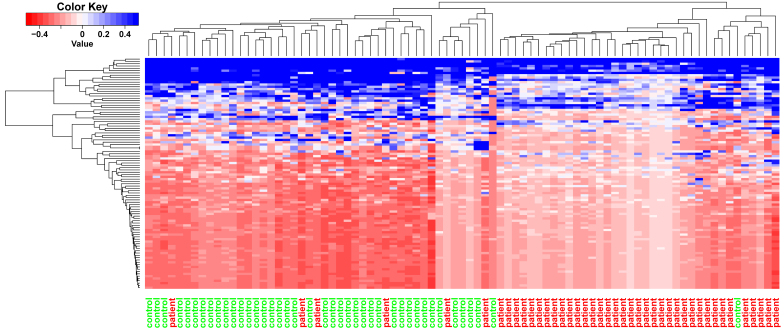
The neighborhood scoring method was employed for the selection of the top 100 feature genes in myocardial infarction samples. The feature genes with a high neighbor score exhibited high expression in the patient samples. The top 10 feature genes are listed in Table I, and included EH domain-binding protein 1, exocyst complex component 6B, growth factor receptor-bound protein 10, AKAP12, SRY-box 4, GLRA3, GLRA2, protein phosphatase 1 regulatory subunit 3A, fatty acid-binding protein (FABP)4 and mediator complex subunit 13-like. Clustering analysis was performed on the top 100 feature genes (Fig. 2), which may allow the classification of myocardial infarction samples to distinguish them from the control samples.
Table I.
Feature genes with top 10 neighbor scores.
| Node | NS_score | Log (fold change) | P-value |
|---|---|---|---|
| EHBP1 | 0.96 | 1.0153 | 0.0004 |
| EXOC6B | 0.96 | 0.9025 | 0.0016 |
| GRB10 | 0.92 | 0.9488 | 0.0009 |
| AKAP12 | 0.91 | 0.9764 | 0.0007 |
| SOX4 | 0.91 | 0.8647 | 0.0026 |
| GLRA3 | 0.91 | −0.8335 | 0.0036 |
| GLRA2 | 0.91 | −0.9855 | 0.0006 |
| PPP1R3A | 0.90 | −1.0402 | 0.0003 |
| FABP4 | 0.90 | 1.0953 | 0.0001 |
| MED13L | 0.90 | 0.7106 | 0.0132 |
NS, neighbor score; EHBP1, EH domain-binding protein 1; EXOC6B, exocyst complex component 6B; GRB10, growth factor receptor-bound protein 10; AKAP12, A-kinase-anchoring protein 12; SOX4, SRY-box 4; GLRA, glycine receptor α; PPP1R3A, protein phosphatase 1 regulatory subunit 3A; FABP4, fatty acid-binding protein 4; MED13L, mediator complex subunit 13-like.
Figure 2.
Clustering analysis results for the top 100 feature genes. The x-axis represents the samples, with control samples marked in green and patient samples marked in red.
Risk genes and SVM classifier
Using the RFE algorithm, a 15-gene combination with a precision of 85% was obtained (Fig. 3) and these genes were recognized as risk genes in myocardial infarction. The expression significance of these risk genes is presented in in Table II, and these risk genes included hes family bHLH transcription factor 5, zinc-finger protein 417, GLRA2, olfactory receptor (OR) family 8 subfamily D member 2 (gene/pseudogene), homeobox A7, FABP6, muscle-associated receptor tyrosine kinase, 5-hydroxytryptamine receptor 6, glutamate receptor-interacting protein 2, OR family 51 subfamily M member 1, OR family 1 subfamily C member 1, killer cell lectin-like receptor K1, vascular endothelial growth factor A, AKAP12 and Ras homolog mTORC1-binding.
Figure 3.
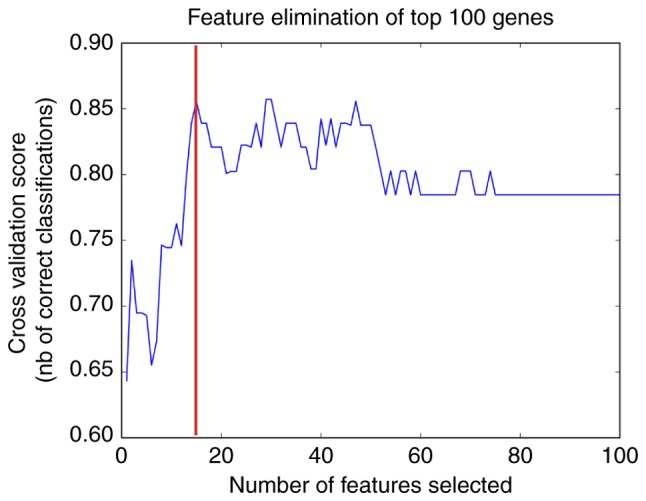
Feature elimination of the top 100 feature genes. The x-axis is the feature gene number and the y-axis indicates the corresponding prediction precision. The gene combination with the highest precision is marked in red, which was a 15-gene combination.
Table II.
Risk genes in myocardial infarction samples.
| Gene | Log (fold change) | P-value |
|---|---|---|
| HES5 | −0.8925 | 0.0018 |
| ZNF417 | −0.8260 | 0.0040 |
| GLRA2 | −0.9855 | 0.0006 |
| OR8D2 | −0.8135 | 0.0045 |
| HOXA7 | 0.7150 | 0.0126 |
| FABP6 | 0.9234 | 0.0013 |
| MUSK | −0.7975 | 0.0054 |
| HTR6 | −0.7651 | 0.0076 |
| GRIP2 | −0.9973 | 0.0005 |
| OR51M1 | −0.8125 | 0.0046 |
| OR1C1 | −0.7755 | 0.0068 |
| KLRK1 | −0.9248 | 0.0013 |
| VEGFA | 0.8442 | 0.0032 |
| AKAP12 | 0.9764 | 0.0007 |
| RHEB | 0.9288 | 0.0012 |
HES5, hes family bHLH transcription factor 5; ZNF417, zinc-finger protein 417; GLRA2, glycine receptor α2; OR, olfactory receptor; OR8D2, OR family 8 subfamily D member 2 (gene/pseudogene); HOXA7, homeobox A7; FABP6, fatty acid-binding protein 6; MUSK, muscle-associated receptor tyrosine kinase; HTR6, 5-hydroxytryptamine receptor 6, GRIP2, glutamate receptor-interacting protein 2, OR51M1, OR family 51 subfamily M member 1; OR1C1, OR family 1 subfamily C member 1; KLRK1, killer cell lectin-like receptor K1; VEGFA, vascular endothelial growth factor A; AKAP12, A-kinase-anchoring protein 12; RHEB, Ras homolog mTORC1-binding.
The average precision of the SVM classifier was 86%, as indicated in the ROC curve (Fig. 4), which was 88% to the patient samples following visualization by a confusion matrix (Fig. 5). The classification effect was also verified using the independent microarray data GSE61144, and the ROC curve is presented in Fig. 6. The predictive precision was 0.92, the average true positive rate was 0.9278 and the average false positive rate was 0.2361.
Figure 4.

ROC curve of the support vector machine classifier. The x-axis represents the false positive rate and the y-axis indicates the true positive rate. The simulation results (mean ROC) is marked by the dotted line. ROC, receiver operating characteristic.
Figure 5.
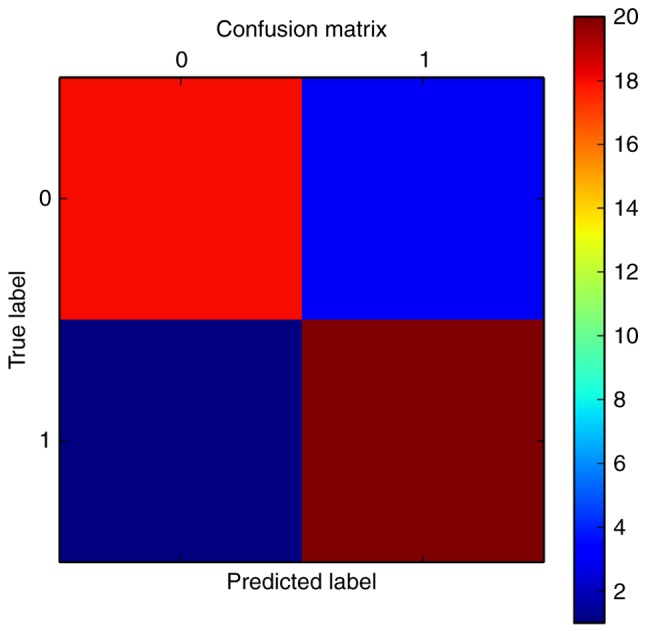
Confusion matrix of the support vector machine classifier.
Figure 6.
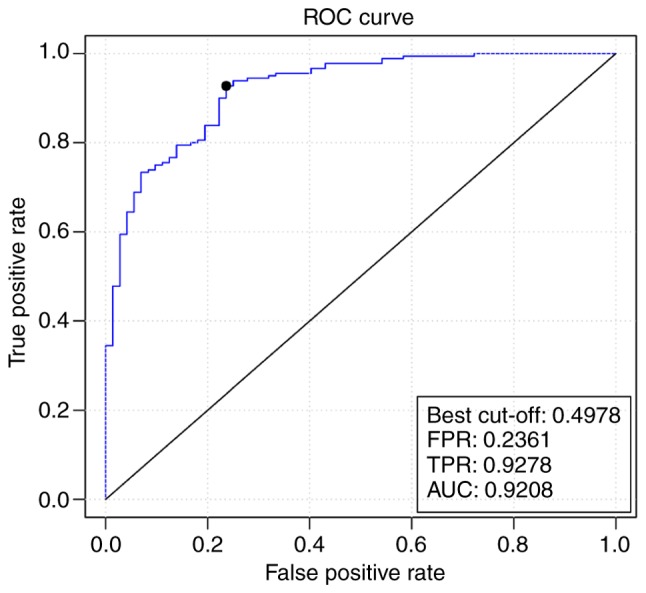
ROC curve of the support vector machine classifier verified by the GSE61144 dataset. The x-axis represents the false positive rate and the y-axis indicates the true positive rate. ROC, receiver operating characteristic; FPR, false positive rate; TPR, true positive rate; AUC, area under the curve.
Discussion
To identify the risk genes in myocardial infarction, the GSE34198 microarray dataset was downloaded from the GEO database, and 724 downregulated and 483 upregulated DEGs were screened in patient samples compared with control samples. The PPI network of myocardial infarction was comprised of 1,083 nodes (genes) and 46,363 lines (connections). Using the neighborhood scoring method, the top 100 feature genes in myocardial infarction samples were identified as the disease feature genes, which allow myocardial infarction samples to be distinguished from the control samples. The RFE algorithm screened 15 risk genes, which were utilized to construct a SVM classifier with an average precision of 88% to the patient samples following visualization by a confusion matrix. The predictive precision of the classifier on another microarray dataset, GSE61144, was 0.92, with average true positive rate of 0.9278 and an average false positive rate of 0.2361. AKAP12 and GLRA2 were two of the risk genes identified.
AKAPs are scaffolding proteins that regulate the cellular cyclic AMP response. Several AKAPs are reported to be expressed in the heart, including AKAP18, AKAP79, AKAP6 and AKAP220 (28,29). AKAPs participate in cardiovascular functions by various mechanisms. For example, AKAPs were reported to anchor protein kinase A (PKA) in the sarcomere for the phosphorylation of myofibril proteins in contractile responses (30). In addition, AKAPs docked APK in proximity of sarcomeric substrates to enhance cardiac contractility (31). AKAPs mediate certain phosphorylation events in the heart, and AKAP6 complex disruption resulted in aberrant Ca2+ cycling, which was associated with arrhythmia (32). Loss of AKAP150 promoted pathological remodeling and heart failure propensity by disrupting Ca2+ cycling and contractile reserve (33). Furthermore, when voltage-gated K+ currents were reduced in ventricular myocytes following myocardial infarction, AKAP150 was reported to be involved in the activation of calcineurin/nuclear factor of activated T-cells (34). PKA is involved in the progression of heart failure (35), therefore, AKAPs, which regulate the activity of PKA, are also risk factors in heart failure. AKAP12 has been associated with various cellular functions, including cytoskeletal architecture and cell cycle regulation (36,37). Activated AKAP12 has been observed in the plasma membrane, cell periphery and perinuclear regions in the cytoplasm (38). Although no associations between AKAP12 and heart disease have been previously reported, its potential role can be inferred based on the functions of other AKAPs.
Glycine is a simple physiological compound whose function in cardiovascular disease is receiving increased attention is research. Glycine was reported to protect against ischemia-reperfusion injury in cells and isolated perfused organs by inhibiting neuronal apoptosis in mice (39,40). Glycine receptors have been identified in the myocardial cell membrane, which aid the cytoprotective effects of glycine in myocardial cells (41). Furthermore, it was reported that the cytoprotective effect of glycine against ATP depletion-induced injury may be mediated by the glycine receptor in renal cells (42). GLRA2 is one type of glycine receptor, and, currently, no direct evidence has revealed its role in cardiovascular disease. However, the present study performed bioinformatics analysis to demonstrated that GLRA2 was a risk gene in myocardial infarction. Although the above result based on bioinformatics analysis is important, confirmation of the above-mentioned results is required by performing functional studies, and the role of AKAP12 and GLRA2 genes in myocardial infarction requires further investigation.
In conclusion, the results of the present study indicate that AKAP12 and GLRA2 exert potential roles in the development of myocardial infarction, potentially by influencing cardiac contractility and protecting against ischemia-reperfusion injury.
References
- 1.Coventry LL, Finn J, Bremner AP. Sex differences in symptom presentation in acute myocardial infarction: A systematic review and meta-analysis. Heart. 2011;40:477–491. doi: 10.1016/j.hrtlng.2011.05.001. [DOI] [PubMed] [Google Scholar]
- 2.Valensi P1, Lorgis L, Cottin Y. Prevalence, incidence, predictive factors and prognosis of silent myocardial infarction: A review of the literature. Arch Cardiovasc Dis. 2011;104:178–188. doi: 10.1016/j.acvd.2010.11.013. [DOI] [PubMed] [Google Scholar]
- 3.Panza JA. Myocardial ischemia and the pains of the heart. N Engl J Med. 2002;346:1934–1935. doi: 10.1056/NEJMp020047. [DOI] [PubMed] [Google Scholar]
- 4.Langenbrunner JC, Marquez PV, Wang S. Human Development Unit; East Asia and Pacific region. Washington, DC: The World Bank; 2011. [Oct 21;2016 ]. Toward a Healthy and Harmonious Life in China: Stemming the Rising Tide of Non-Communicable Diseases. [Google Scholar]
- 5.Velagaleti RS, Pencina MJ, Murabito JM, Wang TJ, Parikh NI, D'Agostino RB, Levy D, Kannel WB, Vasan RS. Long-term trends in the incidence of heart failure after myocardial infarction. Circulation. 2008;118:2057–2062. doi: 10.1161/CIRCULATIONAHA.108.784215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pfeffer MA, Braunwald E. Ventricular remodeling after myocardial infarction. Experimental observations and clinical implications. Circulation. 1990;81:1161–1172. doi: 10.1161/01.CIR.81.4.1161. [DOI] [PubMed] [Google Scholar]
- 7.Go AS, Mozaffarian D, Roger VL, Benjamin EJ, Berry JD, Borden WB, Bravata DM, Dai S, Ford ES, Fox CS, et al. Heart disease and stroke statistics-2013 update: A report from the American Heart Association. Circulation. 2013;127:e6–e245. doi: 10.1161/CIR.0b013e318282ab8f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rosenberg L, Kaufman DW, Helmrich SP, Miller DR, Stolley PD, Shapiro S. Myocardial infarction and cigarette smoking in women younger than 50 years of age. JAMA. 1985;253:2965–2969. doi: 10.1001/jama.253.20.2965. [DOI] [PubMed] [Google Scholar]
- 9.Leong DP, Smyth A, Teo KK, McKee M, Rangarajan S, Pais P, Liu L, Anand SS, Yusuf S. INTERHEART Investigators: Patterns of alcohol consumption and myocardial infarction risk: Observations from 52 Countries in the interheart case-control study. Circulation. 2014;130:390–398. doi: 10.1161/CIRCULATIONAHA.113.007627. [DOI] [PubMed] [Google Scholar]
- 10.Acute myocardial infarction and combined oral contraceptives: Results of an international multicentre case-control study: WHO Collaborative Study of Cardiovascular Disease and Steroid Hormone Contraception. Lancet. 1997;349:1202–1209. doi: 10.1016/S0140-6736(97)02358-1. [DOI] [PubMed] [Google Scholar]
- 11.Cassidy A, Mukamal KJ, Liu L, Franz M, Eliassen AH, Rimm EB. High anthocyanin intake is associated with a reduced risk of myocardial infarction in young and middle-aged women. Circulation. 2013;127:188–196. doi: 10.1161/CIRCULATIONAHA.112.122408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Freiberg MS, Chang CC, Kuller LH, Skanderson M, Lowy E, Kraemer KL, Butt AA, Goetz Bidwell M, Leaf D, Oursler KA, et al. HIV infection and the risk of acute myocardial infarction. JAMA Intern Med. 2013;173:614–622. doi: 10.1001/jamainternmed.2013.3728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Helgadottir A, Manolescu A, Thorleifsson G, Gretarsdottir S, Jonsdottir H, Thorsteinsdottir U, Samani NJ, Gudmundsson G, Grant SF, Thorgeirsson G, et al. The gene encoding 5-lipoxygenase activating protein confers risk of myocardial infarction and stroke. Nat Genet. 2004;36:233–239. doi: 10.1038/ng1311. [DOI] [PubMed] [Google Scholar]
- 14.Do R, Stitziel NO, Won HH, Jørgensen AB, Duga S, Merlini Angelica P, Kiezun A, Farrall M, Goel A, Zuk O, et al. Multiple rare alleles at LDLR and APOA5 confer risk for early-onset myocardial infarction. Nature. 2015;518:102–106. doi: 10.1038/nature13917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Han H, Wang H, Yin Z, Jiang H, Fang M, Han J. Association of genetic polymorphisms in ADH and ALDH2 with risk of coronary artery disease and myocardial infarction: A meta-analysis. Gene. 2013;526:134–141. doi: 10.1016/j.gene.2013.05.002. [DOI] [PubMed] [Google Scholar]
- 16.Valenta Z, Mazura I, Kolár M, Grünfeldová H, Feglarová P, Peleška J, Tomecková M, Kalina J, Slovák D, Zvárová J. Determinants of excess genetic risk of acute myocardial infarction-a matched case-control study. Eur J Biomed Inform. 2012;8:34–43. [Google Scholar]
- 17.Gautier L, Cope L, Bolstad BM, Irizarry RA. affy-analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. 2004;20:307–315. doi: 10.1093/bioinformatics/btg405. [DOI] [PubMed] [Google Scholar]
- 18.Liu Q, Wong L, Li J. Z-score biological significance of binding hot spots of protein interfaces by using crystal packing as the reference state. Biochim Biophys Acta. 2012;1824:1457–1467. doi: 10.1016/j.bbapap.2012.05.014. [DOI] [PubMed] [Google Scholar]
- 19.Paic F, Igwe JC, Nori R, Kronenberg MS, Franceschetti T, Harrington P, Kuo L, Shin DG, Rowe DW, Harris SE, Kalajzic I. Identification of differentially expressed genes between osteoblasts and osteocytes. Bone. 2009;45:682–692. doi: 10.1016/j.bone.2009.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu C, Xuan Z. Prioritization of cancer-related genomic variants by SNP association network. Cancer Inform. 2015;14(Suppl 2):S57–S70. doi: 10.4137/CIN.S17288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu F, Yang Z, Hu X, Sun Y, Lin H, Wang J. Protein complex detection in PPI networks based on data integration and supervised learning method. BMC Bioinformatics. 2015;16(Suppl 12):S3. doi: 10.1186/1471-2105-16-S12-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu AN, Wang LL, Li HP, Gong J, Liu XH. Correlation between posttraumatic growth and posttraumatic stress disorder symptoms based on pearson correlation coefficient: A meta-analysis. J Nerv Ment Dis. 2017;205:380–389. doi: 10.1097/NMD.0000000000000605. [DOI] [PubMed] [Google Scholar]
- 23.Malta DC, Bernal RI, Almeida MC, Ishitani LH, Girodo AM, Paixão LM, Oliveira MT, Junior Pimenta FG, Júnior Silva JB. Inequities in intraurban areas in the distribution of risk factors for non communicable diseases, Belo Horizonte, 2010. Rev Bras Epidemiol. 2014;17:629–641. doi: 10.1590/1809-4503201400030005. (In English, Portuguese) [DOI] [PubMed] [Google Scholar]
- 24.Metsalu T, Vilo J. ClustVis: A web tool for visualizing clustering of multivariate data using principal component analysis and heatmap. Nucleic Acids Res. 2015;43:W566–W570. doi: 10.1093/nar/gkv468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Qureshi MN, Min B, Jo HJ, Lee B. Multiclass classification for the differential diagnosis on the ADHD subtypes using recursive feature elimination and hierarchical extreme learning machine: Structural MRI study. PLoS One. 2016;11:e0160697. doi: 10.1371/journal.pone.0160697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Matwin S, Sazonova V. Direct comparison between support vector machine and multinomial naive Bayes algorithms for medical abstract classification. J Am Med Inform Assoc. 2012;19:917. doi: 10.1136/amiajnl-2012-001072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Park HJ, Noh JH, Eun JW, Koh YS, Seo SM, Park WS, Lee JY, Chang K, Seung KB, Kim PJ, Nam SW. Assessment and diagnostic relevance of novel serum biomarkers for early decision of ST-elevation myocardial infarction. Oncotarget. 2015;6:12970–12983. doi: 10.18632/oncotarget.4001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kapiloff MS, Schillace RV, Westphal AM, Scott JD. mAKAP: An A-kinase anchoring protein targeted to the nuclear membrane of differentiated myocytes. J Cell Sci. 1999;112:2725–2736. doi: 10.1242/jcs.112.16.2725. [DOI] [PubMed] [Google Scholar]
- 29.Redden JM, Dodge-Kafka KL. AKAP phosphatase complexes in the heart. J Cardiovasc Pharmacol. 2011;58:354–362. doi: 10.1097/FJC.0b013e31821e5649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Perino A, Ghigo A, Scott JD, Hirsch E. Anchoring proteins as regulators of signaling pathways. Circ Res. 2012;111:482–492. doi: 10.1161/CIRCRESAHA.111.262899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sumandea CA, Garcia-Cazarin ML, Bozio CH, Sievert GA, Balke CW, Sumandea MP. Cardiac troponin T, a sarcomeric AKAP, tethers protein kinase A at the myofilaments. J Biol Chem. 2011;286:530–541. doi: 10.1074/jbc.M110.148684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lehnart SE, Wehrens XH, Reiken S, Warrier S, Belevych AE, Harvey RD, Richter W, Jin SL, Conti M, Marks AR. Phosphodiesterase 4D deficiency in the ryanodine-receptor complex promotes heart failure and arrhythmias. Cell. 2005;123:25–35. doi: 10.1016/j.cell.2005.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li L, Li J, Drum BM, Chen Y, Yin H, Guo X, Luckey SW, Gilbert ML, McKnight GS, Scott JD, et al. Loss of AKAP150 promotes pathological remodelling and heart failure propensity by disrupting calcium cycling and contractile reserve. Cardiovasc Res. 2017;113:147–159. doi: 10.1093/cvr/cvw221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nieves-Cintrón M, Hirenallur-Shanthappa D, Nygren PJ, Hinke SA, Dell'Acqua ML, Langeberg LK, Navedo M, Santana LF, Scott JD. AKAP150 participates in calcineurin/NFAT activation during the down-regulation of voltage-gated K(+) currents in ventricular myocytes following myocardial infarction. Cell Signal. 2016;28:733–740. doi: 10.1016/j.cellsig.2015.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bockus LB, Humphries KM. cAMP-dependent protein kinase (PKA) signaling is impaired in the diabetic heart. J Biol Chem. 2015;290:29250–29258. doi: 10.1074/jbc.M115.681767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nelson PJ, Moissoglu K, Vargas J, Jr, Klotman PE, Gelman IH. Involvement of the protein kinase C substrate, SSeCKS, in the actin-based stellate morphology of mesangial cells. J Cell Sci. 1999;112:361–370. doi: 10.1242/jcs.112.3.361. [DOI] [PubMed] [Google Scholar]
- 37.Hehnly H, Canton D, Bucko P, Langeberg LK, Ogier L, Gelman I, Santana LF, Wordeman L, Scott JD. A mitotic kinase scaffold depleted in testicular seminomas impacts spindle orientation in germ line stem cells. Elife. 2015;4:e09384. doi: 10.7554/eLife.09384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yan X, Walkiewicz M, Carlson J, Leiphon L, Grove B. Gravin dynamics regulates the subcellular distribution of PKA. Exp Cell Res. 2009;315:1247–1259. doi: 10.1016/j.yexcr.2008.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Petrat F, Boengler K, Schulz R, de Groot H. Glycine, a simple physiological compound protecting by yet puzzling mechanism(s) against ischaemia-reperfusion injury: Current knowledge. Br J Pharmacol. 2012;165:2059–2072. doi: 10.1111/j.1476-5381.2011.01711.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lu Y, Zhang J, Ma B, Li K, Li X, Bai H, Yang Q, Zhu X, Ben J, Chen Q. Glycine attenuates cerebral ischemia/reperfusion injury by inhibiting neuronal apoptosis in mice. Neurochem Int. 2012;61:649–658. doi: 10.1016/j.neuint.2012.07.005. [DOI] [PubMed] [Google Scholar]
- 41.Qi RB, Zhang JY, Lu DX, Wang HD, Wang HH, Li CJ. Glycine receptors contribute to cytoprotection of glycine in myocardial cells. Chin Med J (Engl) 2007;120:915–921. [PubMed] [Google Scholar]
- 42.Pan C, Bai X, Fan L, Ji Y, Li X, Chen Q. Cytoprotection by glycine against ATP-depletion-induced injury is mediated by glycine receptor in renal cells. Biochem J. 2005;390:447–453. doi: 10.1042/BJ20050141. [DOI] [PMC free article] [PubMed] [Google Scholar]