Abstract
Glioblastoma multiforme (GBM), a World Health Organization grade IV glioma, is the most common and aggressive primary brain tumor in humans. microRNAs (miRNAs) are aberrantly expressed in numerous cancer types, including GBM. Abnormally expressed miRNAs are commonly associated with malignant characteristics of GBM, including malignant growth, proliferation, apoptosis, invasion, metastasis and resistance to chemotherapy. miRNA (miR)-376a is abnormally expressed in multiple human cancers; however, the expression pattern and role of miR-376a in GBM, and the underlying molecular mechanisms by which miR-376a exerts its functions remain to be elucidated. Therefore, the aim of this study was to measure miR-376a expression and determine its biological roles in GBM as well as its associated molecular mechanism. In the present study, miR-376a expression was markedly downregulated in GBM tissues and cell lines. Overexpression of miR-376a markedly decreased the proliferation and invasion of GBM cells in vitro. In the present study, specificity protein 1 (SP1) was demonstrated to be a direct target of miR-376a. In addition, a negative association between SP1 mRNA and miR-376a expression was observed in GBM tissues. SP1 upregulation reduced the effects of miR-376a overexpression on GBM cell proliferation and invasion. miR-376a may be a therapeutic target for the treatment of patients with GBM.
Keywords: microRNA-376a, glioblastoma multiforme, specificity protein 1, proliferation, invasion
Introduction
Gliomas are the most common malignant neoplasm of the central nervous system in adults, and accounts for 78% of intracranial primary tumors (1). Based on the degree of malignancy, according to the World Health Organization (WHO) classification, glioma are classified into four histopathologic grades: WHO grades I and II, which are considered low-grade astrocytomas, and WHO grades III and IV, which are considered high-grade astrocytomas (2). Glioblastoma multiforme (GBM), a WHO grade IV glioma, is the most common and aggressive type of primary brain tumor in humans (3). GBM tumors are characterized by an increased degree of invasiveness and rapid growth (4–7). Despite a combination of therapeutic strategies, including surgery, radiotherapy and chemotherapy, the average 5-year survival rate of patients with GBM is <5% (8); the median survival time of patients with GBM is ~1 year (9). Therefore, the molecular mechanisms underlying the tumorigenicity and development of GBM require further study, and novel therapeutic treatments for patients with this disease are necessary.
MicroRNAs (miRNAs) belong to a family of small (19–25 nucleotides long) noncoding RNA molecules (10), that negatively regulate gene expression at the post-transcriptional level by imperfectly base pairing with complementary sites within the 3′-untranslated regions (3′-UTRs) of their target genes, thereby leading to degradation or translational repression of target mRNAs (11). miRNAs have been hypothesized to participate in modulating the expression of more than one-third of human genes (12). Previous studies have reported that a number of miRNAs are aberrantly expressed in various types of human cancer, including GBM (13–15). miRNAs are involved in tumorigenesis and tumor progression through the regulation of a variety of biological processes, including cell proliferation, cell cycle, apoptosis, differentiation, motility and angiogenesis (16–18). Overexpression of miRNAs in human cancers may act as oncogenes by negatively regulating the expression of tumor-suppressor genes. By contrast, downregulated miRNA expression levels may serve roles of tumor suppressors by targeting oncogenes (19,20). Therefore, identifying misregulated miRNAs and investigating their biological roles may provide novel effective therapeutic targets for cancer treatment.
miR-376a, which has been mapped to the 14q32.31 locus, is abnormally expressed in multiple human cancers (21–23). However, the expression pattern and role of miR-376a in GBM, as well as the underlying molecular mechanisms remain to be elucidated. SP1 was previously reported to be upregulated in GBM and associated with the WHO grading and survival status (24). Additionally, SP1 may be involved in initiation and progression of GBM by regulating proliferation, cell cycle and invasion (24–27). The present study demonstrated that miR-376a is expressed at low levels in GBM tissues and cell lines. miR-376a upregulation inhibits GBM cell proliferation and invasion by directly targeting specificity protein 1 (SP1).
Materials and methods
Clinical specimens
The present study was approved by the Ethics Committee of Central Hospital of Linyi (Linyi, China), and written informed consent was obtained from patients with GBM for use of tissues. A total of 26 paired GBM tissues and adjacent normal brain tissues were obtained from the patients with GBM (17 males and 9 females; age range, 42–77 years) who underwent surgical resection at the Central Hospital of Linyi between August 2012 and February 2016. None of the patients were treated with radiotherapy or chemotherapy prior to surgery. Collected specimens were immediately frozen following surgical resection and stored in liquid nitrogen.
Cell lines and transfection
Human GBM cell lines (U138, U251, LN229, T98) were purchased from Shanghai Cell Bank of the Chinese Academy of Sciences (Shanghai, China). GBM cell lines were cultured in Dulbecco's modified Eagle medium (DMEM; Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented with 10% fetal bovine serum (FBS; Invitrogen; Thermo Fisher Scientific, Inc.), 100 U/ml penicillin and 100 mg/ml streptomycin (Invitrogen; Thermo Fisher Scientific, Inc.). Normal human astrocytes (NHAs) were obtained from ScienCell Research Laboratories, Inc. (San Diego, CA, USA) and maintained in Astrocyte Medium (ScienCell Research Laboratories, Inc.). All cell lines were cultured at 37°C in a humidified chamber containing 5% CO2.
miR-376a mimics and miRNA negative control (miR-NC) were obtained from Guangzhou RiboBio Co., Ltd. (Guangzhou, China). The miR-376a mimics sequence was 5′-AUCAUAGAGGAAAAUCCACGU-3′ and the miR-NC sequence was 5′-UUCUCCGAACGUGUCACGUTT-3′. The SP1-overexpression plasmid pcDNA3.1-SP1 and bland pcDNA3.1 plasmid were purchased from Shanghai GenePharma Co., Ltd. (Shanghai, China). For transfection, cells were seeded in a 6-well plate at a density of 5×105 cells/well. When 70% confluence was achieved, cells were transfected with miRNA mimics (100 pmol), miR-NC (100 pmol) or plasmids (2 µg) at room temperature using Lipofectamine 2000 Transfection Reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. Following transfection for 48 h, reverse transcription-quantitative polymerase chain reaction (RT-qPCR) was performed to detect miR-376a or SP1 mRNA level. CCK-8 and Matrigel chamber invasion assays were conducted at 24 and 48 h post transfection. Western blot analysis was carried out at 72 h following transfection.
RNA extraction and RT-qPCR
Total RNA form tissue specimens (100 mg) or cells (1.5×106) was isolated using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol. For the determination of miR-376a expression levels, cDNA was synthesized from 1 µg total RNA using TaqMan MicroRNA Reverse Transcription kit (Applied Biosystems; Thermo Fisher Scientific, Inc.), following the manufacturer's protocol. Relative miR-376a expression was quantified using the TaqMan MicroRNA PCR kit (Applied Biosystems; Thermo Fisher Scientific, Inc.); U6 small nuclear RNA was used as an endogenous control. The following thermocycling conditions were used for the PCR: 50°C for 2 min, 95°C for 10 min; 40 cycles of 95°C for 15 sec and 60°C for 60 sec. For SP1 mRNA expression, total RNA was reverse transcribed into cDNA using PrimeScript RT Reagent kit (Takara Biotechnology Co., Ltd., Dalian, China). A SYBR Premix Ex Taq™ kit (Takara Biotechnology Co., Ltd.) was used to detect SP1 mRNA expression; GAPDH was used as an internal control. The following thermocycling conditions were used for the PCR: 5 min at 95°C, followed by 40 cycles of 95°C for 30 sec and 65°C for 45 sec. qPCR was performed using an ABI-7500 Fast Real-Time PCR system (Applied Biosystems; Thermo Fisher Scientific, Inc.), and data was analyzed using the 2−ΔΔCq method (28). The following primers were used in the present study: miR-376a, forward 5′-GTGCAGGGTCCGAGGT-3′, reverse 5′-ATCATAGAGGAAAATCCACG-3′; U6, forward 5′-ATTGGAACGATACAGAGAAGATT-3′, reverse 5′-GGAACGCTTCACGAATTTG-3′; SP1, forward 5′-TGGTGGGCAGTATGTTGT-3′, reverse 5′-GCTATTGGCATTGGTGAA-3′; and GAPDH, forward 5′-ATGTCGTGGAGTCTACTGGC-3′, reverse 5′-TGACCTTGCCCACAGCCTTG-3′.
Cell Counting Kit-8 (CCK-8) proliferation assay
The proliferative abilities of GBM U251 cells were determined using CCK-8 proliferation assays (Dojindo Molecular Technologies, Inc., Kumamoto, Japan). Transfected cells were seeded in 96-well plates at 3,000 cells/well and cultured for 0, 24, 48 and 72 h at 37°C. At the indicated times, 10 µl CCK-8 Regent was added into each well and, following 2 h incubation at 37°C, the absorbance was measured at a wavelength of 450 nm using an automatic multi-well spectrophotometer (Bio-Rad Laboratories, Inc., Hercules, CA, USA).
Matrigel chamber invasion assay
Transwell filters (BD Biosciences, Franklin Lakes, NJ, USA) coated with Matrigel (BD Biosciences) on the upper surface of the 8 µm pore polycarbonate membrane were used to assess cell invasion, in accordance with the manufacturer's protocol. Transfected cells (1×105 cells/well) were seeded in the upper well of the Transwell filters in FBS-free DMEM. The lower Transwell chamber were filled with DMEM medium containing 10% FBS as a chemoattractant. Following incubation for 24 h at 37°C, cells on the top membranes were removed using cotton swabs. Invasive cells on the lower membrane were fixed with 100% methanol at room temperature for 15 min, stained with 0.5% crystal violet solution at room temperature for 15 min and washed with PBS. Images of invasive cells were captured and cells were counted under an Olympus IX53 inverted fluorescent microscope (Olympus Corporation, Tokyo, Japan) in five independent fields for each well to analyze the invasive ability of cells.
Bioinformatics analysis and luciferase reporter assay
Potential targets of miR-376a were predicted using miRanda (http://www.microrna.org/microrna/home.do) and TargetScan (http://www.targetscan.org). For luciferase reporter assay, reporter plasmids, including pGL3-SP1-3′UTR-wild-type (Wt) and pGL3-SP1-3′UTR-mutant (Mut), were synthesized and verified by Shanghai GenePharma Co., Ltd. Cells were seeded into a 24-well plate at a density of 1.5×105 cells/well and cultured overnight at 37°C prior to co-transfection with pGL3-SP1-3′UTR-Wt or pGL3-SP1-3′UTR-Mut with miR-376a mimics or miR-NC using Lipofectamine® 2000 transfection reagent at 37°C. A total of 48 h following transfection, Firefly and Renilla luciferase activities were determined using the Dual-Luciferase Reporter Assay System (Promega Corporation, Madison, WI, USA), according to the manufacturer's protocol. Renilla luciferase was selected for normalization. Experiments were repeated three times independently.
Protein extraction and western blot analysis
Total protein was extracted from tissue samples (200 mg) and U251 cells (1.5×106) using Total Protein Extraction kit (Nanjing KeyGen Biotech Co., Ltd., Nanjing, China) supplemented with protease inhibitors (EMD Millipore, Billerica, MA, USA). Protein concentrations were measured using a Bicinchoninic Acid Assay kit (Beyotime Institute of Biotechnology, Haimen, China). Equal quantities of protein (20 µg/lane) were separated by 10% SDS-PAGE and transferred to polyvinylidene difluoride membranes (EMD Millipore). The membranes were subsequently blocked with Tris-buffered saline and 0.05% Tween-20 (TBST) containing 5% non-fat milk at room temperature for 1 h, followed by incubation with primary antibodies at 4°C overnight. Primary antibodies used in the present study include mouse anti-SP1 monoclonal antibody (1:1,000; cat. no. sc-420) and mouse anti-GAPDH monoclonal antibody (1:1,000; cat. no. sc-32233), both from Santa Cruz Biotechnology, Inc. (Dallas, TX, USA). Following three washes with TBST, the membranes were incubated with a goat anti-mouse horseradish peroxidase-conjugated secondary antibody (1:5,000 dilution; cat. no. sc-2005; Santa Cruz Biotechnology) at room temperature for 2 h. Protein bands were visualized using an Enhanced Chemiluminescence kit (Beyotime Institute of Biotechnology), according to the manufacturer's protocol, and quantification of band intensities was conducted using ImageJ 1.49 software (National Institutes of Health, Bethesda, MD, USA). GAPDH was used as a loading control.
Statistical analysis
Data are presented as the mean ± standard deviation. Each assay was repeated three times independently. Data were analyzed with two-tailed Student's t-test or one-way analysis of variance using SPSS statistical software (version 19.0; IBM Corp., Armonk, NY, USA). Student-Newman-Keuls method was used to compare differences between two groups in multiple comparison analyses following one-way analysis of variance. P<0.05 was considered to indicate a statistically significant difference.
Results
miR-376a is downregulated in GBM tissue samples and cell lines
Expression levels of miR-376a were detected in GBM tissues and adjacent normal brain tissues to investigate the potential roles of miR-376a in GBM. miR376a expression levels were significantly reduced in GBM tissues compared with adjacent normal brain tissues (P<0.05; Fig. 1A). Subsequently, miR-376a expression was determined in GBM cell lines (U138, U251, LN229 and T98) and normal human astrocytes (NHAs). miR-376a expression levels were significantly lower in all GBM cell lines compared with NHA cells (P<0.05; Fig. 1B). These results suggested that miR-376a may serve a role in GBM progression.
Figure 1.
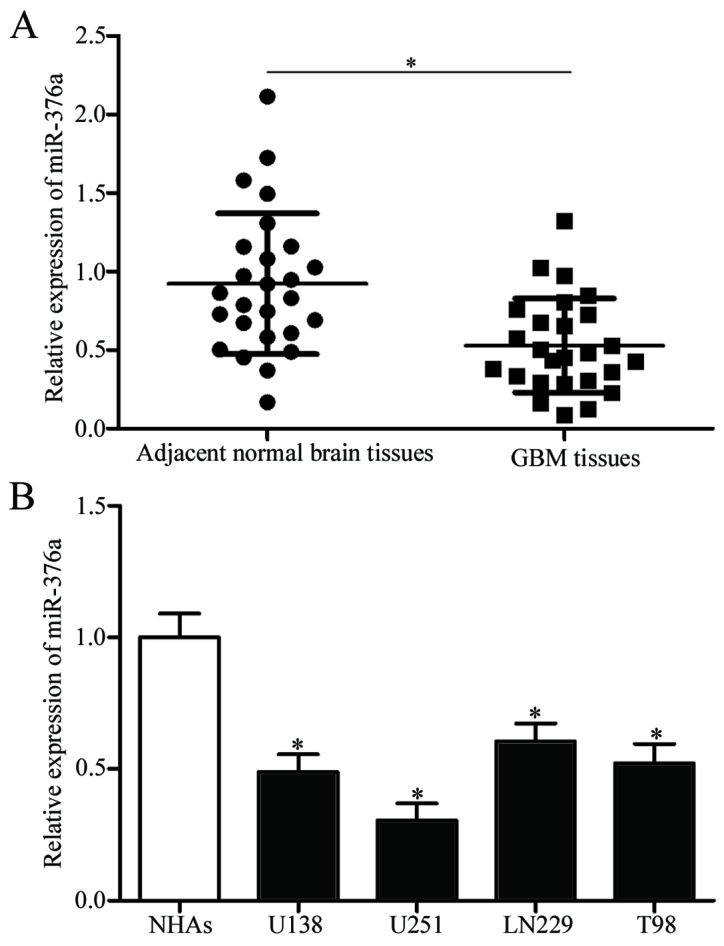
miR-376a expression is decreased in GBM tissues and cell lines. (A) miR-376a expression was downregulated in GBM tissues. Data are presented as the mean ± standard deviation; *P<0.05. (B) Expression levels of miR-376a in GBM cell lines U138, U251, LN229 and T98, and in NHAs were determined using reverse transcription-quantitative polymerase chain reaction. Data are presented as the mean ± standard deviation; *P<0.05 vs. NHA. GBM, glioblastoma multiforme; miR, microRNA; NHA, normal human astrocytes.
miR-376a overexpression inhibits cell proliferation and invasion in GBM cell lines
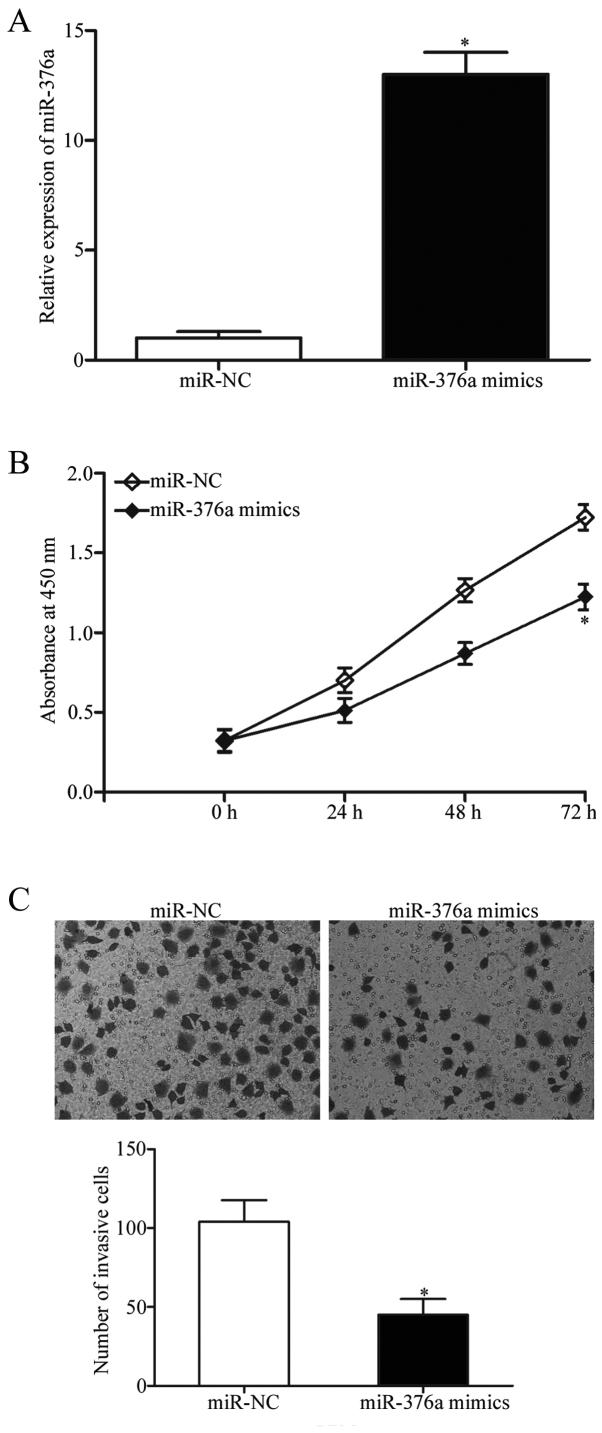
As aforementioned, miR-376a expression was reduced in GBM tissues and cell lines; therefore, it was hypothesized that miR-376a may be a tumor suppressor in GBM. miR-376a mimics or miR-NCs were transfected into U251 cells, and the transfection efficiency of miR-376a was confirmed using RT-qPCR, which demonstrated that miR-376a expression was significantly upregulated in cells transfected with miR-376a mimics compared with cells transfected with miR-NC (P<0.05; Fig. 2A). The effects of miR-376a overexpression on the proliferation of GBM cells were determined by CCK-8 proliferation assay. Upregulation of miR-376a expression levels significantly suppressed U251 cell proliferation at 72 h (P<0.05; Fig. 2B). Matrigel chamber invasion assays were performed to determine whether miR-376a influenced invasive abilities of GBM cells. The results demonstrated that increased miR-376a expression significantly decreased the invasive capacities of U251 cells compared with miR-NC-transfected cells (P<0.05; Fig. 2C). These results suggested that miR-376a may serve a role as a tumor suppressor in GBM progression.
Figure 2.
miR-376a inhibits proliferation and invasion in U251 cells. (A) miR-376a expression was elevated in U251 cells following transfection with miR-376a mimics. (B) Cell proliferation was detected using Cell Counting Kit 8 assay in U251 cells transfected with miR-376a mimics or miR-NC. (C) Cell invasive ability was determined by Matrigel chamber invasion assays in U251 cells following transfection with miR-376a mimics or miR-NC. Data are presented as the mean ± standard deviation; *P<0.05 vs. miR-NC. miR, micro RNA; NC, negative control.
SP1 is a target of miR-376a in GBM
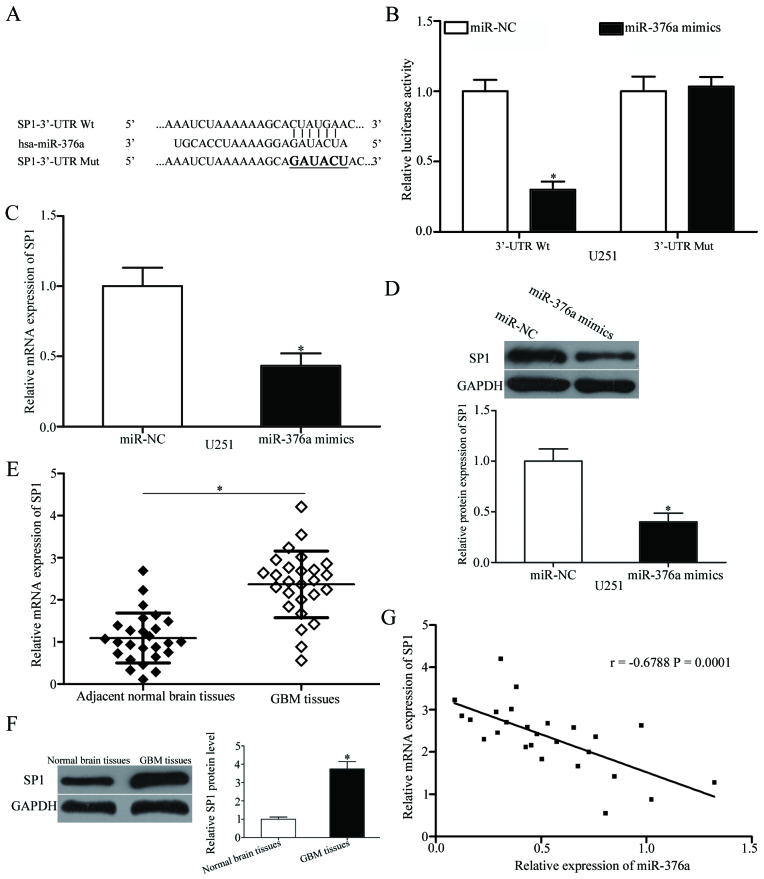
Bioinformatics analysis was used to predict potential targets of miR-376a and to investigate the mechanisms underlying miR-376a-mediated regulation of GBM progression. A number of candidate genes were predicted, including SP1 (Fig. 3A). SP1 was selected for further verification as it was previously demonstrated to function as a proto-oncogene in GBM (24–27). Luciferase reporter assays were performed to confirm this hypothesis. The SP1-3′-UTR-Wt or SP1-3′-UTR-Mut reporter plasmid was co-transfected into U251 cells with miR-376a mimics or miR-NC. The results demonstrated that miR-376a mimics reduced the luciferase activity level of SP1-3′-UTR-Wt compared with miR-NC (P<0.05; Fig. 3B), whereas no significant difference was identified for SP1-3′-UTR-Mut. RT-qPCR and western blot analyses were used to confirm the effects of miR-376a on SP1 expression in GBM cells. The results indicated that both mRNA and protein levels of SP1 were significantly reduced by miR-376a overexpression in U251 cells (P<0.05; Fig. 3C and D, respectively).
Figure 3.
SP1 is a direct target gene of miR-376a in GBM. (A) Predicted miR-376a target sequence in the 3′-UTR of SP1 Wt and Mut containing six mutated nucleotides in the 3′-UTR of SP1. (B) Luciferase reporter assays were performed in U251 cells following co-transfection with pGL3-SP1-3′UTR-Wt or pGL3-SP1-3′UTR-Mut, together with miR-376a mimics or miR-NC. Data are presented as the mean ± standard deviation; *P<0.05 vs. miR-NC. (C) RT-qPCR and (D) western blot analysis of the SP1 mRNA and protein expression levels, respectively, in U251 cells following transfection with miR-376a mimics or miR-NC; *P<0.05 vs. miR-NC. Expression levels of SP1 (E) mRNA and (F) protein in GBM tissues and adjacent normal brain tissues detected using RT-qPCR and western blot analysis, respectively. Data are presented as the mean ± standard deviation; *P<0.05 vs. adjacent normal brain tissues. (G) Spearman's correlation analysis of the association between miR-376a and SP1 mRNA level in GBM tissues. r=−0.6788; P=0.0001. GBM, glioblastoma multiforme; hsa, Homo sapiens RNA miR, microRNA; mut, mutant; NC, negative control; RT-qPCR, reverse transcription-quantitative polymerase chain reaction; SP1, specificity protein 1; UTR, untranslated region; wt, wild-type.
SP1 mRNA and protein levels were also measured in GBM tissues and adjacent normal brain tissues. SP1 mRNA and protein levels were elevated in GBM tissues compared with adjacent normal brain tissues (P<0.05; Fig. 3E and F). Spearman's correlation analysis indicated a significant negative correlation between miR-376a expression and SP1 mRNA levels in GBM tissues (Fig. 3G; r=−0.6788, P=0.0001). These data indicated that SP1 was a direct target of miR-376a in GBM.
SP1 overexpression reduces the suppressive effects of miR-376a on GBM cells
Rescue experiments were performed to confirm whether SP1 mediates the biological role of miR-376a in GBM cells. U251 cells were transfected with miR-376a mimics, with or without the pcDNA3.1-SP1 overexpression vector. Western blot analysis revealed that the miR376a mimics-induced reduction of SP1 protein expression was restored by co-transfection with pcDNA3.1-SP1 (P<0.05; Fig. 4A). Subsequently, CCK-8 proliferation and Matrigel invasion assays demonstrated that overexpression of SP1 in U251 cells reversed the suppressive effects of miR-376a mimics on cell proliferation and invasion (P<0.05; Fig. 4B and C). These results suggested that miR-376a may inhibit GBM progression, at least in part, through negative regulation of SP1.
Figure 4.
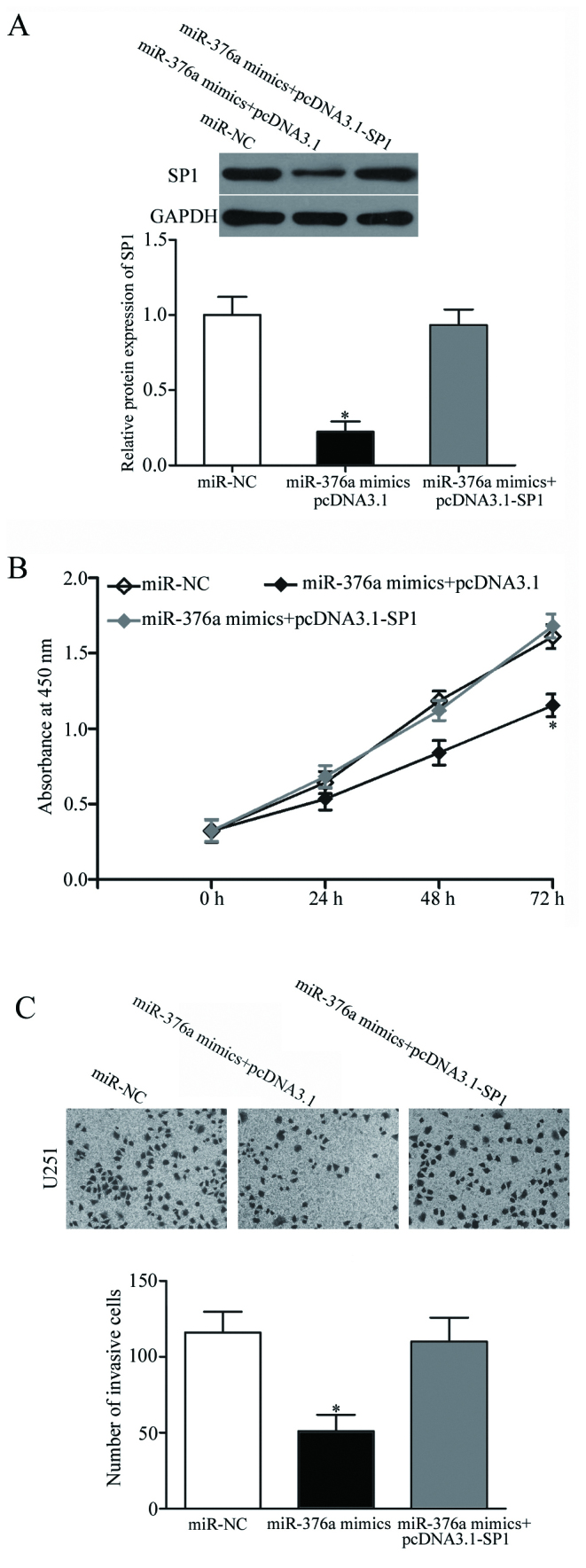
Overexpression of SP1 rescues the effects of miR-376a on U251 cell proliferation and invasion. (A) Western blot analysis of SP1 protein expression levels in U251 cells following transfection with miR-376a mimics with or without co-transfection with a pcDNA3.1-SP1 overexpression plasmid. (B) Cell proliferation was detected by Cell Counting Kit-8 assay in U251 cells transfected with miR-376a mimics with or without pcDNA3.1-SP1. (C) Cell invasion determined by Matrigel invasion assays in U251 cells following transfection with miR-376a mimics with or without pcDNA3.1-SP1. Data are presented as the mean ± standard deviation; *P<0.05 vs. miR-NC, miR-376a mimics+pcDNA3.1-SP1. miR, microRNA; NC, negative control; SP1, specificity protein 1; UTR, untranslated region.
Discussion
A number of previous studies have demonstrated that miRNAs are aberrantly expressed in almost all types of cancers, including GBM (29–31). Abnormally expressed miRNAs are commonly associated with malignant biological behaviors of GBM, including malignant growth, proliferation, apoptosis, invasion, metastasis and chemotherapy resistance (32). In the present study, miR-376a was downregulated in GBM tissues and cell lines. Functional assays revealed that miR-376a overexpression led to decreased GBM cell proliferation and invasion in vitro. Additionally, SP1 was confirmed as a direct target gene of miR-376a in GBM. mRNA and protein expression levels of SP1 in GBM tissues were markedly upregulated compared with the adjacent normal brain tissues. Furthermore, expression of SP1 mRNA was inversely correlated with the expression of miR-376a in GBM tissues. The restored expression of SP1 partially attenuated miR-376a-inhibited cell proliferation and invasion. The results of the present study suggested that miR-376a may be a therapeutic target for treatment of patients with GBM.
miR-376a is abnormally expressed in several human cancers. For example, miR-376a expression is upregulated in ovarian cancer and associated with International Federation of Gynecology and Obstetrics stages of ovarian cancer (21,33); however, miR-376a is downregulated in hepatocellular carcinoma (22). Low miR-376a expression levels are associated with low serum α-fetoprotein levels in hepatocellular carcinoma (22). In prostate cancer, miR-376a was reported to be downregulated in tumor tissues and cell lines, it was associated with an increased incidence of metastatic events and elevated prostate specific antigen levels, and similar trends were observed for lymph node invasion and Gleason score (34). miR-376a downregulation was identified in giant cell tumors of bone (35). These data suggested that miR-376a may be a prognostic marker of certain cancers.
miR-376a may serves roles in carcinogenesis and cancer progression; for example, Yang et al reported that miR-376a overexpression promoted ovarian cancer cell growth and metastasis (21). However, Zheng et al demonstrated that miR-376a serves a role as a tumor suppressor in hepatocellular carcinoma by inhibiting cell proliferation and increasing apoptosis (22). Formosa et al demonstrated that miR-376a overexpression decreases cell growth and motility, and increases apoptosis in prostate cancer (34). A study by Fellenberg et al indicated that restoration of miR-376a expression attenuates cell proliferation, migration, colony formation and spheroid formation of giant cell tumors of bone (35). The aforementioned data appears contradictory as miR-376a acted as an oncogene in certain cancers and as a tumor suppressor in others. This contradiction may be explained by imperfect complementarity of interactions between miRNAs and target genes (36).
Several miR-376a target genes have been previously identified, including Krüppel-like factor 15 and caspase-8 in ovarian cancer, p85α and histone deacetylase in hepatocellular carcinoma, insulin-like growth factor 1 receptor in melanoma, cytochrome c oxidase assembly factor 1 homolog, and nucleoporin GLE1 and protein disulfide-isomerase A6 in giant cell tumors of bone (21–23,35,37). In the present study, SP1 was identified as a novel target of miR-376a. SP1 is a member of the Sp/Krüppel-like factor transcription factor family and is upregulated in several types of human cancer, including gastric, colorectal, prostate, thyroid and liver (38–42). SP1 serves a role in a number of pathophysiological processes, including cell proliferation, cell cycle, apoptosis, angiogenesis and metastasis (43–45). Therefore, SP1 may be a therapeutic target for suppression of tumorigenesis and tumor development.
SP1 was upregulated in glioma tissues and cell lines. Elevated SP1 expression was associated with the WHO grading and survival status of patients with glioma. Patients with lower SP1 expression demonstrated better overall survival compared with patients with increased SP1 expression (24); suggesting that SP1 expression may be a prognostic indicator of survival of patients with glioma (24). Functional experiments demonstrated that SP1 serves a role in glioma progression by regulating proliferation, cell cycle and invasion (24–27). In the present study, miR-376a was demonstrated to target SP1 mRNA to inhibit GBM cell proliferation and invasion, which suggested that miR-376a/SP1-based targeted therapy may be a novel therapeutic strategy for treatment of patients with GBM.
In conclusion, the present study demonstrated that miR-376a expression was reduced in GBM tissues and cell lines. miR-376a overexpression exhibited a suppressive effect on proliferation and invasion of GBM cells, which may occur by directly targeting SP1. miR-376a may be a novel therapeutic target for the treatment of GBM.
References
- 1.Miller CR, Perry A. Glioblastoma. Arch Pathol Lab Med. 2007;131:397–406. doi: 10.5858/2007-131-397-G. [DOI] [PubMed] [Google Scholar]
- 2.Komori T, Sasaki H, Yoshida K. Revised WHO classification of tumours of the central nervous system: Summary of the revision and perspective. No Shinkei Geka. 2016;44:625–635. doi: 10.11477/mf.1436203347. (In Japanese) [DOI] [PubMed] [Google Scholar]
- 3.Easaw JC, Mason WP, Perry J, Laperrière N, Eisenstat DD, Del Maestro R, Bélanger K, Fulton D, Macdonald D. Canadian Glioblastoma Recommendations Committee: Canadian recommendations for the treatment of recurrent or progressive glioblastoma multiforme. Curr Oncol. 2011;18:e126–e136. doi: 10.3747/co.v18i3.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Smith AW, Mehta MP, Wernicke AG. Neural stem cells, the subventricular zone and radiotherapy: Implications for treating glioblastoma. J Neurooncol. 2016;128:207–216. doi: 10.1007/s11060-016-2123-z. [DOI] [PubMed] [Google Scholar]
- 5.Binder DC, Davis AA, Wainwright DA. Immunotherapy for cancer in the central nervous system: Current and future directions. Oncoimmunology. 2016;5:e1082027. doi: 10.1080/2162402X.2015.1082027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Katakowski M, Chopp M. Exosomes as tools to suppress primary brain tumor. Cell Mol Neurobiol. 2016;36:343–352. doi: 10.1007/s10571-015-0280-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Khosla D. Concurrent therapy to enhance radiotherapeutic outcomes in glioblastoma. Ann Transl Med. 2016;4:54. doi: 10.3978/j.issn.2305-5839.2016.01.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kegelman TP, Hu B, Emdad L, Das SK, Sarkar D, Fisher PB. In vivo modeling of malignant glioma: The road to effective therapy. Adv Cancer Res. 2014;121:261–330. doi: 10.1016/B978-0-12-800249-0.00007-X. [DOI] [PubMed] [Google Scholar]
- 9.Van Meir EG, Hadjipanayis CG, Norden AD, Shu HK, Wen PY, Olson JJ. Exciting new advances in neuro-oncology: The avenue to a cure for malignant glioma. CA Cancer J Clin. 2010;60:166–193. doi: 10.3322/caac.20069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alvarez-Garcia I, Miska EA. MicroRNA functions in animal development and human disease. Development. 2005;132:4653–4662. doi: 10.1242/dev.02073. [DOI] [PubMed] [Google Scholar]
- 11.He L, Hannon GJ. MicroRNAs: Small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522–531. doi: 10.1038/nrg1415. [DOI] [PubMed] [Google Scholar]
- 12.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 13.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 14.Wang Z, Yang J, Xu G, Wang W, Liu C, Yang H, Yu Z, Lei Q, Xiao L, Xiong J, et al. Targeting miR-381-NEFL axis sensitizes glioblastoma cells to temozolomide by regulating stemness factors and multidrug resistance factors. Oncotarget. 2015;6:3147–3164. doi: 10.18632/oncotarget.3061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chang JH, Hwang YH, Lee DJ, Kim DH, Park JM, Wu HG, Kim IA. MicroRNA-203 modulates the radiation sensitivity of human malignant glioma cells. Int J Radiat Oncol Biol Phys. 2016;94:412–420. doi: 10.1016/j.ijrobp.2015.10.001. [DOI] [PubMed] [Google Scholar]
- 16.Sampson VB, Yoo S, Kumar A, Vetter NS, Kolb EA. MicroRNAs and potential targets in osteosarcoma: Review. Front Pediatr. 2015;3:69. doi: 10.3389/fped.2015.00069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Luna-Aguirre CM, de la Luz Martinez-Fierro M, Mar-Aguilar F, Garza-Veloz I, Treviño-Alvarado V, Rojas-Martinez A, Jaime-Perez JC, Malagon-Santiago GI, Gutierrez-Aguirre CH, Gonzalez-Llano O, et al. Circulating microRNA expression profile in B-cell acute lymphoblastic leukemia. Cancer Biomark. 2015;15:299–310. doi: 10.3233/CBM-150465. [DOI] [PubMed] [Google Scholar]
- 18.Kagiya T. MicroRNAs and osteolytic bone metastasis: The roles of MicroRNAs in tumor-induced osteoclast differentiation. J Clin Med. 2015;4:1741–1752. doi: 10.3390/jcm4091741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ventura A, Jacks T. MicroRNAs and cancer: Short RNAs go a long way. Cell. 2009;136:586–591. doi: 10.1016/j.cell.2009.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gao B, Gao K, Li L, Huang Z, Lin L. miR-184 functions as an oncogenic regulator in hepatocellular carcinoma (HCC) Biomed Pharmacother. 2014;68:143–148. doi: 10.1016/j.biopha.2013.09.005. [DOI] [PubMed] [Google Scholar]
- 21.Yang L, Wei QM, Zhang XW, Sheng Q, Yan XT. MiR-376a promotion of proliferation and metastases in ovarian cancer: Potential role as a biomarker. Life Sci. 2017;173:62–67. doi: 10.1016/j.lfs.2016.12.007. [DOI] [PubMed] [Google Scholar]
- 22.Zheng Y, Yin L, Chen H, Yang S, Pan C, Lu S, Miao M, Jiao B. miR-376a suppresses proliferation and induces apoptosis in hepatocellular carcinoma. FEBS Lett. 2012;586:2396–2403. doi: 10.1016/j.febslet.2012.05.054. [DOI] [PubMed] [Google Scholar]
- 23.Zheng Y, Chen H, Yin M, Ye X, Chen G, Zhou X, Yin L, Zhang C, Ding B. MiR-376a and histone deacetylation 9 form a regulatory circuitry in hepatocellular carcinoma. Cell Physiol Biochem. 2015;35:729–739. doi: 10.1159/000369733. [DOI] [PubMed] [Google Scholar]
- 24.Guan H, Cai J, Zhang N, Wu J, Yuan J, Li J, Li M. Sp1 is upregulated in human glioma, promotes MMP-2-mediated cell invasion and predicts poor clinical outcome. Int J Cancer. 2012;130:593–601. doi: 10.1002/ijc.26049. [DOI] [PubMed] [Google Scholar]
- 25.Luo J, Wang X, Xia Z, Yang L, Ding Z, Chen S, Lai B, Zhang N. Transcriptional factor specificity protein 1 (SP1) promotes the proliferation of glioma cells by up-regulating midkine (MDK) Mol Biol Cell. 2015;26:430–439. doi: 10.1091/mbc.E14-10-1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dong Q, Cai N, Tao T, Zhang R, Yan W, Li R, Zhang J, Luo H, Shi Y, Luan W, et al. An axis involving SNAI1, microRNA-128 and SP1 modulates glioma progression. PLoS One. 2014;9:e98651. doi: 10.1371/journal.pone.0098651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee WS, Kwon J, Yun DH, Lee YN, Woo EY, Park MJ, Lee JS, Han YH, Bae IH. Specificity protein 1 expression contributes to Bcl-w-induced aggressiveness in glioblastoma multiforme. Mol Cells. 2014;37:17–23. doi: 10.14348/molcells.2014.2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 29.Zhou X, Ren Y, Moore L, Mei M, You Y, Xu P, Wang B, Wang G, Jia Z, Pu P, et al. Downregulation of miR-21 inhibits EGFR pathway and suppresses the growth of human glioblastoma cells independent of PTEN status. Lab Invest. 2010;90:144–155. doi: 10.1038/labinvest.2009.126. [DOI] [PubMed] [Google Scholar]
- 30.Ma R, Yan W, Zhang G, Lv H, Liu Z, Fang F, Zhang W, Zhang J, Tao T, You Y, et al. Upregulation of miR-196b confers a poor prognosis in glioblastoma patients via inducing a proliferative phenotype. PLoS One. 2012;7:e38096. doi: 10.1371/journal.pone.0038096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Qu S, Yao Y, Shang C, Xue Y, Ma J, Li Z, Liu Y. MicroRNA-330 is an oncogenic factor in glioblastoma cells by regulating SH3GL2 gene. PLoS One. 2012;7:e46010. doi: 10.1371/journal.pone.0046010. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 32.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 33.Meng X, Joosse SA, Müler V, Trillsch F, Milde-Langosch K, Mahner S, Geffken M, Pantel K, Schwarzenbach H. Diagnostic and prognostic potential of serum miR-7, miR-16, miR-25, miR-93, miR-182, miR-376a and miR-429 in ovarian cancer patients. Br J Cancer. 2015;113:1358–1366. doi: 10.1038/bjc.2015.340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Formosa A, Markert EK, Lena AM, Italiano D, Finazzi-Agro' E, Levine AJ, Bernardini S, Garabadgiu AV, Melino G, Candi E. MicroRNAs, miR-154, miR-299-5p, miR-376a, miR-376c, miR-377, miR-381, miR-487b, miR-485-3p, miR-495 and miR-654-3p, mapped to the 14q32.31 locus, regulate proliferation, apoptosis, migration and invasion in metastatic prostate cancer cells. Oncogene. 2014;33:5173–5182. doi: 10.1038/onc.2013.451. [DOI] [PubMed] [Google Scholar]
- 35.Fellenberg J, Sähr H, Kunz P, Zhao Z, Liu L, Tichy D, Herr I. Restoration of miR-127-3p and miR-376a-3p counteracts the neoplastic phenotype of giant cell tumor of bone derived stromal cells by targeting COA1, GLE1 and PDIA6. Cancer Lett. 2016;371:134–141. doi: 10.1016/j.canlet.2015.10.039. [DOI] [PubMed] [Google Scholar]
- 36.Yu Z, Ni L, Chen D, Zhang Q, Su Z, Wang Y, Yu W, Wu X, Ye J, Yang S, et al. Identification of miR-7 as an oncogene in renal cell carcinoma. J Mol Histol. 2013;44:669–677. doi: 10.1007/s10735-013-9516-5. [DOI] [PubMed] [Google Scholar]
- 37.Zehavi L, Avraham R, Barzilai A, Bar-Ilan D, Navon R, Sidi Y, Avni D, Leibowitz-Amit R. Silencing of a large microRNA cluster on human chromosome 14q32 in melanoma: Biological effects of mir-376a and mir-376c on insulin growth factor 1 receptor. Mol Cancer. 2012;11:44. doi: 10.1186/1476-4598-11-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kanai M, Wei D, Li Q, Jia Z, Ajani J, Le X, Yao J, Xie K. Loss of Krüppel-like factor 4 expression contributes to Sp1 overexpression and human gastric cancer development and progression. Clin Cancer Res. 2006;12:6395–6402. doi: 10.1158/1078-0432.CCR-06-1034. [DOI] [PubMed] [Google Scholar]
- 39.Hosoi Y, Watanabe T, Nakagawa K, Matsumoto Y, Enomoto A, Morita A, Nagawa H, Suzuki N. Up-regulation of DNA-dependent protein kinase activity and Sp1 in colorectal cancer. Int J Oncol. 2004;25:461–468. [PubMed] [Google Scholar]
- 40.Yin P, Zhao C, Li Z, Mei C, Yao W, Liu Y, Li N, Qi J, Wang L, Shi Y, et al. Sp1 is involved in regulation of cystathionine γ-lyase gene expression and biological function by PI3K/Akt pathway in human hepatocellular carcinoma cell lines. Cell Signal. 2012;24:1229–1240. doi: 10.1016/j.cellsig.2012.02.003. [DOI] [PubMed] [Google Scholar]
- 41.Sankpal UT, Goodison S, Abdelrahim M, Basha R. Targeting Sp1 transcription factors in prostate cancer therapy. Med Chem. 2011;7:518–525. doi: 10.2174/157340611796799203. [DOI] [PubMed] [Google Scholar]
- 42.Chiefari E, Brunetti A, Arturi F, Bidart JM, Russo D, Schlumberger M, Filetti S. Increased expression of AP2 and Sp1 transcription factors in human thyroid tumors: A role in NIS expression regulation? BMC Cancer. 2002;2:35. doi: 10.1186/1471-2407-2-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Beishline K, Azizkhan-Clifford J. Sp1 and the ‘hallmarks of cancer’. FEBS J. 2015;282:224–258. doi: 10.1111/febs.13148. [DOI] [PubMed] [Google Scholar]
- 44.Wang YT, Yang WB, Chang WC, Hung JJ. Interplay of posttranslational modifications in Sp1 mediates Sp1 stability during cell cycle progression. J Mol Biol. 2011;414:1–14. doi: 10.1016/j.jmb.2011.09.027. [DOI] [PubMed] [Google Scholar]
- 45.Yen WH, Ke WS, Hung JJ, Chen TM, Chen JS, Sun HS. Sp1-mediated ectopic expression of T-cell lymphoma invasion and metastasis 2 in hepatocellular carcinoma. Cancer Med. 2016;5:465–477. doi: 10.1002/cam4.611. [DOI] [PMC free article] [PubMed] [Google Scholar]