Abstract
Hypertension is frequently associated with metabolic disorders. The present study was designed to investigate the long-term effect of prehypertensive losartan therapy on metabolic disorders in high-fat-fed spontaneously hypertensive rats (SHRs), and to examine the role of epigenetic regulation of angiotensin II type 1 receptor (AT1R) and AT1 receptor-associated protein (ATRAP) expression in adipose tissue. A total of 32 4-week-old male SHRs were divided into four groups (n=8 rats/group): Standard chow; standard chow + losartan; high-fat diet; and high-fat diet + losartan. At 10 weeks of age, treatment with losartan was discontinued. Rats were followed up until 26 weeks of age. Obesity, dyslipidemia, hyperglycemia, abnormal adipokine secretion, larger adipocytes and decreased expression of markers of adipocyte differentiation were present in high-fat-fed SHRs, and were attenuated in losartan-treated rats. The increased expression and promoter hypomethylation of AT1R subtype a (AT1aR) in the adipose tissue of high-fat-fed SHRs were reversed by treatment with losartan. No difference was observed in the expression and promoter methylation of AT1R subtype b (AT1bR) among the four groups. Decreased expression and promoter hypermethylation of ATRAP were demonstrated in the adipose tissue of high-fat-fed SHRs. However, losartan made no difference to the expression and promoter methylation of ATRAP. Prehypertensive losartan therapy may relieve metabolic disorders in the later life of high-fat-fed SHRs. Differential epigenetic regulation of AT1aR and ATRAP expression through DNA methylation in adipose tissue may be involved in the long-term beneficial effect.
Keywords: losartan, metabolic disorders, methylation, AT1R, ATRAP, adipose tissue, epigenetics, SHR
Introduction
Hypertension is frequently associated with metabolic disorders, characterized by obesity, dyslipidemia and hyperglycemia. The clustering of these disorders contributes to the increased morbidity and mortality of cardiovascular diseases (1). Spontaneously hypertensive rats (SHRs) are commonly-used rat models of essential hypertension. Antihypertensive treatment with losartan during the prehypertensive period, a specific stage in the early life of SHRs, has been demonstrated to be able to inhibit the increased in the blood pressure of the rats long-term. In addition, the expression of angiotensin II type 1 receptor (AT1R), an important component of the renin-angiotensin system (RAS), was decreased in the heart and kidney of the rats, suggesting that RAS inactivation in hypertension-associated tissues is involved in the benefit of early treatment with losartan (2). However, it has yet to be determined whether this therapeutic strategy is beneficial to relieve metabolic disorders and inhibit the activation of RAS in metabolism-associated tissues.
Metabolic disorders are associated with adipose tissue dysfunction. Adipose tissue functions as a storage place of fatty acids and an endocrine organ, producing a large number of hormones and adipokines (3). RAS is considered to be an important regulator of blood pressure and fluid homeostasis. Accumulating evidence has demonstrated that RAS is additionally present in adipose tissue. The local adipose tissue RAS is reported to be able to modulate adipocyte differentiation, adipokine production, local blood flow, inflammation and oxidative stress (4,5). These newly verified roles provided a novel perspective on the pathophysiology and possible interventional targets for metabolic disorders.
It has been established that the expression of RAS genes in specific tissues may be affected by low-protein diet, nicotine and high-salt diet exposure in early life, and that alterations in gene expression are associated with certain chronic diseases in later life, a phenomenon attributed to ‘developmental programming’ (6–8). Epigenetic mechanisms serve a role in these programmed diseases (6,7). DNA methylation is considered to be one of the most fundamental epigenetic modifications. It refers to covalent binding of a methyl group to the 5′ carbon of cytosine, and this binding primarily occurs at CpG dinucleotide sequences in the mammalian genome (9,10). In general, promoter methylation leads to long-term repression of gene expression by altering the chromatin structure or blocking the binding of transcription factors to the DNA sequence (11).
Losartan, one of the typical RAS inhibitors, is commonly used as an antihypertensive agent. RAS inhibitors have been demonstrated to have a beneficial effect in the regulation of metabolism (12,13). As a common chronic disease, metabolic disorders may be induced by suboptimal exposure in early life (14). Therefore, it may be hypothesized that prehypertensive treatment with losartan may be able to relieve metabolic disorders in a model of high-fat-fed SHRs long-term, and that methylation regulation of RAS gene expression in adipose tissue may be involved in the long-term effect elicited by the early treatment with losartan.
Materials and methods
Animals, diet and pharmacological treatment
Prior to beginning the experiment, specific criteria of humane endpoints were established. The criteria included: i) A marked reduction in food or water intake; ii) shaggy coat; iii) labored breathing; iv) an inability to remain upright; and v) unconsciousness or unresponsiveness to external stimuli, including picking up, handling and sound (15,16). All signs were validated by an animal specialist. If rats presented one or more of the above signs, they were considered to have reached the humane endpoint, and were anesthetized with sodium pentobarbital (50 mg/kg, administered intraperitoneally) and sacrificed by cervical dislocation immediately. A total of 32 4-week-old male SHRs [body weight (BW) 122±11 g] were obtained from Vital River Laboratory Animal Technology Co., Ltd. (Beijing, China). Animals were housed (5 rats/cage) under controlled conditions (temperature, 22–24°C; humidity, 40–60%; 12/12-h dark/light cycle), with free access to food and water. Rats were randomly divided into four groups based on diets and treatment (n=8 rats/group): Standard chow, standard chow + losartan, high-fat diet, and high-fat diet + losartan. The standard chow consisted of 10% Kcal as fat, 70% Kcal as carbohydrate, and 20% Kcal as protein. The high-fat diet consisted of 45% Kcal as fat, 35% Kcal as carbohydrate and 20% Kcal as protein. Additionally, 25% soybean oil and 0.23% NaCl were mixed into the diets. The diets were manufactured by the Laboratory Animal Center of Fujian Medical University (Fuzhou, China). Losartan was administrated at the dosage of 30 mg/kg/day by gavage. Rats without treatment with losartan were administrated with 1 ml saline. At 10 weeks of age, treatment with losartan was discontinued. The rats were followed up. At 26 weeks of age, all rats were sacrificed following the measurement of blood pressure and BW. The study protocol was approved by the Institutional Animal Care Committee of the First Affiliated Hospital of Fujian Medical University (approval no. 001).
Measurement of BW and systolic blood pressure (SBP)
BW was determined using a balance at 2–4 week intervals. At 26 weeks of age, SBP was measured under conscious conditions using the tail-cuff method (Softron BP-98A; Softron Co., Ltd., Tokyo, Japan) as previously described (17,18). Prior to the measurement, rats were placed inside a warming chamber for 15 min to calm the animals and dilate the tail blood vessels. The rats were subsequently removed to plastic restrainers, and a cuff with a pneumatic pulse sensor was attached to the tail blood vessels. Mean SBP was calculated from three consecutive SBP readings.
Plasma and tissue collection
The rats were deprived of food for 6 h prior to sacrifice, and anesthetized with sodium pentobarbital (50 mg/kg, administered intraperitoneally) at 26 weeks of age. Blood samples were rapidly collected via abdominal aortic puncture and the rats were immediately sacrificed by cervical dislocation. Plasma was separated by centrifugation (1,200 × g for 10 min) at 4°C, and stored at −20°C until analysis. Inguinal, epididymal, retroperitoneal and mesenteric fat pads were dissected and weighed. The percentage of each fat pad weight compared with the total BW was calculated to determine the alterations in white adipose tissue (WAT) mass. A portion of the epididymal fat pad was stored in a fixative solution (formaldehyde 4% w/v; 0.1 M phosphate buffer; pH 7.2) for morphological assessment, and the remaining fat pad was rapidly frozen at −80°C for ELISA and molecular analyses.
Biochemical analysis and ELISA
The concentrations of plasma total cholesterol (TC), triglycerides (TG), high density lipoprotein cholesterol (HDL-C), low density lipoprotein cholesterol (LDL-C) and glucose were analyzed using an automated autoanalyzer (Olympus Au2700; Olympus Corporation, Tokyo, Japan). A total of 30 mg epididymal fat was homogenized at 4°C following the addition of 1 ml PBS. The supernatant was carefully collected following centrifugation for 10 min at 1,200 × g at 4°C. Leptin (cat. no. KRC2281), adiponectin (cat. no. KRP0041), interleukin-6 (IL-6; cat. no. BMS625), tumor necrosis factor-α (TNF-α; cat. no. 88-7340-86) and monocyte chemoattractant protein-1 (MCP-1; cat. no. BMS631INST) in the plasma and the supernatant of the epididymal fat were determined using commercially available ELISA kits (Thermo Fisher Scientific, Inc., Waltham, MA, USA).
Morphological analysis in WAT
Epididymal fat was fixed in 10% (v/v) formaldehyde/PBS for 48 h at room temperature, followed by dehydration with acetone for 24 h. The dehydrated tissue was embedded in paraffin and sliced into 10-µm-thick sections. The sections were stained with hematoxylin for 10 min and eosin for 15 sec at room temperature. A total of four sections were selected from each sample. A total of four different microscopic fields (upper left, upper right, lower left and lower right) per section were observed under a light microscope (CX31; Olympus Corp., Tokyo, Japan). At least 25 adjacent adipocytes per microscopic field were analyzed. The diameter and cross-sectional area of each adipocyte was determined using Image Pro Plus 6.0 (Media Cybernetics. Inc., Rockville, MD, USA). The mean diameter and area were calculated.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR) analysis in WAT
The mRNA expression levels of peroxisome proliferator-activated receptor γ (PPARγ), transcription factor adaptin 2 (aP2), CCAAT/enhancer binding protein α (CEBP/α), CEBP/β, AT1R subtype a (AT1aR), AT1bR and AT1R-associated protein (ATRAP) were assessed via RT-qPCR, as previously described (19,20). Total RNA was extracted from epididymal fat using TRIzol (Invitrogen; Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol. A total of 2 µg RNA was reverse-transcribed into cDNA using a RevertAid First Strand cDNA Synthesis kit (Thermo Fisher Scientific, Inc.). Primers were designed according to the PubMed Database (www.ncbi.nlm.nih.gov/pubmed; Table I). qPCR was performed using SYBR Green PCR Master Mix (Takara Bio, Inc., Otsu, Japan), and detected using an ABI Prism 7000 sequence detection system (Applied Biosystems; Thermo Fisher Scientific, Inc.). The following qPCR conditions were used: 95°C (30 min), followed by a three-step PCR program of 95°C (10 sec), 60°C (31 sec) and 72°C (30 sec) for 40 cycles, and a final extension step of 72°C for 5 min. The mRNA level was quantified by calculating the values of the Δ quantification cycle (ΔCq) by normalizing the average Cq value compared with the internal control (GAPDH), and calculating 2−ΔΔCq (19).
Table I.
Primer sequences used in reverse transcription-quantitative polymerase chain reaction analysis of genes.
| Gene | Forward primer (5′→3′) | Reverse primer (5′→3′) |
|---|---|---|
| PPARγ | ATGACAGACCTCAGGCAGATTG | TGTCAGCGACTGGGACTTTTC |
| aP2 | ATGAAAGAAGTGGGAGTTGGC | CAGTTTGAAGGAAATCTCGGTGT |
| CEBP/α | TCAAGGGCTTGGCTGGTCC | CGCGATGTTGTTGCGTTCC |
| CEBP/β | CACCGGGTTTCGGGACTTG | CCCGCAGGAACATCTTTAAGTG |
| AT1aR | ATTCGTGGCTTGAGTCCTGT | GTTAACTCAGGGAATGTGGCA |
| AT1bR | GATTTTTTTTTATAATTTTTTTAAGGTGG | CAAATAAACCTATATCAAATAAATAACAC |
| ATRAP | ACTCTGTTGATGCCATTG | GAAGATGCTAATGTAGATAATGTC |
| GAPDH | CCTGCACCACCAACTGCTTA | AGTGATGGCATGGACTGTGG |
PPARγ, peroxisome proliferator-activated receptor γ; aP2, transcription factor adaptin 2; CEBP, CCAAT/enhancer binding protein; AT1aR, angiotensin II type 1 receptor subtype a; ATRAP, angiotensin II type 1 receptor-associated protein.
AT1R and ATRAP western blot analysis in WAT
Western blotting was performed as previously described, with minor modifications (21). The total protein was extracted from epididymal fat using a cold radioimmunuoprecipitation assay cell lysis buffer (Shanghai Sunred Biological Technology Co., Ltd, Shanghai, China) supplemented with a protease and phosphatase inhibitor cocktail, and phenylmethane sulfonyl fluoride. The supernatant was separated by centrifugation for 10 min at 800 × g at 4°C, followed by centrifugation for 15 min at 14,000 × g at 4°C. The concentration of protein was determined using a bicinchoninic acid protein assay kit (Beyotime Institute of Biotechnology, Haimen, China). Aliquots of 80 µg protein were separated using 10% SDS-PAGE gels, and electro-transferred onto polyvinylidene fluoride membranes. The membranes were blocked with 5% non-fat milk in TBS containing 0.05% Tween 20 (TBST). The protein bands were incubated overnight with anti-AT1R antibody (1:200; cat. no. sc-1173; Santa Cruz Biotechnology, Inc., Dallas, TX, USA), anti-ATRAP antibody (1:200; cat. no. sc134652; Santa Cruz Biotechnology, Inc.) or anti-β-actin antibody (1:1,000; cat. no. sc1616; Santa Cruz Biotechnology, Inc.) overnight at 4°C. Following three washes with TBST, the blot was hybridized with horseradish peroxidase-conjugated goat anti-mouse or rabbit immunoglobulin G (1:5,000; cat. nos. ZB-2301 and ZB-2305; OriGene Technologies, Inc., Beijing, China) for 1 h at room temperature. Following a further three washes with TBST for 10 min, the membrane was visualized using enhanced chemiluminescence (Xiamen Lulong Biotech Co., Ltd., Xiamen, China) with a high-performance chemiluminescence film and quantified using Image Pro Plus version 6.0 (Media Cybernetics, Inc.).
Promoter methylation of AT1aR, AT1bR and ATRAP: Bisulfite-specific polymerase chain reaction (BSP) and DNA sequencing
The methylation levels of the AT1aR, AT1bR and ATRAP promoters were determined as described previously (22). Genomic DNA was extracted from epididymal fat using a GenElute Mammalian Genomic DNA mini-prep kit (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany), according to the manufacturer's protocol. DNA was subjected to bisulfate treatment using EpiTect Bisulfite kit (Qiagen GmbH, Hilden, Germany). The process specifically converts only unmethylated cytosine residues of CpG to uracil residues. PCR analysis of the target regions of interest was performed to amplify the bisulfite-modified products. The primer sequences for the PCR and the sequencing strategy are presented in Table II. The conditions of the PCR cycle were as follows: 95°C for 3 min, followed by 50 cycles at 95°C for 15 sec, 54°C for 30 sec and 72°C for 30 sec, and a final step of 72°C for 5 min. The reaction mixture contained 10X PCR buffer (4 µl), dNTP (10 mM; 0.8 µl), primer F (10 µM; 0.5 µl), primer R (10 µM; 0.5 µl), Takara Hotstart Taq (0.5 µl; 2.5 units; Takara Bio, Inc.), DNA template (2 µl) and H2O (31.7 µl). PCR products were separated using 3% agarose gels, and the bands were visualized by ethidium bromide staining (Thermo Fisher Scientific, Inc.), and were excised using the QIAquick Gel Extraction kit (Qiagen GmbH). Purified PCR products were mixed with sequencing buffer containing 0.5 µM sequencing primer. Following denaturation at 80°C for 2 min, the products were sequenced using a PyroMark Q96 ID sequencer (Qiagen GmbH).
Table II.
Primer sequences used in bisulfite-specific polymerase chain reaction and DNA sequencing for CpG methylation in gene promoters.
| Gene | Forward primer (5′→3′) | Reverse primer (5′→3′) | Sequencing primer (5′→3′) |
|---|---|---|---|
| AT1aR | TGGGAGGGATTGGATGATGT | Biotin-AAACCCCTAAACTAAAACTCACCAA | GGAGAGGATTAGTGGTTTG |
| AT1bR | TTTGTTAAGGGAGGGGTTAGGA | Biotin-CCTACTACCTAAAATCCAAACTACCTACA | GGGAGGGGTTAGGAG |
| ATRAP | TGGTAGAGGTTTAGGTAGTAGTAGGAGT | Biotin-CCAACTCCAAAACAAACTTCCT | GTTTTGTAGTAAGGGTAAGT |
AT1aR, angiotensin II type 1 receptor subtype a; ATRAP, angiotensin II type 1 receptor-associated protein.
Statistical analysis
Statistical analysis was performed using SPSS version 17.0 (SPSS, Inc., Chicago, IL, USA). Continuous data are presented as the mean ± standard deviation. Differences between groups were detected by one-way analysis of variance, followed by the least significant difference post hoc test for multiple comparisons. P<0.05 was considered to indicate a statistically significant difference.
Results
BW and percentage of WAT mass compared with BW
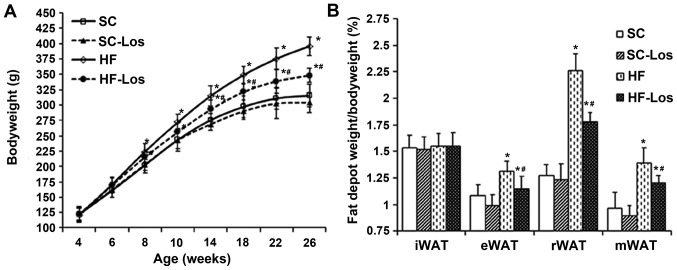
During the experiment, no rats met the criteria of humane endpoints, and all of the rats were followed up until the planned experimental endpoints. BW increased with age in all rats. No difference was observed in BW between standard chow-fed SHRs (SC) and losartan-treated SHRs on standard chow (SC-Los) throughout the study period (P>0.05). BW was increased in high-fat-fed SHRs (HF) compared with SC from 8 weeks of age onwards (26 weeks old, HF 396±15 g vs. SC 316±18 g; P<0.05). Losartan-treated SHRs on a high-fat diet (HF-Los) exhibited a significantly decreased BW compared with age-matched HF animals from 10 weeks of age onwards (26 weeks old, HF-Los 348±12 g vs. HF 396±15 g; P<0.05) (Fig. 1A). As presented in Fig. 1B, there was no difference in the percentage of inguinal fat pad weight compared with BW among the four groups (P>0.05). The percentage of epididymal, retroperitoneal and mesenteric fat pad weights compared with BW was similar between the SC and SC-Los groups (P>0.05), whereas they were significantly increased in HF compared with SC (P<0.05), and significantly decreased in HF-Los compared with HF (P<0.05).
Figure 1.
Effect of prehypertensive losartan therapy on body weight and WAT in high-fat-fed SHRs. (A) Bodyweight, (B) the percentage of white adipose tissue weight to bodyweight in rats at 26 weeks of age. *P<0.05 vs. age-matched SC; #P<0.05 vs. age-matched HF. The data are presented as the mean ± standard deviation. n=8. SHRs, spontaneously hypertensive rats; SC, standard chow-fed SHRs; HF, high-fat-fed SHRs; SC-Los, losartan-treated SHRs on standard chow; HF-Los, losartan-treated SHRs on high-fat diet. WAT, white adipose tissue; iWAT, inguinal WAT; eWAT, epididymal WAT; rWAT, retroperitoneal WAT; mWAT, mesenteric WAT.
Blood pressure, metabolic profiles and levels of plasma adipokines
SBP was decreased in SC-Los compared with SC (26 weeks old, SC-Los 185±22 mmHg vs. SC 215±15 mmHg; P<0.05). Although a slight increase in SBP was observed in HF compared with SC, it did not reach statistical significance (P>0.05). SBP in HF-Los was decreased compared with HF (26 weeks old, HF-Los 192±18 mmHg vs. HF 222±19 mmHg; P<0.05). There was no difference in the expression levels of plasma TC, TG, HDL-C, LDL-C, IL-6, MCP-1, TNF-α, leptin or adiponectin (P>0.05) between SC and SC-Los. The levels of HDL-C and adiponectin were significantly decreased, and the levels of TC, TG, LDL-C, glucose, IL-6 and MCP-1 were significantly increased in HF compared with SC (P<0.05). The levels of TC, TG, LDL-C, IL-6 and MCP-1 were decreased, and the levels of HDL-C, fasting blood glucose and adiponectin were increased in HF-Los compared with HF. There was no difference in the level of TNF-α among the four groups (Table III).
Table III.
Effect of prehypertensive losartan therapy on the metabolic profiles and plasma adipokine levels of high-fat-fed SHRs at 26 weeks of age (mean ± standard deviation; n=8).
| Group | ||||
|---|---|---|---|---|
| Variable | SC | SC-Los | HF | HF-Los |
| SBP, mmHg | 215±15 | 185±22a | 222±19 | 192±18a,b |
| TC, mmol/l | 2.11±0.30 | 2.05±0.27 | 3.88±0.72a | 3.40±0.78a,b |
| TG, mmol/l | 0.73±0.08 | 0.72±0.11 | 1.36±0.22a | 1.18±0.17a,b |
| HDL-C, mmol/l | 1.03±0.09 | 1.07±0.10 | 0.61±0.07a | 0.79±0.06a,b |
| LDL-C, mmol/l | 0.84±0.10 | 0.82±0.16 | 3.18±0.56a | 2.57±0.36a,b |
| Glucose, mmol/l | 6.22±0.49 | 6.11±0.62 | 6.92±0.31a | 6.34±0.43a,b |
| Leptin, ng/ml | 5.31±0.79 | 5.11±0.74 | 10.11±1.66a | 8.60±1.89a,b |
| Adiponectin, µg/ml | 5.73±2.24 | 5.36±1.67 | 2.23±0.80a | 4.24±1.63a,b |
| IL-6, pg/ml | 21.38±4.02 | 20.90±3.22 | 29.44±3.80a | 23.41±3.22a,b |
| MCP-1, pg/ml | 78.63±15.08 | 93.38±9.47 | 176.25±18.33a | 156.38±21.13a,b |
| TNF-α, pg/ml | 22.12±4.80 | 21.41±4.58 | 21.41±4.18 | 20.72±4.56 |
SHR, spontaneously hypertensive rat; SC, standard chow-fed SHRs; HF, high-fat-fed SHRs; SC-Los, losartan-treated SHRs on standard chow; HF-Los, losartan-treated SHRs on high-fat diet; SBP, systolic blood pressure; TC, total cholesterol; TG, triglyceride; HDL-C, high density lipoprotein cholesterol; LDL-C, low density lipoprotein cholesterol; IL-6, interleukin-6; MCP-1, monocyte chemoattractant protein-1; TNF-α, tumor necrosis factor-α.
P<0.05 vs. SC
P<0.05 vs. HF.
Adipocyte differentiation and adipokine production in adipose tissue
There was no difference in mean diameter and area of adipocytes between SC and SC-Los. The mean diameter and area of adipocytes was increased in HF compared with SC (P<0.05), and significantly decreased in HF-Los compared with HF (mean diameter of adipocytes, HF-Los 43.63±7.93 µm vs. HF 62.38±10.31 µm; mean area of adipocytes, HF-Los 1,537±529 µm2 vs. HF 3,127±1,043 µm2; P<0.05) (Fig. 2A and B). The mRNA expression levels of PPARγ, aP2, CEBP/α, and CEBP/β in adipose tissue were similar between SC and SC-Los (P<0.05), and decreased in HF compared with SC (P<0.05). The mRNA levels of PPAR and CEBP/β were significantly higher in HF-Los compared with HF (P<0.05). However, no difference was observed in the mRNA levels of aP2 and CEBP/α between HF and HF-Los (P>0.05; Fig. 2C). There was no difference in the levels of IL-6, MCP-1 TNF-α, leptin or adiponectin in adipose tissue between SC and SC-Los (P>0.05). The level of adiponectin was decreased, and the levels of IL-6, MCP-1 and leptin were increased in HF compared with SC (P<0.05). HF-Los exhibited a significantly higher level of adiponectin and significantly lower levels of IL-6, MCP-1 and leptin compared with HF (P<0.05). There was no difference in the level of TNF-α among the four groups (P>0.05; Table IV).
Figure 2.
Effect of prehypertensive losartan therapy on adipocyte size and differentiation in high-fat-fed SHRs at 26 weeks of age. (A) Adipocyte diameter, (B) adipocyte area and (C) the mRNA expression levels of PPARγ, CEBP/α, CEBP/β and aP2 in epididymal adipose tissue were analyzed. *P<0.05 vs. SC; #P<0.05 vs. HF. The data are presented as the mean ± standard deviation. n=8. SHRs, spontaneously hypertensive rats; SC, standard chow-fed SHRs; HF, high-fat-fed SHRs; SC-Los, losartan-treated SHRs on standard chow; HF-Los, losartan-treated SHRs on high-fat diet; PPARγ, peroxisome proliferator-activated receptor γ; CEBP, CCAAT/enhancer binding protein; aP2, transcription factor adaptin 2.
Table IV.
Effect of prehypertensive treatment with losartan on adipokine production in the adipose tissue of high-fat-fed SHRs at 26 weeks of age (mean ± standard deviation; n=8).
| Group | ||||
|---|---|---|---|---|
| Variable | SC | SC-Los | HF | HF-Los |
| Leptin, ng/ml | 19.26±2.72 | 20.45±3.13 | 74.46±8.23a | 45.46±5.99a,b |
| Adiponectin, µg/ml | 25.56±3.44 | 24.66±1.64 | 8.34±0.91a | 18.44±2.65a,b |
| IL-6, pg/ml | 33.37±8.72 | 30.94±3.55 | 58.49±9.80a | 42.46±8.29a,b |
| MCP-1, pg/ml | 96.66±19.08 | 97.35±12.44 | 245.25±24.35a | 175.38±21.44a,b |
| TNF-α, pg/ml | 42.38±6.22 | 44.23±7.58 | 39.39±6.76 | 41.23±6.22 |
SHR, spontaneously hypertensive rat; SC, standard chow-fed SHRs; HF, high-fat-fed SHRs; SC-Los, losartan-treated SHRs on standard chow; HF-Los, losartan-treated SHRs on high-fat diet. IL-6, interleukin-6; MCP-1, monocyte chemoattractant protein-1; TNF-α, tumor necrosis factor-α.
P<0.05 vs. SC
P<0.05 vs. HF.
Expression of AT1aR, AT1bR and ATRAP in WAT
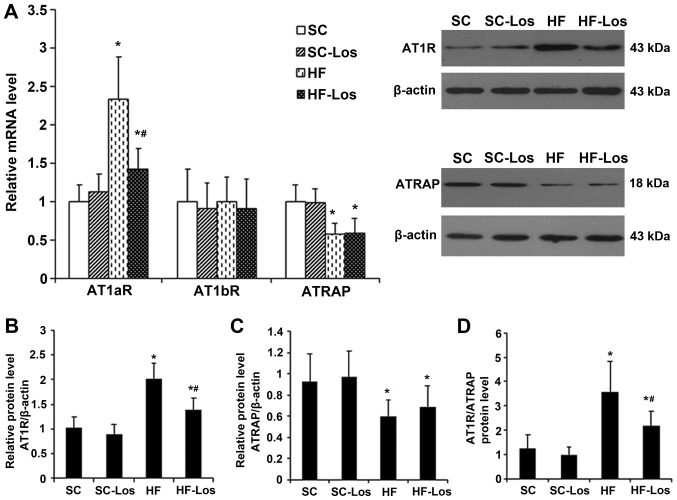
The mRNA expression level of AT1aR and the protein expression level of AT1R in adipose tissue were similar between SC and SC-Los (P>0.05). The mRNA expression level of AT1aR and protein expression level of AT1R were increased in HF compared with SC (P<0.05), and were decreased in HF-Los compared with HF (P<0.05) (Fig. 3A and B). There was no difference in the mRNA expression of AT1b among the four groups (P>0.05; Fig. 3A). No difference was observed in the mRNA and protein expression levels of ATRAP between SC and SC-Los. The mRNA and protein expression levels of ATRAP were decreased in HF compared with SC (P<0.05), and were similar between HF and HF-Los (P>0.05) (Fig. 3A and C). As a result, the protein ratio of AT1R to ATRAP was increased in HF compared with SC (HF 3.58±1.23 vs. SC 1.24±0.56; P<0.05), and was decreased in HF-Los compared with HF (HF-Los 2.16±0.62 vs. HF 3.58±1.23; P<0.05) (Fig. 3D).
Figure 3.
Effect of prehypertensive losartan therapy on the expression of AT1aR, AT1bR and ATRAP in the adipose tissue of high-fat-fed SHRs at 26 weeks of age. (A) The mRNA expression levels of AT1aR, AT1bR and ATRAP, (B) the protein expression level of AT1R, (C) the protein expression level of ATRAP and (D) the protein ratio of AT1R to ATRAP were measured. *P<0.05 vs. SC; #P<0.05 vs. HF. The data are presented as the mean ± standard deviation. n=8. AT1aR, angiotensin II type 1 receptor subtype a; ATRAP, AT1R-associated protein; SHRs, spontaneously hypertensive rats; SC, standard chow-fed SHRs; HF, high-fat-fed SHRs; SC-Los, losartan-treated SHRs on standard chow; HF-Los, losartan-treated SHRs on high-fat diet.
DNA methylation in AT1aR, AT1bR and ATRAP promoters
A total of seven CpG sites (+70, +77, +84, +93, +99, +108 and +113 bp) were examined in the AT1aR promoter (Fig. 4A). The methylation levels of the +77, +84, +99, +108 and +113 bp sites were decreased in HF compared with SC (P<0.05), and were increased in HF-Los compared with HF (P<0.05; Fig. 4B). A total of three CpG sites (−77, −72 and −42 bp) were examined in the AT1bR promoter (Fig. 4C). There was no difference in the methylation levels of these sites among the four groups (P>0.05) (Fig. 4D). A total of nine CpG sites (−120, −117, −103, −91, −85, −83, −81, −68 and −65 bp) were examined in the ATRAP promoter (Fig. 4E). The methylation levels at the −120, −117, −85 and −65 bp sites were higher in HF compared with SC (P<0.05), and were similar between HF and HF-Los (P>0.05; Fig. 4F).
Figure 4.
Effect of prehypertensive losartan therapy on the methylation level of the AT1aR, AT1bR and ATRAP promoters in the adipose tissue of high-fat-fed SHRs at 26 weeks of age. (A) CpG sites of AT1aR gene promoter as the targets of methylation status analysis, (B) methylation level of AT1aR gene promoter, (C) CpG sites of AT1bR gene promoter as the targets of methylation status analysis, (D) methylation level of AT1bR gene promoter, (E) CpG sites of ATRAP gene promoter as the targets of methylation status analysis. (F) Methylation level of ATRAP gene promoter. *P<0.05 vs. SC; #P<0.05 vs. HF. The data are represented as the mean ± standard deviation. n=8. AT1aR, angiotensin II type 1 receptor subtype a; ATRAP, AT1R-associated protein; SHRs, spontaneously hypertensive rats; SC, standard chow-fed SHRs; HF, high-fat-fed SHRs; SC-Los, losartan-treated SHRs on standard chow; HF-Los, losartan-treated SHRs on high-fat diet.
Discussion
In the present study, it was demonstrated that prehypertensive treatment with losartan was able to attenuate metabolic disorders in high-fat-fed SHRs, persisting up to 26 weeks of age. It was additionally observed that the long-term effect may be associated with differential epigenetic regulation of AT1aR and ATRAP expression in adipose tissue.
It is well known that hypercaloric diets, particularly a high-fat diet, are potential contributors to metabolic disorders in human (23). Knight et al (24) and Cao et al (25) demonstrated that when fed with high-fat-diet, SHRs experienced certain symptoms, including obesity, excessive adipose deposition, dyslipidemia, which were similar to metabolic disorders in obese patients with hypertension. The present study successfully duplicated this previous animal model and a number of alterations were further revealed in adipose tissue. Adipocytes became larger, the expression of PPARγ, aP2, CEBP/α and CEBP/β was decreased, the production of IL-6, MCP-1 and leptin was increased, and the production of adiponectin was decreased. PPARγ is an important transcription factor that mediates adipocyte differentiation, and it is considered to be a marker of adipocyte differentiation in addition to aP2, CEBP/α and CEBP/β (26). IL-6 and MCP-1 may be produced by adipocytes and generally serve as proinflammatory cytokines. Leptin and adiponectin are typical adipokines produced by adipose tissue. The action of leptin is primarily to inhibit food intake and decrease body weight. However, an increase in leptin production generally indicates obesity due to of leptin resistance. Adiponectin possesses antiatherogenic, antiflammatory and antioxidant properties (3). Due to the roles of these molecules, it was inferred that abnormal adipocyte differentiation and adipose tissue dysfunction were elicited in high-fat-fed SHRs in the present study. In addition, the protein ratio of AT1R to ATRAP was increased, which was consistent with alterations observed in other models of metabolic disorders (13). AT1R has been identified to be the primary mediator of the pathophysiology of angiotensin II, an important effector of RAS (27). In rodents, AT1R is divided into two subtypes: AT1aR and AT1bR (27). ATRAP is an AT1R-binding protein, and it may selectively downregulate AT1R-mediated signaling by promoting AT1R internalization (28). An imbalance of AT1R and ATRAP in the cardiovascular system was associated with the development of hypertension and cardiac remodeling (29). Therefore, it is reasonable to hypothesize that the increase in the protein ratio of AT1R to ATRAP in adipose tissue was involved in abnormal adipocyte differentiation and adipose tissue dysfunction in high-fat-fed SHRs. Additionally, hypomethylation of AT1aR and hypermethylation of ATRAP were observed to be associated with the increase in AT1R mRNA expression and the decrease in ATRAP mRNA expression, suggesting that the expression of these two genes may be regulated by DNA methylation. To the best of our knowledge, this is the first report on the DNA methylation of RAS genes in adipose tissue.
A previous study suggested that early life is a critical period for adipose tissue development (30). The results of the present study demonstrated that treatment with losartan during the prehypertensive period attenuated metabolic disorders, abnormal adipocyte differentiation and adipose tissue dysfunction in high-fat-fed SHRs. Notably, the observed benefit lasted for at least 16 weeks following treatment withdrawal. The early treatment with losartan was revealed to be able to reverse the increased expression and promoter hypomethylation of AT1aR in adipose tissue, suggesting that epigenetic regulation of AT1aR expression through DNA methylation may serve a role in the long-term effect elicited by the early treatment. However, it was demonstrated in the present study that the treatment with losartan failed to mitigate the decreased expression and hypermethylation of ATRAP in the adipose tissue of high-fat-fed SHRs. By contrast, a previous study reported that promoter hypermethylation of ATRAP in the myocardium of SHRs may be reversed by early treatment with losartan (22). Therefore, the present and previous results suggested that the effect of the early treatment on DNA methylation is gene- and tissue- specific. Li et al (31) additionally reported that the alteration in DNA methylation was gender-specific. Further studies are required to examine the mechanism underlying the specificity of DNA methylation.
Based on the role of epigenetics in the development of chronic disease, it is presumed that epigenetic intervention may be an attractive strategy for the treatment of these diseases. Typical epigenetic drugs, including histone deacetylase inhibitors and DNA methyltransferase inhibitors, have previously been investigated in animal models of cardiovascular diseases (32,33). However, these typical epigenetic drugs have been demonstrated to confer a beneficial effect in certain tissues and to cause a deleterious effect in other tissues (33,34). A possible interpretation is that these drugs alter epigenetic modification without tissue specificity. By contrast, the present and previous data suggested that early treatment with losartan may inhibit the increase in blood pressure, and protect against cardiac and renal damage, and metabolic disorders. In addition, the effect of losartan on epigenetic modification is gene- and tissue- specific, and this specificity may be due to the intrinsic properties of losartan, serving as a potential inducer of epigenetic modification, and not as a direct inhibitor or activator of epigenetic enzymes. Pentaerythritol tetranitrate has been reported to exhibit similar characteristic to losartan (35). Losartan and pentaerythritol tetranitrate may be regarded as atypical epigenetic drugs, and atypical epigenetic drugs may be potential alternatives to typical epigenetic drugs in the treatment of chronic disease.
The results of the present study demonstrated that prehypertensive treatment with losartan was able to relieve metabolic disorders in the later life of high-fat-fed SHRs. The long-term effects may be associated with the differential epigenetic regulation of RAS gene expression in adipose tissue via DNA methylation. The present study provided evidence for the additional benefits of early treatment with losartan, beyond the prolongation of a decrease in blood pressure.
Acknowledgements
The present study was supported by grants from the National Natural Science Foundation of China (grant no. 81570446), Natural Science Foundation of Fujian province (grant no. 2017J01193) and Middle and Young Aged Backbone of Fujian health department (grant no. 2017-ZQN-44).
Glossary
Abbreviations
- SHRs
spontaneously hypertensive rats
- AT1R
angiotensin II type 1 receptor
- ATRAP
AT1 receptor-associated protein
- AT1aR
AT1R subtype a
- RAS
renin-angiotensin system
- WAT
white adipose tissue
- BW
body weight
- SBP
systolic blood pressure
- TC
total cholesterol
- TG
triglycerides
- HDL-C
high-density lipoprotein cholesterol
- LDL-C
low-density lipoprotein cholesterol
- IL-6
interleukin-6
- MCP-1
monocyte chemoattractant protein-1
- TNF-α
tumor necrosis factor-α
- PPAR-γ
peroxisome proliferator-activated receptor-γ
- aP2
transcription factor adaptin 2
- CEBP
CCAAT/enhancer binding protein
References
- 1.Galassi A, Reynolds K, He J. Metabolic syndrome and risk of cardiovascular disease: A meta-analysis. Am J Med. 2006;119:812–819. doi: 10.1016/j.amjmed.2006.02.031. [DOI] [PubMed] [Google Scholar]
- 2.Peng F, Lin J, Lin L, Tang H. Transient prehypertensive treatment in spontaneously hypertensive rats: A comparison of losartan and amlodipine regarding long-term blood pressure, cardiac and renal protection. Int J Mol Med. 2012;30:1376–1386. doi: 10.3892/ijmm.2012.1153. [DOI] [PubMed] [Google Scholar]
- 3.Maury E, Brichard SM. Adipokine dysregulation, adipose tissue inflammation and metabolic syndrome. Mol Cell Endocrinol. 2010;314:1–16. doi: 10.1016/j.mce.2009.07.031. [DOI] [PubMed] [Google Scholar]
- 4.Jing F, Mogi M, Horiuchi M. Role of renin-angiotensin-aldosterone system in adipose tissue dysfunction. Mol Cell Endocrinol. 2013;378:23–28. doi: 10.1016/j.mce.2012.03.005. [DOI] [PubMed] [Google Scholar]
- 5.Lastra G, Sowers JR. Obesity and cardiovascular disease: Role of adipose tissue, inflammation, and the renin-angiotensin-aldosterone system. Horm Mol Biol Clin Investig. 2013;15:49–57. doi: 10.1515/hmbci-2013-0025. [DOI] [PubMed] [Google Scholar]
- 6.Bogdarina I, Welham S, King PJ, Burns SP, Clark AJ. Epigenetic modification of the renin-angiotensin system in the fetal programming of hypertension. Circ Res. 2007;100:520–526. doi: 10.1161/01.RES.0000258855.60637.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xiao D, Dasgupta C, Li Y, Huang X, Zhang L. Perinatal nicotine exposure increases angiotensin II receptor-mediated vascular contractility in adult offspring. PLoS One. 2014;9:e108161. doi: 10.1371/journal.pone.0108161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mao C, Liu R, Bo L, Chen N, Li S, Xia S, Chen J, Li D, Zhang L, Xu Z. High-salt diets during pregnancy affected fetal and offspring renal renin-angiotensin system. J Endocrinol. 2013;218:61–73. doi: 10.1530/JOE-13-0139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schübeler D. Function and information content of DNA methylation. Nature. 2015;517:321–326. doi: 10.1038/nature14192. [DOI] [PubMed] [Google Scholar]
- 10.Jones PA. Functions of DNA methylation: Islands, start sites, gene bodies and beyond. Nat Rev Genet. 2012;13:484–492. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- 11.Deaton AM, Bird A. CpG islands and the regulation of transcription. Genes Dev. 2011;25:1010–1022. doi: 10.1101/gad.2037511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Effects of ramipril on cardiovascular and microvascular outcomes in people with diabetes mellitus: Results of the HOPE study and MICRO-HOPE substudy. Heart Outcomes Prevention Evaluation Study Investigators. Lancet. 2000;355:253–259. doi: 10.1016/S0140-6736(99)12323-7. [DOI] [PubMed] [Google Scholar]
- 13.Maeda A, Tamura K, Wakui H, Ohsawa M, Azushima K, Uneda K, Kobayashi R, Tsurumi-Ikeya Y, Kanaoka T, Dejima T, et al. Effects of Ang II receptor blocker irbesartan on adipose tissue function in mice with metabolic disorders. Int J Med Sci. 2014;11:646–651. doi: 10.7150/ijms.8577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fernandez-Twinn DS, Ozanne SE. Early life nutrition and metabolic programming. Ann N Y Acad Sci. 2010;1212:78–96. doi: 10.1111/j.1749-6632.2010.05798.x. [DOI] [PubMed] [Google Scholar]
- 15.Lindl T, Gross U, Ruhdel I, von Aulock S, Völkel M. Guidance on determining indispensability and balancing potential benefits of animal experiments with costs to the animals with specific consideration of EU directive 2010/63/EU. Altex. 2012;29:219–228. doi: 10.14573/altex.2012.2.219. [DOI] [PubMed] [Google Scholar]
- 16.Ray MA, Johnston NA, Verhulst S, Trammell RA, Toth LA. Identification of markers for imminent death in mice used in longevity and aging research. J Am Assoc Lab Anim Sci. 2010;49:282–288. [PMC free article] [PubMed] [Google Scholar]
- 17.Widdop RE, Li XC. A simple versatile method for measuring tail cuff systolic blood pressure in conscious rats. Clin Sci (Lond) 1997;93:191–194. doi: 10.1042/cs0930191. [DOI] [PubMed] [Google Scholar]
- 18.Xie LD, Chen DG, Zhang S, Wang HJ, Chen HJ. Sympatholytic effect of captopril in regression of cardiovascular remodeling in spontaneously hypertensive rats. Zhongguo Yao Li Xue Bao. 1994;15:123–128. [PubMed] [Google Scholar]
- 19.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Method. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 20.Xie LD, Lin PS, Xie H, Xu CS. Effects of atorvastatin and losartan on monocrotaline-induced pulmonary artery remodeling in rats. Clin Exp Hypertens. 2011;32:547–554. doi: 10.3109/10641963.2010.503295. [DOI] [PubMed] [Google Scholar]
- 21.Chen HF, Xie LD, Xu CS. The signal transduction pathways of heat shock protein 27 phosphorylation in vascular smooth muscle cells. Mol Cell Biochem. 2010;333:49–56. doi: 10.1007/s11010-009-0203-5. [DOI] [PubMed] [Google Scholar]
- 22.Wang TJ, Lin X, Lian GL, Zhong HB, Xie LD. The effect of early losartan treatment on methylation of angiotensin II type 1 receptor subtype b and angiotensin II type 1 receptor associated protein genes in the myocardium of spontaneously hypertensive rats. Chin J Hypertens. 2016;24:141–146. doi: 10.1097/01.hjh.0000500264.88110.ff. [DOI] [Google Scholar]
- 23.Lee AM, Gurka MJ, DeBoer MD. Trends in metabolic syndrome severity and lifestyle factors among adolescents. Pediatrics. 2016;137:e20153177. doi: 10.1542/peds.2015-3177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Knight SF, Quigley JE, Yuan J, Roy SS, Elmarakby A, Imig JD. Endothelial dysfunction and the development of renal injury in spontaneously hypertensive rats fed a high-fat diet. Hypertension. 2008;51:352–359. doi: 10.1161/HYPERTENSIONAHA.107.099499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cao J, Inoue K, Sodhi K, Puri N, Peterson SJ, Rezzani R, Abraham NG. High-fat diet exacerbates renal dysfunction in SHR: Reversal by induction of HO-1-adiponectin axis. Obesity (Silver Spring) 2012;20:945–953. doi: 10.1038/oby.2011.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tontonoz P, Spiegelman BM. Fat and beyond: The diverse biology of PPARgamma. Annu Rev Biochem. 2008;77:289–312. doi: 10.1146/annurev.biochem.77.061307.091829. [DOI] [PubMed] [Google Scholar]
- 27.Singh KD, Karnik SS. Angiotensin receptors: Structure, function, signaling and clinical applications. J Cell Signal. 2016;1 doi: 10.4172/jcs.1000111. pii: 111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tamura K, Wakui H, Maeda A, Dejima T, Ohsawa M, Azushima K, Kanaoka T, Haku S, Uneda K, Masuda S, et al. The physiology and pathophysiology of a novel angiotensin receptor-binding protein ATRAP/Agtrap. Curr Pharm Des. 2013;19:3043–3048. doi: 10.2174/1381612811319170010. [DOI] [PubMed] [Google Scholar]
- 29.Shigenaga A, Tamura K, Wakui H, Masuda S, Azuma K, Tsurumi-Ikeya Y, Ozawa M, Mogi M, Matsuda M, Uchino K, et al. Effect of olmesartan on tissue expression balance between angiotensin II receptor and its inhibitory binding molecule. Hypertension. 2008;52:672–678. doi: 10.1161/HYPERTENSIONAHA.108.117341. [DOI] [PubMed] [Google Scholar]
- 30.Spalding KL, Arner E, Westermark PO, Bernard S, Buchholz BA, Bergmann O, Blomqvist L, Hoffstedt J, Naslund E, Britton T, et al. Dynamics of fat cell turnover in humans. Nature. 2008;453:783–787. doi: 10.1038/nature06902. [DOI] [PubMed] [Google Scholar]
- 31.Li Y, Xiao D, Yang S, Zhang L. Promoter methylation represses AT2R gene and increases brain hypoxic-ischemic injury in neonatal rats. Neurobiol Dis. 2013;60:32–38. doi: 10.1016/j.nbd.2013.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McKinsey TA. Therapeutic potential for HDAC inhibitors in the heart. Annu Rev Pharmacol Toxicol. 2012;52:303–319. doi: 10.1146/annurev-pharmtox-010611-134712. [DOI] [PubMed] [Google Scholar]
- 33.Xiao D, Dasgupta C, Chen M, Zhang K, Buchholz J, Xu Z, Zhang L. Inhibition of DNA methylation reverses norepinephrine-induced cardiac hypertrophy in rats. Cardiovasc Res. 2014;101:373–382. doi: 10.1093/cvr/cvt264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cho HM, Lee HA, Kim HY, Han HS, Kim IK. Expression of Na+−K+ −2Cl− cotransporter 1 is epigenetically regulated during postnatal development of hypertension. Am J Hypertens. 2011;24:1286–1293. doi: 10.1038/ajh.2011.136. [DOI] [PubMed] [Google Scholar]
- 35.Wu Z, Siuda D, Xia N, Reifenberg G, Daiber A, Münzel T, Förstermann U, Li H. Maternal treatment of spontaneously hypertensive rats with pentaerythritol tetranitrate reduces blood pressure in female offspring. Hypertension. 2015;65:232–237. doi: 10.1161/HYPERTENSIONAHA.114.04416. [DOI] [PubMed] [Google Scholar]