Abstract
The identification of optimal methylation biomarkers to achieve maximum diagnostic ability remains a challenge. The present study aimed to elucidate the potential molecular mechanisms underlying osteosarcoma (OS) using DNA methylation analysis. Based on the GSE36002 dataset obtained from the Gene Expression Omnibus database, differentially methylated genes were extracted between patients with OS and controls using t-tests. Subsequently, hierarchical clustering was performed to segregate the samples into two distinct clusters, OS and normal. Gene Ontology (GO) and pathway enrichment analyses for differentially methylated genes were performed using the Database for Annotation, Visualization and Integrated Discovery tool. A protein-protein interaction (PPI) network was established, followed by hub gene identification. Using the cut-off threshold of ≥0.2 average β-value difference, 3,725 unique CpGs (2,862 genes) were identified to be differentially methylated between the OS and normal groups. Among these 2,862 genes, 510 genes were differentially hypermethylated and 2,352 were differentially hypomethylated. The differentially hypermethylated genes were primarily involved in 20 GO terms, and the top 3 terms were associated with potassium ion transport. For differentially hypomethylated genes, GO functions principally included passive transmembrane transporter activity, channel activity and metal ion transmembrane transporter activity. In addition, a total of 10 significant pathways were enriched by differentially hypomethylated genes; notably, neuroactive ligand-receptor interaction was the most significant pathway. Based on a connectivity degree >90, 7 hub genes were selected from the PPI network, including neuromedin U (NMU; degree=103) and NMU receptor 1 (NMUR1; degree=103). Functional terms (potassium ion transport, transmembrane transporter activity, and neuroactive ligand-receptor interaction) and hub genes (NMU and NMUR1) may serve as potential targets for the treatment and diagnosis of OS.
Keywords: molecular mechanisms, osteosarcoma, enrichment analysis
Introduction
Osteosarcoma (OS) is the most common type of malignant bone carcinoma, which occurs primarily in the metaphyseal regions of long bones in adolescents and young adults (1). The 5-year survival rates of patients with OS without metastases is 60% (2). Although the survival rate has improved considerably following the introduction of neoadjuvant chemotherapy, it has reached a plateau and novel biological therapies are required to achieve further improvement. Although genetic alterations in OS have been extensively studied, understanding of the etiology of OS remains limited. Notably, epigenetics is emerging as an promising strategy for the study of OS.
DNA methylation, one of the most important mechanisms involved in microRNA expression regulation (3), gene silencing (4) and alternative gene splicing (5), exerts important functions in the early stage of carcinoma. As DNA methylation is stable and easily detected qualitatively or quantitatively, it has been identified to be a promising diagnostic marker for the early detection of cancer (6), compared with copy number variations (7), single nucleotide polymorphisms (SNPs)/mutations (8) and gene/microRNA expression (9). Previously, numerous instances of abnormal DNA methylation in the early stage of OS have been identified. For example, previous studies have indicated that Ras association domain family member 1A, death associated protein kinase, O-6-methylguanine-DNA methyltransferase, TIMP metallopeptidase inhibitor 3, and RB transcriptional corepressor 1 are hypermethylated in OS (10–12). In addition, Sonaglio et al (13) demonstrated that the hypermethylation of cyclin dependent kinase inhibitor 2A is associated with the absence of OS metastases, although estrogen receptor 1 hypermethylation is associated with decreased overall survival in OS. In addition, hypomethylation of iroquois homeobox 1 has been suggested to promote OS metastasis by inducing C-X-C motif chemokine ligand 14/nuclear factor-κB signaling (14). Despite the fact that a number of diagnostic panels have been developed, to the best of our knowledge, the causes of OS remain to be completely elucidated.
Previously, microarray analysis has been used to detect genetic alterations and identify potential targets in human OS cell lines. Kresse et al (15) combined the genetic and epigenetic profiles of 19 OS samples on the basis of microarray technologies, and deposited the OS-associated DNA methylation dataset in the Gene Expression Omnibus (GEO) database (no. GSE36002). In the previous study, the differentially methylated genes were extracted and functional enrichment analysis was implemented. However, the interactions among the differentially methylated genes were not measured.
Therefore, in order to better understand the molecular mechanisms of OS, the present study aimed to extract the differentially methylated genes from OS and normal samples from GSE36002. Hierarchical clustering was performed based on Euclidian distance and average linkage criteria. Functional enrichment analysis was respectively implemented for differentially hypomethylated/hypermethylated genes, to further identify the potential biological processes based on the Database for Annotation, Visualization and Integrated Discovery (DAVID). Subsequently, a protein-protein interaction (PPI) network was established to analyze the interactions between differentially methylated genes, followed by hub gene identification. The results of the present study provided evidence of the cumulative roles of epigenetic mechanisms in OS.
Materials and methods
Collection of DNA methylation data
DNA methylation data for OS (accession no. GSE36002) (15) was obtained from the GEO in the National Center for Biotechnology Information database (www.ncbi.nlm.nih.gov/gds), which was deposited in the GPL8490 platform (Illumina Human Methylation27 BeadChip; Illumina Inc., San Diego, CA, USA). GSE36002 included 25 samples (19 OS cell lines, and six normal samples derived from two osteoblast and four normal bone samples). Specifically, the 19 OS cell lines were obtained from EuroBoNeT (eurobonet.pathobiology.eu/cd/index.php). The two normal bone samples were collected from patients with cancer (one with OS and one with renal cell cancer) at the Norwegian Radium Hospital (Oslo, Norway). The normal bone was obtained from a site as distant as possible from the tumor site. The other two normal bone samples from different donors were purchased from Capital Biosciences, Inc. (Gaithersburg, MD, USA). The two human osteoblast cultures separated from the calvariae of different donors were purchased from ScienCell Research Laboratories, Inc. (San Diego, CA, USA).
DNA methylation data quality control and identification of differentially methylated genes
The methylation-identification algorithm in Genelibs (www.genelibs.com/gb) was utilized in the present study. The raw methylation status of 27,578 CpG sites was downloaded. Probes were eliminated from the dataset when they met the following criteria: i) Probes with SNP-CpG distances ≤2; ii) probes on X and Y chromosomes; iii) minimum allelic frequency <0.05; and iv) cross-hybridizing probes. Subsequently, DNA methylation data from the 25,628 CpG sites were kept for subsequent analysis.
The DNA methylation microarray data were processed using the Lumi package (bioconductor.org/packages/release/bioc/html/lumi.html) (16,17) of Bioconductor software. Data were normalized via the β-mixture quantile normalization method (18). Subsequently, β values (percentage methylation changes) were utilized in graphical representations of the data and demonstrated the percentage of methylation counted using the formula methylated/(methylated + unmethylated), ranging between 0 and 1; 0 represented fully unmethylated, while 1 represented fully methylated. In the present study, the β values of the OS and normal groups were calculated. Subsequently, the absolute value of the difference in mean β values between OS and normal groups was calculated, termed A. A Student's t-test was employed in the analysis to identify the differentially methylated CpGs between the two conditions. Differentially methylated CpGs were identified based on P<0.05 and A>0.01. SPSS version 18.0 (SPSS Inc., Chicago, IL, USA) was used for statistical analysis.
Refinement of the basic differential methylation analysis
In order to decrease the number of non-variable sites to enhance the statistical power of the following analyses, further filtering steps were performed to promote a more stringent analysis. All sites with β values ≤0.2 and ≥0.8 were deleted from all 25 samples, in order to decrease the number of non-variable sites to further promote the statistical power of the subsequent analyses (19). Additionally, only CpGs with a mean β-value difference ≥0.2 were kept. The absolute β-values in each matched pair were examined. A cut-off threshold of ≥0.2 average β-values difference was applied to detect CpGs with substantial methylation differences.
Hierarchical clustering analysis
Hierarchical clustering is a common approach utilized to determine clusters of similar data in multidimensional spaces (20). Generally, cancers having similar methylation profiles clustered together. To analyze whether these differentially methylated CpGs could segregate the samples into two distinct clusters including OS and normal samples, unsupervised hierarchical clustering was conducted using Euclidian distance and average linkage criteria (21). The matrix of mean β-value levels of differential methylated CpGs was formed between OS and normal samples.
Gene ontology (GO) analysis of differentially methylated genes
GO analysis has been widely utilized for large-scale functional enrichment research (22). In the present study, GO functional enrichment analysis was implemented for differentially methylated genes using DAVID (david.ncifcrf.gov) which is a software tool providing a comprehensive set of functional annotation for researchers to understand the biological meaning behind a large number of genes (23). Specifically, Fisher's exact test was used to classify the GO category. Subsequently, P-values were corrected using the false discovery rate (FDR) with the Benjamini & Hochberg method (24). Functional terms with FDR <0.01 and gene count >10 were considered to be statistically significant.
Pathway enrichment analysis of differentially methylated genes
The Kyoto Encyclopedia of Genes and Genomes (KEGG; www.genome.jp/kegg/pathway) is a knowledge base used to systematically analyze gene functions (25). In the present study, pathway analysis was performed according to the KEGG using the DAVID tool. Fisher's exact test was used to extract the significant pathways, and the threshold of significance was defined by FDR. Significant pathways were selected according to the thresholds of FDR <0.01 and gene count >10.
PPI network construction and hub genes identification
The Search Tool for the Retrieval of Interacting Genes (STRING) database (www.bork.embl-heidelberg.de/STRING) is a global resource for analyzing PPI information (26). In the present study, the STRING tool was used to identify the PPIs of differentially methylated genes. The differentially methylated genes with required confidence (combined score) >0.8 were extracted, and the PPI network was established and displayed using Cytoscape software (cytoscape.org) (27). Given that the networks were scale-free, the hub genes were identified with degree >90.
Results
Identification of differentially methylated genes
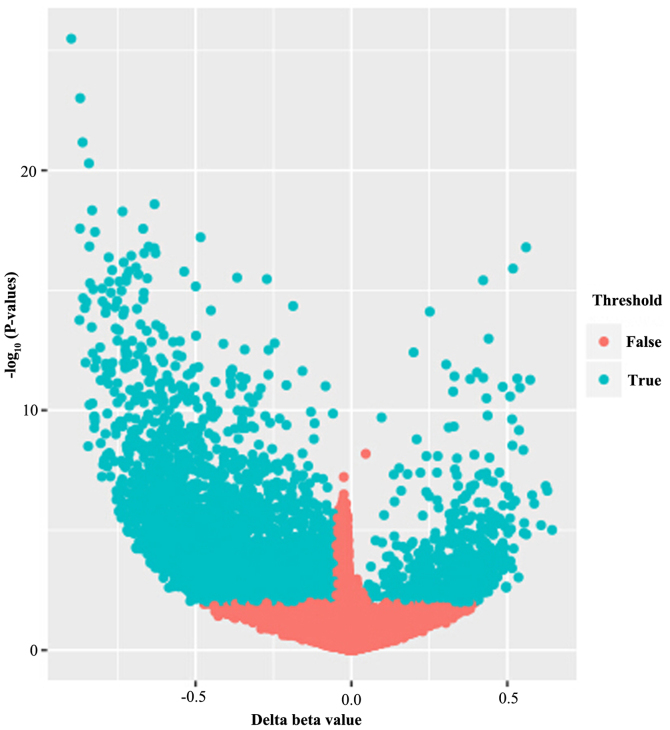
Following quality control and normalization to remove probes with SNP-CpG distance ≤2, on the X and Y chromosomes, with minimum allelic frequency <0.05, and cross-hybridizing, a total of 25,628 methylated CpGs remained in the final dataset of 25 samples. A volcano plot exhibiting the distribution of the 25,628 analyzed methylated CpGs was produced, as presented in Fig. 1. Among these 25,628 methylated CpGs, 5,889 CpGs (representing 4,677 genes) were differentially methylated, when the absolute value of the mean β-value difference between the OS and normal groups was >0.01 and the P-value was <0.05. A total of 572 of the CpGs were hypermethylated and 5,317 CpGs were hypomethylated in the OS group compared with the normal group.
Figure 1.
Volcano plot exhibiting the methylation data of 19 osteosarcoma and 6 normal samples. X axis, mean methylation differences between osteosarcoma and normal. Y axis, log transformed P-values. A total of 5,889 CpG sites were considered to be significantly differently methylated, exhibited in blue.
Subsequently, these 5,889 methylated CpGs initially extracted as differentially methylated sites were subjected to further filtering. Using the cut-off threshold of ≥0.2 average β-values difference, 3,725 unique CpGs (covering 2,862 genes) were detected to be differentially methylated between the OS and normal groups. Among these 2,862 genes, 510 gene were differentially hypermethylated genes and 2,352 were differentially hypomethylated genes.
Cluster analysis of the differentially methylated genes
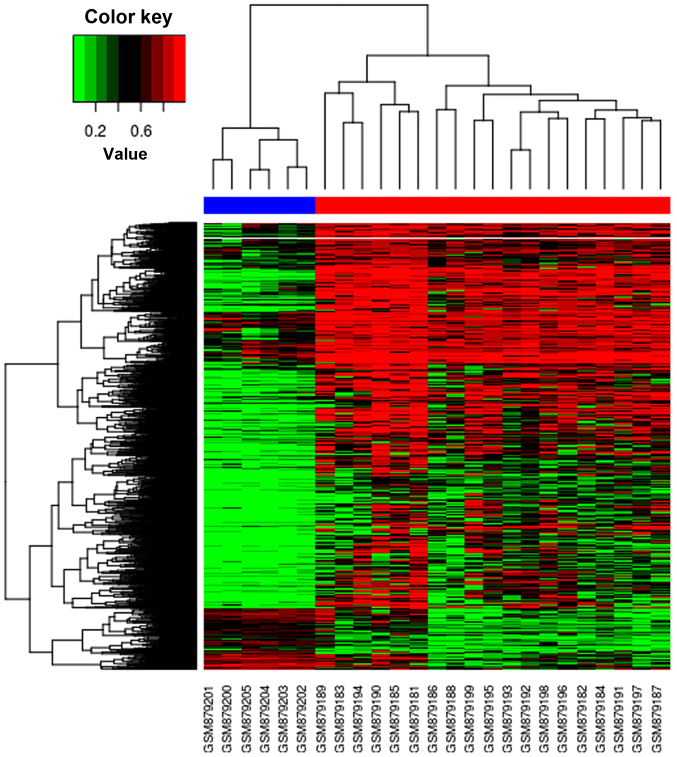
The cluster heat map is presented in Fig. 2. In this figure, it was observed that there were distinctive methylation patterns in OS and normal samples, which segregated samples into two distinct groups comprising those from OS and normal populations.
Figure 2.
Hierarchical clustering analysis of significantly differentially methylated CpGs between osteosarcoma and normal samples. DNA methylation across the 3,725 sites in each of the samples was analyzed by hierarchical clustering. Each row is an individual CpG site and each column is a different sample. Color gradation from green to red denotes low to high DNA methylation, with β-values ranging from 0 (no methylation; green) to 1 (complete methylation; red).
GO enrichment of differentially methylated genes
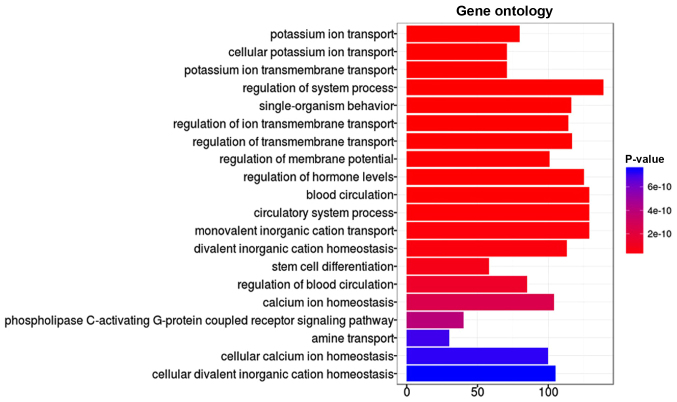
In order to better understand the potential biological functions of the differentially methylated genes, all the genes were annotated using GO annotation based on DAVID software. GO categories with FDR <0.01 and gene count >10 were regarded as significantly enriched. Overall, 20 GO terms were significantly enriched by differentially hypermethylated genes, as presented in Fig. 3. These GO terms were sorted in ascending order based on FDR value, and the top 3 most significantly enriched differentially hypermethylated genes were associated with potassium ion transport. For differentially hypomethylated genes, GO functions primarily included passive transmembrane transporter activity, channel activity and metal ion transmembrane transporter activity, as described in Fig. 4.
Figure 3.
Differentially hypermethylated genes identified between osteosarcoma and normal samples were functionally classificated via gene ontology analysis. The most enriched functional terms, which satisfied the FDR <0.01 and gene count >10 criteria, are presented.
Figure 4.
Differentially hypomethylated genes identified between osteosarcoma and normal samples were functionally classified via gene ontology analysis. The most enriched functional terms, which satisfied the FDR <0.01 and gene count >10 criteria, are presented.
KEGG pathway analysis for differentially methylated genes
Pathway enrichment analysis of all differentially methylated genes was performed based on the KEGG automatic annotation server. Based on FDR <0.01 and gene count >10, no KEGG pathways were enriched by the differentially hypermethylated genes. Notably, differentially hypomethylated genes were enriched in 10 KEGG pathways, including neuroactive ligand-receptor interaction (FDR=3.5×10−9; gene count=80), pathway in cancer (FDR=3.2×10−7; gene count=96) and Rap1 signaling pathway (FDR=6.5×10−6; gene count=58). The specific enrichment results are presented in Table I.
Table I.
KEGG pathway analysis for differentially methylated genes, based on FDR <0.01 and gene count >10.
| Terms | Gene count | FDR |
|---|---|---|
| Neuroactive ligand-receptor interaction | 80 | 3.5×10−9 |
| Pathways in cancer | 96 | 3.2×10−7 |
| Rap1 signaling pathway | 58 | 6.5×10−6 |
| Calcium signaling pathway | 58 | 1.9×10−4 |
| Morphine addiction | 31 | 2.1×10−3 |
| MAPK signaling pathway | 36 | 2.4×10−3 |
| Cholinergic synapse | 46 | 3.9×10−3 |
| Circadian entrainment | 50 | 6.4×10−3 |
| cAMP signaling pathway | 30 | 6.5×10−3 |
| Cell adhesion molecules | 27 | 7.1×10−3 |
FDR, false discovery rate; KEGG, Kyoto Encyclopedia of Genes and Genomes; cAMP, cyclic adenosine monophosphate; MAPK, mitogen-activated protein kinase; Rap1, telomeric repeat-binding factor 2-interacting protein 1.
PPI network construction and hub genes identification
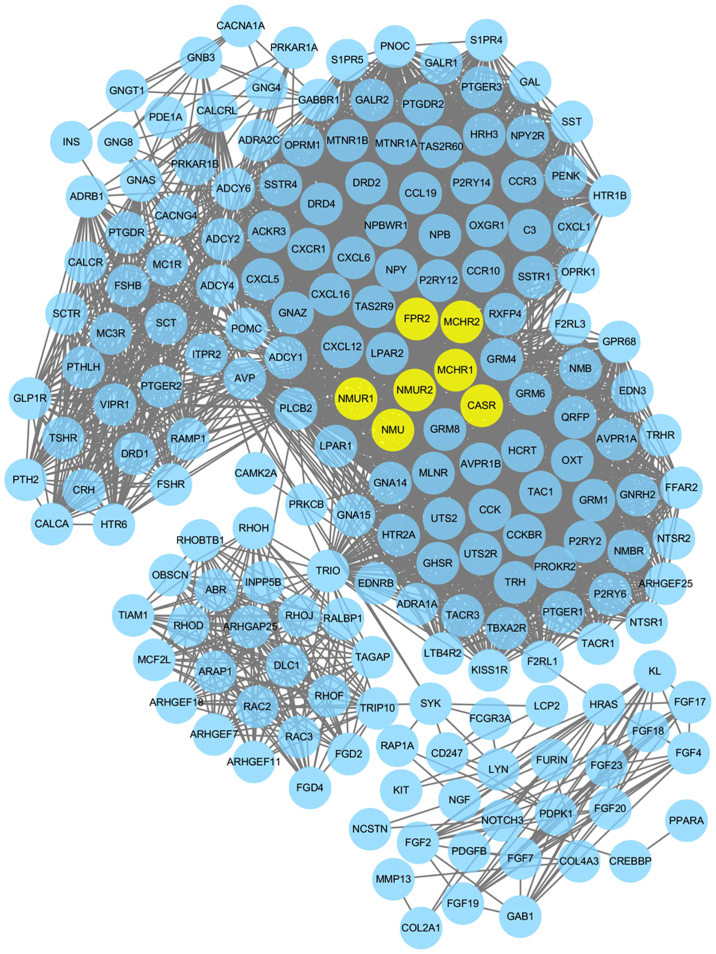
With the goal of analyzing the association between differentially methylated genes, STRING software was used to establish the PPI network. When the differentially methylated genes with required confidence (combined score) >0.8 were submitted into STRING, a total of 3,775 PPI interactions (covering 195 nodes) were obtained, as presented in Fig. 5. The hub genes in the networks with connectivity degree >90 were identified. A total of 7 hub genes were selected from the PPI network, which included neuromedin U (NMU; degree=103), NMU receptor 1 (NMUR1; degree=103), NMUR2 (degree=103), calcium sensing receptor (degree=103), formyl peptide receptor 2 (degree =92), melanin concentrating hormone receptor (MCHR) 2 (degree=91) and MCHR1 (degree=91).
Figure 5.
Protein-protein interaction network constructed from differentially methylated genes with the required confidence (combined score) >0.8. Yellow nodes represent hub genes.
Discussion
The analysis of DNA methylation data has been widely used to identify abnormally methylated genes associated with OS and has enabled the extraction of targets for therapeutic strategies. In the present study, OS pathogenesis was analyzed using bioinformatics, including the detection of differentially methylated genes, functional analyses of the differentially methylated genes, PPI construction, and hub genes identification. According to the results, the potential mechanisms of OS were revealed, which provided novel insights into OS diagnosis and therapy.
Following implementation of the pathway functional analyses for differentially hypomethylated genes, the pathway of neuroactive ligand-receptor interaction was selected as the most significant pathway. Neuroactive steroids act as mediators of neurotransmitter receptors to regulate neuronal activity (28). The effect of steroids indicates a ligand-receptor interaction. Previous studies have reported that neuroactive steroids have been suggested to affect the modulation of γ-aminobutyric acid (GABA) receptors (29,30). Notably, studies have reported that GABA receptors control cell proliferation and have suggested that there may be an association between the GABAergic system and oncogenesis (31,32). In particular, GABA has been observed to be overexpressed in a number of types of tumors, including gastric, colon, ovarian, and breast cancer (32–34). At present, the pathway of neuroactive ligand-receptor interaction has not been demonstrated to be directly involved in OS. According to the results of the present study, it may be inferred that neuroactive ligand-receptor interaction might serve a role in OS, partially by regulating the expression of GABA.
Notably, the GO results in the present study indicated that the top 3 functions enriched by differentially hypermethylated genes were associated with potassium ion transport. A previous study suggested that ion channels may be involved in the progression of cancer (35). Potassium channels, a class of ion channel, have been demonstrated to be aberrantly expressed in tumor cells and to be involved in carcinogenesis (36,37). Potassium channels serve diverse roles in cancer-associated processes, including cell survival, proliferation, and migration (38,39). Notably, a previous study demonstrated the roles of potassium channels in the control of glioma cell survival, growth and migration (40). Potassium chloride cotransporter 2 has been indicated to increase cervical cancer cell invasion through an ion transport-based mechanism (41). A high level of potassium expression has been reported in OS (42,43). Accordingly, the results of the present study indicated a further link between potassium ion transport and OS progression.
For differentially hypomethylated genes, GO functions were associated with transmembrane transporter activity. Facilitated glucose transporters regulate the energy-associated transport of glucose across the plasma membrane (44). A previous study demonstrated that the suppression of glucose transport was associated with apoptosis (45). Apoptosis has been implicated to exert an important role in the progression of OS. Increasing evidence has suggested that the induction of apoptosis may be an effective means of inhibiting tumor formation and development (46–48). As reported, one cellular mechanism which produces resistance to antineoplastic therapy involves the efflux of drugs from cancer cells through specific transmembrane transporters (49). Therefore, it may be inferred that transmembrane transporter activity is associated with OS progression.
Based on the degree distribution in the PPI network, NMU and NMUR1 exhibited the highest degrees. NMU, a neuropeptide, has potent activity in energy homeostasis (50). A previous study demonstrated that NMU may cause the release of inflammatory cytokines from T cells or macrophages (51). Notably, cytokine-releasing immune cells may stimulate neovascularization to promote the growth of human neoplasms. Studies have indicated that NMU is associated with tumorigenesis. For example, NMU has been reported to stimulate the migration of renal cancer cells (52,53), and acts as a potential growth factor for myeloid leukemia (54). NMUR1, a receptor of NMU, has been demonstrated to be expressed in renal cancer cells (52). The potential role of NMU and NMUR1 in OS has not been documented. According to the results of the present study, it may be inferred that NMU and NMUR1 may serve an important role in OS.
In the present study, the data were recruited from the DNA methylation dataset GSE36002, which was analyzed by Kresse et al (15). Consistent with the study of Kresse et al, differentially methylated genes between OS and normal samples were extracted in the present study, and GO functional and pathway enrichment analyses were implemented to examine the underlying mechanism of OS. However, there remain certain discrepancies. On the basis of the study of Kresse et al, further analyses using bioinformatics were performed in the present study, including the construction of the PPI network and hub gene analysis.
However, the present study had certain limitations. There was a small amount of sample data. Additionally, the data used in the present study were recruited from the GEO database, and not collected specifically for the present study. Since GEO is a large data repository, a meta-analysis of the relevant datasets for OS may be performed in the future. Additionally, the OS samples were obtained from cell lines, while normal samples were obtained from the human bones of different donors. In addition, the findings were bioinformatics-based identification and were not validated using biological experiments. Therefore, further experimental studies are required.
In conclusion, the present study provided a comprehensive bioinformatics analysis of differentially methylated genes which may be involved in the development and progression of OS. The results of the present study may provide an insight into the potential pathogenesis of OS. Additionally, functional terms (potassium ion transport, transmembrane transporter activity and neuroactive ligand-receptor interaction) and hub genes (NMU and NMUR1) may serve as potential therapeutic targets for OS. However, further work is required to improve the diagnosis and treatment of OS by regulating functional terms.
References
- 1.Kobayashi E, Hornicek FJ, Duan Z. MicroRNA Involvement in Osteosarcoma. Sarcoma. 2012;2012:359739. doi: 10.1155/2012/359739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Posthumadeboer J, Witlox MA, Kaspers GJ, van Royen BJ. Molecular alterations as target for therapy in metastatic osteosarcoma: A review of literature. Clin Exp Metastasis. 2011;28:493–503. doi: 10.1007/s10585-011-9384-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.He Y, Cui Y, Wang W, Gu J, Guo S, Ma K, Luo X. Hypomethylation of the hsa-miR-191 locus causes high expression of hsa-mir-191 and promotes the epithelial-to-mesenchymal transition in hepatocellular carcinoma. Neoplasia. 2011;13:841–853. doi: 10.1593/neo.11698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Geiman TM, Robertson KD. Chromatin remodeling, histone modifications and DNA methylation-how does it all fit together? J Cell Biochem. 2002;87:117–125. doi: 10.1002/jcb.10286. [DOI] [PubMed] [Google Scholar]
- 5.Flores K, Wolschin F, Corneveaux JJ, Allen AN, Huentelman MJ, Amdam GV. Genome-wide association between DNA methylation and alternative splicing in an invertebrate. BMC Genomics. 2012;13:480. doi: 10.1186/1471-2164-13-480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhao Y, Sun J, Zhang H, Guo S, Gu J, Wang W, Tang N, Zhou X, Yu J. High-frequency aberrantly methylated targets in pancreatic adenocarcinoma identified via global DNA methylation analysis using methylCap-seq. Clin Epigenetics. 2014;6:18. doi: 10.1186/1868-7083-6-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jiang F, Todd NW, Li R, Zhang H, Fang H, Stass SA. A panel of sputum-based genomic marker for early detection of lung cancer. Cancer Prev Res (Phila) 2010;3:1571–1578. doi: 10.1158/1940-6207.CAPR-10-0128. [DOI] [PubMed] [Google Scholar]
- 8.Guo S, Wang YL, Li Y, Jin L, Xiong M, Ji QH, Wang J. Significant SNPs have limited prediction ability for thyroid cancer. Cancer Med. 2014;3:731–735. doi: 10.1002/cam4.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhu J, Yao X. Use of DNA methylation for cancer detection: Promises and challenges. Int J Biochem Cell Biol. 2009;41:147–154. doi: 10.1016/j.biocel.2008.09.003. [DOI] [PubMed] [Google Scholar]
- 10.Hou P, Ji M, Yang B, Chen Z, Qiu J, Shi X, Lu Z. Quantitative analysis of promoter hypermethylation in multiple genes in osteosarcoma. Cancer. 2006;106:1602–1609. doi: 10.1002/cncr.21762. [DOI] [PubMed] [Google Scholar]
- 11.Lim S, Yang MH, Park JH, Nojima T, Hashimoto H, Unni KK, Park YK. Inactivation of the RASSF1A in osteosarcoma. Onco Rep. 2003;10:897–901. [PubMed] [Google Scholar]
- 12.Patiño-García A, Piñeiro ES, Díez MZ, Iturriagagoitia LG, Klüssmann FA, Ariznabarreta LS. Genetic and epigenetic alterations of the cell cycle regulators and tumor suppressor genes in pediatric osteosarcomas. J Pediatr Hematol Oncol. 2003;25:362–367. doi: 10.1097/00043426-200305000-00003. [DOI] [PubMed] [Google Scholar]
- 13.Sonaglio V, de Carvalho AC, Toledo SR, Salinas-Souza C, Carvalho AL, Petrilli AS, de Camargo B, Vettore AL. Aberrant DNA methylation of ESR1 and p14ARF genes could be useful as prognostic indicators in osteosarcoma. OncoTargets Ther. 2013;6:713–723. doi: 10.2147/OTT.S44918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lu J, Song G, Tang Q, Zou C, Han F, Zhao Z, Yong B, Yin J, Xu H, Xie X, et al. IRX1 hypomethylation promotes osteosarcoma metastasis via induction of CXCL14/NF-κB signaling. J Clin Invest. 2015;125:1839–1856. doi: 10.1172/JCI78437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kresse SH, Rydbeck H, Skårn M, Namløs HM, Barragan-Polania AH, Cleton-Jansen AM, Serra M, Liestøl K, Hogendoorn PC, Hovig E, et al. Integrative analysis reveals relationships of genetic and epigenetic alterations in osteosarcoma. PLoS One. 2012;7:e48262. doi: 10.1371/journal.pone.0048262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Du P, Kibbe WA, Lin SM. Lumi: A pipeline for processing Illumina microarray. Bioinformatics. 2008;24:1547–1548. doi: 10.1093/bioinformatics/btn224. [DOI] [PubMed] [Google Scholar]
- 17.Du P, Zhang X, Huang CC, Jafari N, Kibbe WA, Hou L, Lin SM. Comparison of Β-value and M-value methods for quantifying methylation levels by microarray analysis. BMC Bioinformatics. 2010;11:587. doi: 10.1186/1471-2105-11-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Teschendorff AE, Marabita F, Lechner M, Bartlett T, Tegner J, Gomez-Cabrero D, Beck S. A β-mixture quantile normalization method for correcting probe design bias in Illumina Infinium 450 k DNA methylation data. Bioinformatics. 2013;29:189–196. doi: 10.1093/bioinformatics/bts680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu P, Farrell WE, Haworth KE, Emes RD, Kitchen MO, Glossop JR, Hanna FW, Fryer AA. Maternal genome-wide DNA methylation profiling in gestational diabetes shows distinctive disease-associated changes relative to matched healthy pregnancies. Epigenetics. 2016 Mar 16; doi: 10.1080/15592294.2016.1166321. (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Olson CF. Parallel Algorithms for Hierarchical Clustering. Pattern An Mach Int IEEE Trans. 1990;12:1088–1092. doi: 10.1109/34.61708. [DOI] [Google Scholar]
- 21.Sturn A, Quackenbush J, Trajanoski Z. Genesis: Cluster analysis of microarray data. Bioinformatics. 2002;18:207–208. doi: 10.1093/bioinformatics/18.1.207. [DOI] [PubMed] [Google Scholar]
- 22.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.da Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 24.Benjamini Y, Drai D, Elmer G, Kafkafi N, Golani I. Controlling the false discovery rate in behavior genetics research. Behav Brain Res. 2001;125:279–284. doi: 10.1016/S0166-4328(01)00297-2. [DOI] [PubMed] [Google Scholar]
- 25.Kanehisa M, Goto S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.von Mering C, Huynen M, Jaeggi D, Schmidt S, Bork P, Snel B. STRING: A database of predicted functional associations between proteins. Nucleic Acids Res. 2003;31:258–261. doi: 10.1093/nar/gkg034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smoot ME, Ono K, Ruscheinski J, Wang PL, Ideker T. Cytoscape 2.8: New features for data integration and network visualization. Bioinformatics. 2011;27:431–432. doi: 10.1093/bioinformatics/btq675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smith SS. Female sex steroid hormones: From receptors to networks to performance-actions on the sensorimotor system. Prog Neurobiol. 1994;44:55–86. doi: 10.1016/0301-0082(94)90057-4. [DOI] [PubMed] [Google Scholar]
- 29.Chisari M, Eisenman LN, Krishnan K, Bandyopadhyaya AK, Wang C, Taylor A, Benz A, Covey DF, Zorumski CF, Mennerick S. The influence of neuroactive steroid lipophilicity on GABAA receptor modulation: Evidence for a low-affinity interaction. J Neurophysiol. 2009;102:1254–1264. doi: 10.1152/jn.00346.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smith SS. Withdrawal properties of a neuroactive steroid: Implications for GABA A receptor gene regulation in the brain and anxiety behavior. Steroids. 2002;67:519–528. doi: 10.1016/S0039-128X(01)00170-2. [DOI] [PubMed] [Google Scholar]
- 31.Watanabe M, Maemura K, Oki K, Shiraishi N, Shibayama Y, Katsu K. Gamma-aminobutyric acid (GABA) and cell proliferation: Focus on cancer cells. Histol Histopathol. 2006;21:1135–1141. doi: 10.14670/HH-21.1135. [DOI] [PubMed] [Google Scholar]
- 32.Szczaurska K, Mazurkiewicz M, Opolski A. The role of GABA-ergic system in carcinogenesis. Postepy Hig Med Dosw. 2003;57:485–500. [PubMed] [Google Scholar]
- 33.Moon MS, Cho EW, Byun HS, Jung IL, Kim IG. GAD 67KD antisense in colon cancer cells inhibits cell growth and sensitizes to butyrate and pH reduction and H2O2 and gamma-radiation. Arch Biochem Biophys. 2004;430:229–236. doi: 10.1016/j.abb.2004.07.015. [DOI] [PubMed] [Google Scholar]
- 34.Kleinrok Z, Matuszek M, Jesipowicz J, Matuszek B, Opolski A, Radzikowski C. GABA content and GAD activity in colon tumors taken from patients with colon cancer or from xenografted human colon cancer cells growing as s.c. Tumors in athymic nu/nu mice. J Physiol Pharmacol. 1998;49:303–310. [PubMed] [Google Scholar]
- 35.Kunzelmann K. Ion Channels and Cancer. J Membr Biol. 2005;205:159–173. doi: 10.1007/s00232-005-0781-4. [DOI] [PubMed] [Google Scholar]
- 36.Pardo LA, Stuhmer W. Eag1: An emerging oncological target. Cancer Res. 2008;68:1611–1613. doi: 10.1158/0008-5472.CAN-07-5710. [DOI] [PubMed] [Google Scholar]
- 37.Voloshyna I, Besana A, Castillo M, Matos T, Weinstein IB, Mansukhani M, Robinson RB, Cordon-Cardo C. TREK-1 is a novel molecular target in prostate cancer. Cancer Res. 2008;68:1197–1203. doi: 10.1158/0008-5472.CAN-07-5163. [DOI] [PubMed] [Google Scholar]
- 38.Weaver AK, Bomben VC, Sontheimer H. Expression and function of calcium-activated potassium channels in human glioma cells. Glia. 2006;54:223–233. doi: 10.1002/glia.20364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pluznick JL, Sansom SC. BK channels in the kidney: Role in K(+) secretion and localization of molecular components. Am J Physiol Renal Physiol. 2006;291:F517–F529. doi: 10.1152/ajprenal.00118.2006. [DOI] [PubMed] [Google Scholar]
- 40.Weaver AK, Liu X, Sontheimer H. Role for calcium-activated potassium channels (BK) in growth control of human malignant glioma cells. J Neurosci Res. 2004;78:224–234. doi: 10.1002/jnr.20240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wei WC, Akerman CJ, Newey SE, Pan J, Clinch NW, Jacob Y, Shen MR, Wilkins RJ, Ellory JC. The potassium-chloride cotransporter 2 promotes cervical cancer cell migration and invasion by an ion transport-independent mechanism. J Physiol. 2011;589:5349–5359. doi: 10.1113/jphysiol.2011.214635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rezzonico R, Schmid-Alliana A, Romey G, Bourget-Ponzio I, Breuil V, Breittmayer V, Tartare-Deckert S, Rossi B, Schmid-Antomarchi H. Prostaglandin E2 induces interaction between hSlo potassium channel and Syk tyrosine kinase in osteosarcoma cells. J Bone Miner Res. 2002;17:869–878. doi: 10.1359/jbmr.2002.17.5.869. [DOI] [PubMed] [Google Scholar]
- 43.Wu J, Zhong D, Wu X, Sha M, Kang L, Ding Z. Voltage-gated potassium channel Kv1.3 is highly expressed in human osteosarcoma and promotes osteosarcoma growth. Int J Mol Sci. 2013;14:19245–19256. doi: 10.3390/ijms140919245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Macheda ML, Rogers S, Best JD. Molecular and cellular regulation of glucose transporter (GLUT) proteins in cancer. J Cell Physiol. 2005;202:654–662. doi: 10.1002/jcp.20166. [DOI] [PubMed] [Google Scholar]
- 45.Moley KH, Mueckler MM. Glucose transport and apoptosis. Apoptosis. 2000;5:99–105. doi: 10.1023/A:1009697908332. [DOI] [PubMed] [Google Scholar]
- 46.Lorenz HM, Herrmann M, Winkler T, Gaipl U, Kalden JR. Role of apoptosis in autoimmunity. Apoptosis. 2000;5:443–449. doi: 10.1023/A:1009692902805. [DOI] [PubMed] [Google Scholar]
- 47.Schulze-Bergkamen H, Krammer PH. Apoptosis in cancer-implications for therapy. Semin Oncol. 2004;31:90–119. doi: 10.1053/j.seminoncol.2003.11.006. [DOI] [PubMed] [Google Scholar]
- 48.Wyllie AH. Apoptosis and the regulation of cell numbers in normal and neoplastic tissues: An overview. Cancer Metastasis Rev. 1992;11:95–103. doi: 10.1007/BF00048057. [DOI] [PubMed] [Google Scholar]
- 49.Deeley RG, Westlake C, Cole SP. Transmembrane transport of endo-and xenobiotics by mammalian ATP-binding cassette multidrug resistance proteins. Physiol Rev. 2006;86:849–899. doi: 10.1152/physrev.00035.2005. [DOI] [PubMed] [Google Scholar]
- 50.Fujii R, Hosoya M, Fukusumi S, Kawamata Y, Habata Y, Hinuma S, Onda H, Nishimura O, Fujino M. Identification of neuromedin U as the cognate ligand of the orphan G protein-coupled receptor FM-3. J Biol Chem. 2000;275:21068–21074. doi: 10.1074/jbc.M001546200. [DOI] [PubMed] [Google Scholar]
- 51.Moriyama M, Sato T, Inoue H, Fukuyama S, Teranishi H, Kangawa K, Kano T, Yoshimura A, Kojima M. The neuropeptide neuromedin U promotes inflammation by direct activation of mast cells. J Exp Med. 2005;202:217–224. doi: 10.1084/jem.20050248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Harten SK, Esteban MA, Shukla D, Ashcroft M, Maxwell PH. Inactivation of the von Hippel-Lindau tumour suppressor gene induces Neuromedin U expression in renal cancer cells. Mol Cancer. 2011;10:89. doi: 10.1186/1476-4598-10-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kohno Y. Oral cancer in vivo gene expression profiling assisted by laser capture microdissection and microarray analysis. Oncogene. 2001;20:6196–6204. doi: 10.1038/sj.onc.1204685. [DOI] [PubMed] [Google Scholar]
- 54.Shetzline SE, Rallapalli R, Dowd KJ, Zou S, Nakata Y, Swider CR, Kalota A, Choi JK, Gewirtz AM. Neuromedin U: A Myb-regulated autocrine growth factor for human myeloid leukemias. Blood. 2004;104:1833–1840. doi: 10.1182/blood-2003-10-3577. [DOI] [PubMed] [Google Scholar]