Abstract
It has been reported that oligodendrocyte precursor cells (OPCs) may be used to treat contusive spinal cord injury (SCC), and may alter microRNA (miRNA/miR) expression following SCC in rats. However, the association between miRNA expression and the treatment of rats with SCC with OPC transplantation remain unclear. The present study transplanted OPCs into the spinal cord of rats with SCC and subsequently used the Basso, Beattie and Bresnahan (BBB) score to assess the functional recovery and pain scores. An miRNA assay was performed to detect differentially expressed miRNAs in the spinal cord of SCC rats transplanted with OPCs, compared with SCC rats transplanted with medium. Quantitative polymerase chain reaction was used to verify significantly altered miRNA expression levels. The results demonstrated that OPC transplantation was able to improve motor recovery and relieve mechanical allodynia in rats with SCC. In addition, through a miRNA assay, 45 differentially expressed miRNAs (40 upregulated miRNAs and 5 downregulated miRNAs) were detected in the spinal cord of rats in the OPC group compared with in the Medium group. Differentially expressed miRNAs were identified according to the following criteria: Fold change >2 and P<0.05. Furthermore, quantitative polymerase chain reaction was used to verify the most highly upregulated (miR-375-3p and miR-1-3p) and downregulated (miR-363-3p, miR-449a-5p and miR-3074) spinal cord miRNAs that were identified in the miRNA assay. In addition, a bioinformatics analysis of these miRNAs indicated that miR-375 and miR-1 may act primarily to inhibit cell proliferation and apoptosis via transcriptional and translational regulation, whereas miR-363, miR-449a and miR-3074 may act primarily to inhibit cell proliferation and neuronal differentiation through transcriptional regulation. These results suggested that OPC transplantation may promote functional recovery of rats with SCC, which may be associated with the expression of various miRNAs in the spinal cord, including miR-375-3p, miR-1-3p, miR-363-3p, miR-449a-5p and miR-3074.
Keywords: contusive spinal cord injury, oligodendrocyte precursor cell transplantation, microRNAs
Introduction
Spinal cord injury (SCI) is a severe condition of the central nervous system, which represents a severe health problem that is associated with life-long disabilities and high mortality (1–3). The WHO reported in November 2013 that worldwide, 250,000–500,000 people annually suffer a spinal cord injury (4). It is well known that in addition to white matter axon damage, oligodendrocyte cell death (5) and demyelination in spared axons (6–9) may lead to the initial deficits associated with human SCC and impair functional recovery, which represents a challenge for the effective treatment of SCI. Oligodendrocytes are cells that produce the myelin sheath, which have been observed to die hours to weeks after SCI, near and distant to the epicenter, leading to demyelination (10–16). Therefore, therapeutic measures are required to reduce oligodendrocyte death and/or to enhance remyelination. Replacing lost oligodendrocytes, preventing the progression of demyelination or promoting remyelination is critical for functional recovery following SCC. In previous years, the advancement of cell therapy for SCI has been encouraging (17,18). Cell transplantation has been reported to promote neuroprotection by releasing trophic factors, providing a growth-promoting matrix for regeneration of axons or replacing lost cells (19). However, mature oligodendrocytes are unable to remyelinate naked axons in SCI, and also inhibit axonal regeneration by releasing myelin-associated inhibitors, including Nogo, myelin-associated glycoprotein and oligodendrocyte myelin glycoprotein (20,21). Therefore, to the best of our knowledge, no studies regarding the treatment of SCI with oligodendrocyte transplantation have been conducted. However, with the progress made regarding neural development, more studies have reported the possibility of using oligodendrocyte precursor cell (OPC) transplantation for the treatment of demyelinating diseases, including SCC (22,23). Therefore, OPC transplantation may be considered a potential therapy for the treatment of SCI (24).
It has been reported that human embryonic stem cell (hESC)-derived OPC transplantation may attenuate lesion pathogenesis and improve recovery of forelimb function in rats with cervical SCC (25). In addition, hESC-derived OPC transplantation into SCI sites in adult rats resulted in remyelination and functional repair (22). Combined treatment with OPC grafts expressing ciliary neurotrophic factor (CNTF) can also enhance remyelination and facilitate functional recovery following SCC (26). However, the molecular mechanism underlying the effects of OPC transplantation on SCI treatment, particularly regarding microRNA (miRNA) expression, is not fully understood.
miRNAs are ~23 nucleotide long endogenous ribonucleic acid molecules that specifically pair with the 3′ untranslated region of target mRNAs post-transcription, in order to direct mRNA degradation and suppress protein translation (27–29). Accumulating evidence has indicated that miRNAs serve crucial roles in various human diseases, including cancer, metabolic diseases, cardiovascular diseases, viral infections and contusive neurological injuries (30,31). miRNAs are also involved in numerous cellular processes, including development, proliferation and differentiation (28,32). Furthermore, numerous miRNAs have been detected in the mammalian central nervous system, including the brain and spinal cord, where they have key roles in neurodevelopment and are likely to be important mediators of plasticity (33–36). In addition, altered miRNA expression has been revealed in the spinal cord following SCC through microarray analysis (37,38). However, whether miRNAs are involved in the regulatory effects of OPC transplantation on the spinal cord of rats with SCC remains unclear.
In the present study, following OPC transplantation into the spinal cord of rats with SCC, the Basso, Beattie and Bresnahan (BBB) score and pain value were determined, in order to assess functional recovery of rats. Subsequently, a miRNA assay was performed to detect the differentially expressed miRNAs in the spinal cord of rats with SCC and OPC transplantation compared with in rats with SCC and medium transplantation. Furthermore, quantitative polymerase chain reaction (qPCR) was used to verify the significantly altered miRNA expression. These results may identify the regulatory effects of miRNA in rats with SCC following OPC transplantation, which may provide a novel potential molecular target for the clinical treatment of SCI in the future.
Materials and methods
Induced pluripotent stem cell (iPSC) preparation
The present study was approved by the ethics committee of Kunming Medical University (Kunming, China; KMMU2015013). Two pregnant mice were purchased from the Experimental Animal Center of Kunming Medical University. Mice were housed in cages at 22±2°C with 55±5% humidity under a 12-h light/dark cycle with free access to food and water. The skin of green fluorescent protein (GFP)-transgenic fetuses, obtained from 13.5-day-pregnant mice, was cut into small pieces and the skin samples were digested using 0.25% trypsin. Subsequently, cells were cultured in Dulbecco's modified Eagle's medium (DMEM; Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented with 10% fetal bovine serum (Gibco; Thermo Fisher Scientific, Inc.) at 37°C with 5% CO2. On the same day, fetal skin cells were co-transfected with 100 µl Moloney-based retroviral vectors (provided by Su Liu, Johns Hopkins University, Baltimore, MD, USA) that expressed octamer-binding transcription factor 3/4 (Oct3/4), sex determining region Y box-2 Sox2), c-Myc and Kruppel-like factor 4 (Klf4) (Oct3/4: Sox2: c-Myc: Klf4=1:1:1:1) which were transfected into PlatE clles using FuGENE 6 transfection reagent (Roche Diagnostics, Basel, Switzerland), twice within 2 days. On day 5 post-transfection, the transfected cells (1×105) were inoculated into mouse embryonic fibroblasts (MEFs; MTI-GlobalStem, Gaithersburg, MD, USA) that were pre-processed with 10 µg/ml mitomycin (cat. no. 01035, BBI Solutions, Cardiff, UK) for 1 day. Subsequently, the medium was replaced with embryonic stem cell growth medium (Invitrogen; Thermo Fisher Scientific, Inc.) every day. On day 16, iPSC clones were picked, digested using trypsin and inoculated into radiated MEFs that served as a feeder layer to obtain amplification.
iPSC differentiation
The iPSCs were cultured as aforementioned. iPSCs were separated by 0.25% trypsin every 4 days until passage 3 and were incubated with collagenase IV (1% in DMEM/F12; Invitrogen; Thermo Fisher Scientific, Inc.) at 37°C for 15 min in order to separate them from MEFs. After 4 days, iPSCs cultured in stem cell differentiation medium became embryonic bodies. Subsequently, the embryonic bodies were incubated with tretinoin (all-trans-retinoic acid, 500 nM; Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) and purmorphamin (1 µm; Calbiochem; EMD Millipore, Billerica, MA, USA) for 4 days at 37°C. The stem cell differentiation medium consisted of 50% DMEM/F12 (Gibco; Thermo Fisher Scientific, Inc.), 50% Neurobasal medium (Gibco; Thermo Fisher Scientific, Inc.), 1% N2 (Gibco; Thermo Fisher Scientific, Inc.), 1% B27 (Gibco; Thermo Fisher Scientific, Inc.), 10% serum fluid replacement, 0.1 mM mercaptoethanol (Sigma-Aldrich; Merck KGaA), 2 mM Glutamax (Gibco; Thermo Fisher Scientific, Inc.) and 2 µg/ml heparin (Sigma-Aldrich; Merck KGaA). A total of 24 days were required for iPSC preparation and differentiation.
Culture of iPSC-induced OPCs
At the 8th day of differentiation stage, the medium was replaced with OPC+ medium supplemented with platelet-derived growth factor (PDGF) and the cells were continually cultured for 8–10 days. Subsequently, cells were inoculated into plates that were coated with poly-D-lysine/laminin (BioCoat; BD Biosciences, Franklin Lakes, NJ, USA) at 37°C to proliferate for 2–3 days. The OPC+ medium contained 48% neurobasal medium (Gibco; Thermo Fisher Scientific, Inc.), 48% ml DMEM/F12 (Gibco; Thermo Fisher Scientific, Inc.), 50X OPC medium (Johns Hopkins University, Baltimore, MD, USA) 1/50 ml, 1% penicillin-streptomycin (Gibco; Thermo Fisher Scientific, Inc.), 0.5% Glutamax (Invitrogen; Thermo Fisher Scientific, Inc.), 1% non-essential amino acids and 0.02% fibroblast growth factor.
Animal care and grouping
A total of 32 male Sprague-Dawley rats (7 weeks-old, weight, 200–250 g) were purchased from the Experimental Animal Center of Kunming Medical University and were randomly divided into four groups: Sham, SCC, SCC + medium (Medium) and SCC + OPC transplantation (OPC) groups (Table I). All animal care, breeding and testing procedures conformed to the principles of guidance suggested in the National Institutes of Health Guide for the Care and Use of Laboratory Animals and supported by the Use Committee of Kunming Medical University. Rats were housed in cages at 22±2°C with 55±5% humidity under a 12-h light/dark cycle with free access to food and water.
Table I.
Animals and grouping.
| Group | BBB score (no. of rats) | miRNA chip (no. of rats) | qPCR (no. of rats) |
|---|---|---|---|
| Sham | 5 | 0 | 0 |
| SCC | 5 | 0 | 0 |
| Medium | 5 | 3 | 3 |
| OPC | 5 | 3 | 3 |
BBB score was measured on days 1, 3, 7, 14, 21 and 28 post transplantation; miRNA chip analysis and qPCR were conducted on day 7. BBB, Basso, Beattie and Bresnahan; miRNA, microRNA; OPC, oligodendrocyte precursor cells; SCC, contusive spinal cord injury; qPCR, quantitative polymerase chain reaction.
Animal model
SCC was performed to induce SCI in the present study. The rats were anesthetized by intraperitoneal injection with 3.6% chloral hydrate (380 mg/kg) and were placed in the prone position. A surgical incision was made in the thoracic region (T9-T11), and the paravertebral muscles and supraspinal ligaments were separated. Subsequently, the rats underwent a T9-T11 laminectomy, and were subjected to SCC at T10 level using a 10 g weight that was dropped from a 30 mm height. In addition, rats in the Sham group underwent the laminectomy only. Finally, the surgical wounds were sutured with a 3–0 silk suture. The rats were then injected with 5 ml saline, and received 0.5 ml cefotaxime sodium for 7 days and 0.05 ml Ciclosporin A injection (Novartis International AG, Basel, Switzerland) for 28 days. Their bladders were manually compressed three times a day until recovery of the micturition reflex.
Immunofluorescence staining
Immunofluorescence staining of the differentiated cells was performed as follows. Briefly, after OPCs were fixed with 4% paraformaldehyde for 10 min, they were washed three times (5 min each) with PBS (0.01 mol/l). OPCs were subsequently incubated with 0.3% Triton X-100 for 30 min at 37°C and 5% goat serum for 30 min at 37°C, respectively, and incubated with the primary antibody against PDGF (1:100; Neomarkers, Inc., Fremont, CA) to identify OPCs (39), at 4°C for 24 h. Finally, OPCs were washed using 0.01 mol/l PBS three times and incubated with Cy3 (1:100; Abcam, Cambridge, UK) for 60 min at 37°C. The nuclei were stained using DAPI and images were captured under a Leica DMI6000B (LAS AF system; Leica Microsystems, Inc., Buffalo Grove, IL, USA).
OPC transplantation
Rats in each group were placed in the prone position and their vertebral laminae were reopened 7 days after SCC surgery. Subsequently, 5 µl cell suspensions (2×105 cells/µl) were transplanted into each rat at two different sites at the rostral and tail end of the injury with a constant speed of 2.5 µl/min using a micropipette. The microinjector was maintained in the injection site for 5 min prior to removal. The Medium group was treated with OPC+ culture medium using the same procedure, while the vertebral laminae of rats in the sham and SCC control groups were re-opened without transplanting OPC or OPC+ culture medium.
Assessment of locomotion recovery
Locomotion recovery of rats was evaluated using the BBB Locomotion Rating Scale (40). The BBB scores range between 0 and 21. Score 0 indicates flaccid paralysis, whereas score 21 indicates normal gait. Three investigators, blinded to the experimental groups, performed behavioral experiments for rats in the Sham, SCC, Medium and OPC groups, in order to reduce the error of subjectivity. The assessments were performed on days 1, 3, 7, 14, 21 and 28 days following the transplantation operation in an open field (41). The average score from the three investigators was considered the final score.
Detection of mechanical pain threshold
Mechanical pain threshold assessments were performed on days 1, 3, 7, 14, 21 and 28 following the operation. Briefly, prior to the paw withdrawal threshold test, the value of the instrument was adjusted to zero. As the weight increased, the pressure that the rat felt was gradually increased. When the maximum pressure was reached it was recorded. This test was repeated three times and interval time was maintained at ≥10 min.
Tissues collection
After the rats were anesthetized by intraperitoneal injection with 3.6% chloral hydrate (380 mg/kg), spinal cord tissues were harvested from Medium and OPC group rats 7 days following the OPC/medium transplantation operation. The samples were stored at −80°C for miRNA microarray analysis and qPCR.
miRNA microarray analysis
Samples were transported to Genminix Informatics Co., Ltd. (Shanghai, China) for miRNA microarray analysis. Briefly, total RNA was extracted from the spinal cord samples using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.), quantified and assessed for quality using a Nanodrop ND-1000 (Thermo Fisher Scientific, Inc., Wilmington, DE, USA), and reverse transcribed into cDNA using SuperScript II Reverse Transcriptase (cat. no. 18064014, Invitrogen; Thermo Fisher Scientific, Inc.). Subsequently, the cDNA samples were hybridized to the Affymetrix Gene 2.0 Array (Affymetrix; Thermo Fisher Scientific, Inc.) to detect differentially expressed miRNAs. Subsequently, the positive signal was recorded using an Axon GenePix 4000B scanner (Molecular Devices, LLC, Sunnyvale, CA, UAS). The scanned images were imported into GenePix Pro 6.0 software (Axon Instruments; Molecular Devices, LLC) for grid alignment and data extraction. All microarray signals were analyzed by subtracting the background and were normalized using median normalization. Following normalization, differentially expressed miRNAs were identified according to fold change. In all samples, only miRNAs with a fold change >2 and P<0.05 were defined as significant.
Bioinformatics analysis of differentially expressed miRNAs
Initially, the target genes of the miRNAs were identified using the following prediction databases: miRecords (c1.accurascience.com/miRecords/), TargetScan (www.targetscan.org/vert_71/), MicroCosm (www.ebi.ac.uk/enright-srv/microcosm/htdocs/targets/v5/), miRanda (www.microrna.org/microrna/home.do) and PicTar (pictar.mdc-berlin.de/), and genes that were identified across two or more databases were selected for further analysis. Subsequently, a Gene Ontology (GO; www.geneontology.org/) enrichment analysis was conducted to determine the significantly enriched biological processes associated with the target genes of the differentially expressed miRNAs. Furthermore, predicted target genes of the differentially expressed miRNAs were analyzed using a functional method mapping genes to Kyoto Encyclopedia of Genes and Genomes (KEGG; www.genome.jp/kegg/) pathways.
qPCR
Total RNA was extracted from the spinal cord samples of rats in the Medium and SCC groups using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.), after which RNA was reverse transcribed to produce cDNA using an reverse transcribed reagent kit (Takara Bio, Inc.). Subsequently, the expression levels of miR-375-3p, miR-1-3p, miR-363-3p, miR-449a-5p and miR-3074 were analyzed using an SYBR Green RT-PCR Master Mix Kit (Takara Bio, Inc.). The primers were provided by Guangzhou RiboBio Co., Ltd. (Guangzhou, China) and the catalogue numbers are provided in Table II. β-actin was used as an internal control; the primer sequences and annealing temperature for β-actin were as follows: Forward 5′-GCCAACACAGTGCTGTCT-3′ and reverse 5′-GGAGCAATGATCTTGATCTT-3′; annealing temperature, 53°C. The reaction was performed at 95°C for 2 min and 40 cycles of 95°C for 20 sec, 53°C for 30 sec and 60°C for 40 sec. Data were analyzed using the quantification cycle (Cq) method (42), and target gene expression was normalized to internal control expression.
Table II.
Primer sequence catalogue numbers for miRNAs.
| miRNA | Number |
|---|---|
| rno-miR-375-5p | RmiRQP3367 |
| rno-miR-1-3p | RmiRQP1188 |
| rno-miR-363-3p | RmiRQP3254 |
| rno-miR-449a-5p | RmiRQP0504 |
| rno-miR-3074 | RmiRQP1482 |
miRNA, micro RNA.
Statistical analysis
All data are presented as the mean ± standard deviation. SPSS 17.0 (SPSS, Inc., Chicago, IL, USA) was used to analyze the data. Differences between two groups were assessed using a t-test and among multiple groups using one-way analysis of variance followed by least significant difference. P<0.05 was considered to indicate a statistically significant difference.
Results
Identification of GFP-OPCs induced from iPSCs derived from MEFs
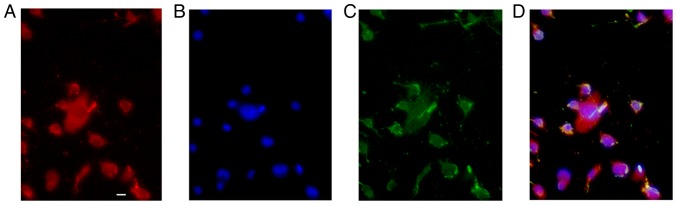
To identify the characteristics of GFP-OPCs induced from iPSCs derived from MEFs, they were cultured for 24 days in differentiation medium (Fig. 1). When GFP-OPCs were cultured in basal-OPC-medium supplemented with PDGF, almost all of the cells displayed the typical morphology of OPCs (Fig. 1C), and >90% of cells expressed PDGF (Fig. 1A and D).
Figure 1.
Identification of GFP-OPCs. GFP-OPCs induced from induced pluripotent stem cells derived from mouse embryonic fibroblasts were cultured in differentiation medium for 24 days. Subsequently, cells were cultured in basal-OPC-medium containing PDGF. (A) PDGF staining was conducted, and >90% of cells expressed PDGF. (B) DAPI nuclear staining was used to counterstain the cells (blue staining). (C) GFP detection indicated that cells displayed bipolar or tripolar morphology, which is typical of OPCs. (D) Merged image. Scale bar: 25 µm. GFP, green fluorescent protein; OPCs, oligodendrocyte precursor cells; PDGF, platelet-derived growth factor.
Transplantation of OPCs promotes motor and sensory recovery of rats subjected to SCC
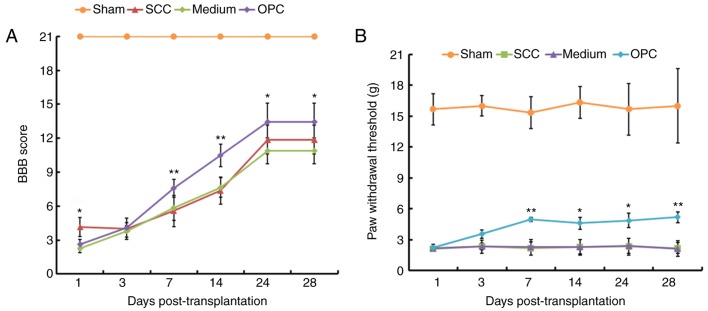
To investigate the effects of OPC transplantation on the motor and sensory function of rats subjected to SCC, BBB and paw withdrawal threshold analyses were conducted on rats from the Sham, SCC, Medium and OPC groups. As presented in Fig. 2A, with the exception of day 1 and 3, post-transplantation, the BBB score demonstrated that rats in the OPC group exhibited significant motor improvement compared with those in the Medium group. Results of the paw withdrawal threshold analysis indicated that OPCs attenuated SCC-induced mechanical allodynia. Days 1 and 3 post-transplantation, there were no significant differences between the average paw withdrawal threshold for rats in the Medium and OPC groups. However, 7, 14, 21 and 28 days following the transplantation, the OPC group exhibited a marked attenuation of mechanical allodynia (i.e. an increase in paw-withdrawal threshold) compared with in the Medium group (Fig. 2B).
Figure 2.
Results of behavioral analyses. (A) With the exception of day 1 and 3, treatment with OPCs resulted in significantly higher BBB scores compared with in the Medium group. (B) Mechanical allodynia data from the paw withdrawal threshold analysis. On days 1 and 3 post-transplantation, the OPC, Medium and SCC groups exhibited a decreased paw withdrawal threshold, indicating mechanical allodynia, compared with in the Sham group. Rats in the OPC group exhibited significant attenuation of paw withdrawal threshold compared with in the Medium group. *P<0.05, **P<0.01 vs. the Medium group. BBB, Basso, Beattie and Bresnahan; OPC, oligodendrocyte precursor cell; SCC, contusive spinal cord injury.
miRNA expression profile in the spinal cord of rats in the OPC group compared with the Medium group
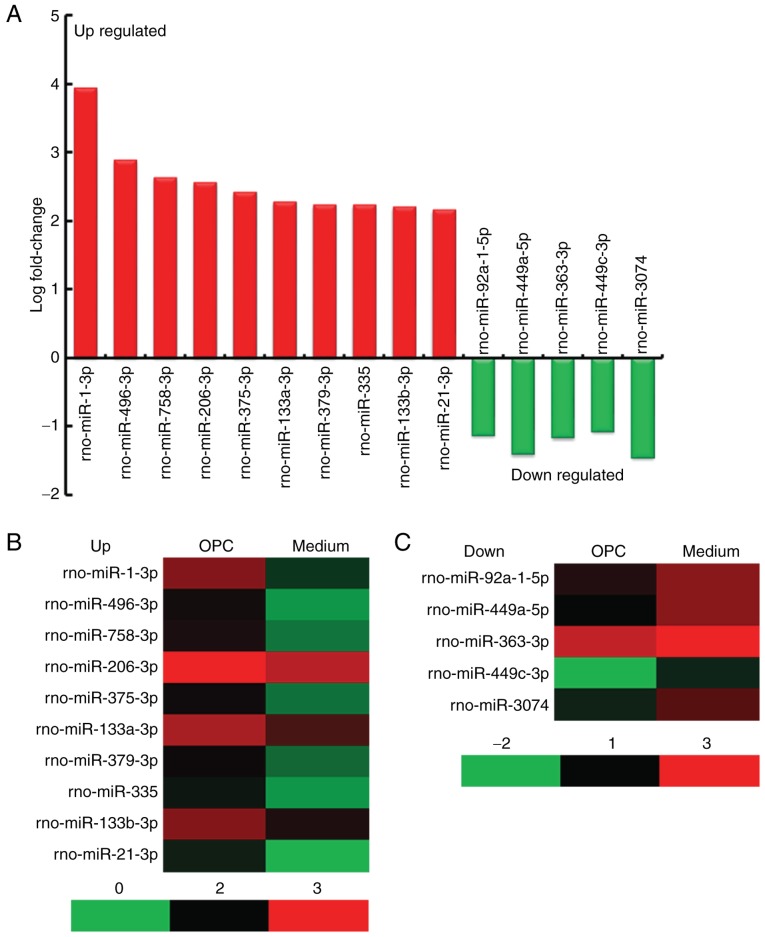
A miRNA assay was conducted on spinal cords obtained from rats in the OPC and Medium groups. The results indicated that there were 45 differentially expressed miRNAs (40 upregulated and 5 downregulated) in the spinal cord of rats from the OPC group compared with in the Medium group, according to the following criteria: Fold change >2 and P<0.05 (Table III). The top 10 upregulated miRNAs were rno-miR-1-3p, rno-miR-496-3p, rno-miR-758-3p, rno-miR-206-3p, rno-miR-375-3p, rno-miR-133a-3p, rno-miR-379-3p, rno-miR-335, rno-miR-133b-3p and rno-miR-21-3p, the 5 downregulated miRNAs were rno-miR-92a-1-5p, rno-miR-449a-5p, rno-miR-363-3p, rno-miR-449c-3p and rno-miR-3074 (Fig. 3A). A heat map of the miRNAs is presented in Fig. 3B.
Table III.
Differentially expressed miRNAs in the spinal cord of rats from the oligodendrocyte precursor cell transplantation group compared with the Medium group.
| miRNA | log2 (fold change) | Expression | Species | Sequence length (bp) | Sequence (5′-3′) |
|---|---|---|---|---|---|
| rno-miR-1-3p | 3.932485 | Up | Rattus norvegicus | 22 | UGGAAUGUAAAGAAGUGUGUAU |
| rno-miR-496-3p | 2.874668 | Up | Rattus norvegicus | 21 | AGUAUUACAUGGCCAAUCUCC |
| rno-miR-758-3p | 2.632376 | Up | Rattus norvegicus | 22 | UUUGUGACCUGGUCCACUAACC |
| rno-miR-206-3p | 2.552146 | Up | Rattus norvegicus | 22 | UGGAAUGUAAGGAAGUGUGUGG |
| rno-miR-375-3p | 2.413651 | Up | Rattus norvegicus | 22 | UUUGUUCGUUCGGCUCGCGUGA |
| rno-miR-133a-3p | 2.264416 | Up | Rattus norvegicus | 22 | UUUGGUCCCCUUCAACCAGCUG |
| rno-miR-379-3p | 2.227997 | Up | Rattus norvegicus | 22 | CUAUGUAACAUGGUCCACUAAC |
| rno-miR-335 | 2.224635 | Up | Rattus norvegicus | 23 | UCAAGAGCAAUAACGAAAAAUGU |
| rno-miR-133b-3p | 2.198147 | Up | Rattus norvegicus | 22 | UUUGGUCCCCUUCAACCAGCUA |
| rno-miR-21-3p | 2.15513 | Up | Rattus norvegicus | 22 | CAACAGCAGUCGAUGGGCUGUC |
| rno-miR-134-3p | 1.966194 | Up | Rattus norvegicus | 22 | CUGUGGGCCACCUAGUCACCAA |
| rno-let-7a-2-3p | 1.875046 | Up | Rattus norvegicus | 22 | CUGUACAGCCUCCUAGCUUUCC |
| rno-miR-764-5p | 1.757712 | Up | Rattus norvegicus | 22 | GGUGCUCACAUGUCCUCCUCCA |
| rno-miR-369-5p | 1.688398 | Up | Rattus norvegicus | 22 | AGAUCGACCGUGUUAUAUUCGC |
| rno-miR-485-3p | 1.6806526 | Up | Rattus norvegicus | 22 | CAUACACGGCUCUCCUCUCUUC |
| rno-miR-376a-3p | 1.67571 | Up | Rattus norvegicus | 21 | AUCGUAGAGGAAAAUCCACGU |
| rno-miR-133a-5p | 1.5515371 | Up | Rattus norvegicus | 22 | AGCUGGUAAAAUGGAACCAAAU |
| rno-miR-124-5p | 1.505714 | Up | Rattus norvegicus | 22 | CGUGUUCACAGCGGACCUUGAU |
| rno-miR-218a-5p | 1.499168 | Up | Rattus norvegicus | 21 | UUGUGCUUGAUCUAACCAUGU |
| rno-miR-323-3p | 1.491767 | Up | Rattus norvegicus | 21 | CACAUUACACGGUCGACCUCU |
| rno-miR-361-3p | 1.343701 | Up | Rattus norvegicus | 24 | CCCCCAGGUGUGAUUCUGAUUCGU |
| rno-miR-500-5p | 1.336195 | Up | Rattus norvegicus | 24 | AAUCCUUGCUAUCUGGGUGCUUAG |
| rno-miR-20a-3p | 1.3328006 | Up | Rattus norvegicus | 21 | ACUGCAUUACGAGCACUUACA |
| rno-miR-493-3p | 1.332116 | Up | Rattus norvegicus | 21 | UGAAGGUCUACUGUGUGCCAG |
| rno-miR-551b-3p | 1.316522 | Up | Rattus norvegicus | 23 | GGCGACCCAUACUUGGUUUCAGU |
| rno-miR-122-5p | 1.3071 | Up | Rattus norvegicus | 22 | UGGAGUGUGACAAUGGUGUUUG |
| rno-miR-764-3p | 1.268782 | Up | Rattus norvegicus | 23 | GAGGAGGCCAUAGUGGCAACUGU |
| rno-miR-433-5p | 1.248092 | Up | Rattus norvegicus | 22 | UACGGUGAGCCUGUCAUUAUUC |
| rno-miR-411-5p | 1.203803 | Up | Rattus norvegicus | 21 | UAGUAGACCGUAUAGCGUACG |
| rno-miR-200b-5p | 1.201743 | Up | Rattus norvegicus | 22 | CAUCUUACUGGGCAGCAUUGGA |
| rno-miR-511-3p | 1.200015 | Up | Rattus norvegicus | 21 | AAUGUGUAGCAAAAGACAGGA |
| rno-miR-380-3p | 1.17496 | Up | Rattus norvegicus | 21 | UAUGUAGUAUGGUCCACAUCU |
| rno-miR-137-5p | 1.142088 | Up | Rattus norvegicus | 22 | ACGGGUAUUCUUGGGUGGAUAA |
| rno-miR-292-3p | 1.134055 | Up | Rattus norvegicus | 23 | AAGUGCCGCCAGGUUUUGAGUGU |
| rno-miR-381-5p | 1.117327 | Up | Rattus norvegicus | 22 | AGCGAGGUUGCCCUUUGUAUAU |
| rno-miR-873-3p | 1.064982 | Up | Rattus norvegicus | 21 | GAGACUGACAAGUUCCCGGGA |
| rno-miR-10b-3p | 1.063514 | Up | Rattus norvegicus | 21 | ACAGAUUCGAUUCUAGGGGAA |
| rno-miR-434-5p | 1.02408 | Up | Rattus norvegicus | 23 | AGCUCGACUCAUGGUUUGAACCA |
| rno-miR-205 | 1.015991 | Up | Rattus norvegicus | 23 | UCCUUCAUUCCACCGGAGUCUGU |
| rno-miR-154-5p | 1.003104 | Up | Rattus norvegicus | 22 | UAGGUUAUCCGUGUUGCCUUCG |
| rno-miR-92a-1-5p | −1.138072 | Down | Rattus norvegicus | 23 | AGGUUGGGAUUUGUCGCAAUGCU |
| rno-miR-449a-5p | −1.414279 | Down | Rattus norvegicus | 22 | UGGCAGUGUAUUGUUAGCUGGU |
| rno-miR-363-3p | −1.172769 | Down | Rattus norvegicus | 21 | AAUUGCACGGUAUCCAUCUGU |
| rno-miR-449c-3p | −1.0767871 | Down | Rattus norvegicus | 17 | UAGAACUUCGUCCCAAC |
| rno-miR-3074 | −1.463293 | Down | Rattus norvegicus | 22 | GAUAUCAGCUCAGUAGGCACCG |
miR/miRNA, microRNA.
Figure 3.
Significant differentially expressed miRNAs. (A) Top 10 upregulated miRNAs and 5 downregulated miRNAs. (B and C) Heat map of the 15 significant differentially expressed miRNAs. miR/miRNA, microRNA; OPC, oligodendrocyte precursor cell.
GO analysis of the target genes of altered miRNAs
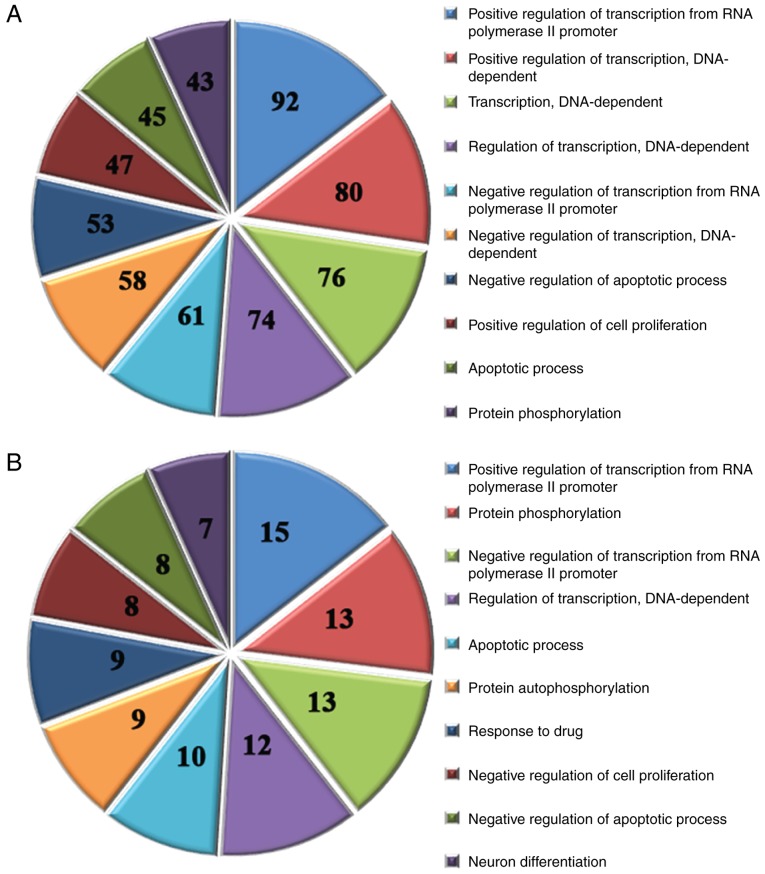
It has previously been reported that miRNAs regulate gene expression by controlling protein translation or destabilizing mRNA transcripts (29,43). Therefore, using prediction databases (miRecords, TargetScan, MicroCosm, Miranda and Pictar), the target genes of all of the altered miRNAs were screened. Subsequently, the main functional categories associated with the target genes were identified by GO analysis. The results identified 578 significantly enriched GO terms by analyzing the target genes of the 40 upregulated miRNAs (P<0.05); the top 10 GO terms are presented in Fig. 4A. In addition, 185 significantly enriched GO terms were identified by analyzing the target genes of the 5 downregulated miRNAs (P<0.05); the top 10 GO terms are shown in Fig. 4B.
Figure 4.
GO analysis of the target genes of differentially expressed miRNAs. (A) Top 10 significantly enriched GO terms associated with the target genes of the 40 upregulated miRNAs. (B) Top 10 significantly enriched GO terms associated with the target genes of the 5 downregulated miRNAs. Numbers indicate the number of genes enriched in that GO term. GO, Gene Ontology; miRNA, microRNA.
KEGG pathways associated with the target genes of altered miRNAs
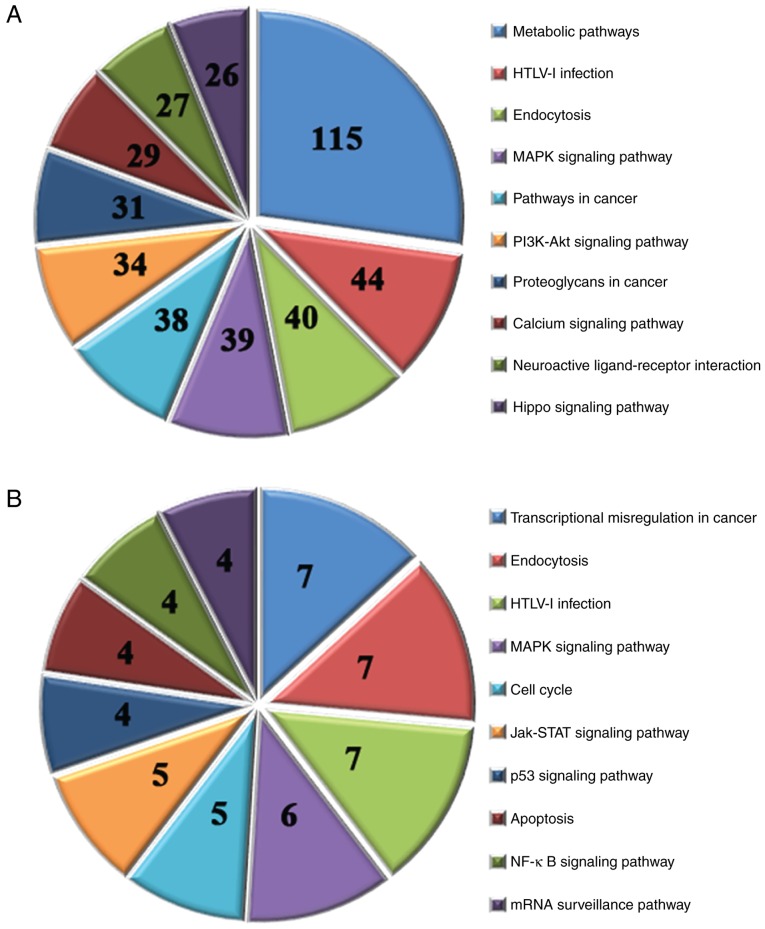
A KEGG pathway analysis was performed for the target genes of all of the differentially expressed miRNAs. The results identified 107 associated pathways by analyzing the target genes of the 40 upregulated miRNAs (P<0.05); the top 10 pathways are presented in Fig. 5A. In addition, 14 associated pathways were identified by analyzing the target genes of the 5 downregulated miRNAs (P<0.05); the top 10 pathways are presented in Fig. 5B.
Figure 5.
Kyoto Encyclopedia of Genes and Genomes pathway analysis of the target genes of differentially expressed miRNAs. (A) Top 10 significant pathways of the target genes of the 40 upregulated miRNAs. (B) Top 10 significant pathways of the target genes of the 5 downregulated miRNAs. Numbers indicate the number of genes enriched in that pathway. miRNA, microRNA.
qPCR verification of OPC transplantation-responsive spinal cord miRNAs from rats subjected to SCC
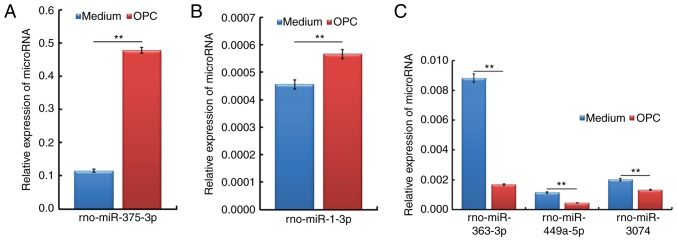
In order to validate the expression profiles generated from the miRNA assay, a qPCR analysis was performed to identify the association between the two datasets. As presented in Fig. 6, the most highly upregulated (miR-375-3p and miR-1-3p) and downregulated (miR-363-3p, miR-449a-5p and miR-3074) spinal cord miRNAs identified in the miRNA assay were further verified, and showed consistent alterations, in a qPCR analysis.
Figure 6.
Verification of miRNA expression profiles in the spinal cord of rats in the OPC group compared with in the Medium group. The relative expression levels of the most significantly altered miRNAs were examined using quantitative polymerase chain reaction in the spinal cord of rats from the Medium and OPC groups. (A and B) Relative expression levels of rno-miR-375-3p and rno-miR-1-3p, which were revealed to be upregulated in a miRNA assay. **P<0.01. (C) Relative expression levels of rno-miR-363-3p, rno-miR-449a-5p and rno-miR-3074, which were revealed to be downregulated in a miRNA assay. miR/miRNA, microRNA; OPC, oligodendrocyte precursor cell.
Bioinformatics analysis of the important and verified miRNAs
The results of the qPCR analysis demonstrated that miR-375 and miR-1 were upregulated in the OPC group compared with in the Medium group, and a bioinformatics analysis indicated that they targeted similar types of genes; therefore, miR-375 and miR-1 were grouped together for functional analysis. The results suggested that miR-375 and miR-1 may act primarily to inhibit cell proliferation and apoptosis through the regulation of transcription and translation. Furthermore, due to their association within the same annotation cluster (such as cell proliferation), it is possible that the primary method by which these two miRNAs regulate cell proliferation is via the inhibition of GO terms, including the mitogen-activated protein kinases signaling pathway (Table IV). miR-363, miR-449a and miR-3074 were downregulated in the OPC group compared with in the Medium group, as confirmed by qPCR, and a bioinformatics analysis demonstrated that they targeted similar types of genes; therefore, miR-363, miR-449a and miR-3074 were grouped together for functional analysis. The results indicated that miR-363, miR-449a and miR-3074 may primarily inhibit cell proliferation and neuronal differentiation through the regulation of transcription (Table IV).
Table IV.
Statistical annotation of miRNA function.
| GO ID/pathway function term | Gene count | P-value |
|---|---|---|
| miR-375 and miR-1 common targets | ||
| GO:0045893-positive regulation of transcription, DNA-dependent | 2 | 1.96792×10−24 |
| GO:0045944-positive regulation of transcription from RNA polymerase II promoter | 2 | 9.15325×10−21 |
| GO:0006351-transcription, DNA-dependent | 1 | 2.48966×10−15 |
| GO:0000122-negative regulation of transcription from RNA polymerase II promoter | 1 | 5.12873×10−13 |
| GO:0006355-regulation of transcription, DNA-dependent | 1 | 5.9179×10−13 |
| GO:0006417-regulation of translation | 1 | 0.000151831 |
| GO:0006486-protein glycosylation | 1 | 1.21081×10−10 |
| GO:0006915-apoptotic process | 3 | 3.19378×10−10 |
| GO:0043066-negative regulation of apoptotic process | 3 | 7.94504×10−10 |
| GO:0043065-positive regulation of apoptotic process | 1 | 2.34005×10−6 |
| GO:0051402-neuron apoptotic process | 1 | 1.43174×10−5 |
| GO:0043524-negative regulation of neuron apoptotic process | 2 | 7.17984×10−6 |
| GO:0008283-cell proliferation | 3 | 0.001320952 |
| GO:0008285-negative regulation of cell proliferation | 1 | 3.71268×10−10 |
| GO:0008284-positive regulation of cell proliferation | 4 | 4.37506×10−10 |
| GO:0042552-myelination | 2 | 0.0003695 |
| GO:0035556-intracellular signal transduction | 3 | 1.05697×10−10 |
| GO:0000165-MAPK cascade | 1 | 0.000318552 |
| Path_id:1100-Metabolic pathways | 4 | 7.89723×10−14 |
| Path_id:4144-Endocytosis | 1 | 2.5492×10−13 |
| Path_id:4010-MAPK signaling pathway | 1 | 4.94224×10−11 |
| Path_id:4020-Calcium signaling pathway | 2 | 1.24247×10−8 |
| Path_id:4070-Phosphatidylinositol signaling system | 1 | 1.86855×10−7 |
| Path_id:4310-Wnt signaling pathway | 1 | 2.71935×10−7 |
| Path_id:4151-PI3K-Akt signaling pathway | 5 | 1.92934×10−5 |
| Path_id:3015-mRNA surveillance pathway | 1 | 0.000876288 |
| Path_id:4064-NF-κB signaling pathway | 2 | 0.008087159 |
| Path_id:4514-Cell adhesion molecules (CAMs) | 1 | 0.048139487 |
| miR-363, miR-449a and miR-3074 common targets | ||
| GO:0000122-negative regulation of transcription from RNA polymerase II promoter | 2 | 9.32468×10−5 |
| GO:0045944-positive regulation of transcription from RNA polymerase II promoter | 3 | 0.000267853 |
| GO:0006355-regulation of transcription, DNA-dependent | 1 | 0.006384786 |
| GO:0006468-protein phosphorylation | 1 | 2.21252×10−6 |
| GO:0015031-protein transport | 1 | 0.032924475 |
| GO:0030182-neuron differentiation | 2 | 6.76381×10−5 |
| GO:0008285-negative regulation of cell proliferation | 1 | 0.003549981 |
| GO:0007264-small GTPase mediated signal transduction | 1 | 0.014162188 |
| GO:0007165-signal transduction | 2 | 0.044607167 |
| Path_id:4630-Jak-STAT signaling pathway | 2 | 0.009890945 |
GO, Gene Ontology; Jak, Janus kinase; MAPK, mitogen-activated protein kinase; miR/miRNA, microRNA; NF-κB, nuclear factor-κB; PI3K, phosphoinositide 3-kinase; STAT, signal transducer and activator of transcription.
Discussion
Human SCC can cause severe and long-term disability (3). At present, there is no effective clinical treatment for functional recovery following SCI. However, it has been reported that OPC transplantation may have potential for the treatment of SCI (44,45), and abnormal miRNA expression may contribute to the pathogenesis of SCI (37). However, to the best of our knowledge, there are no reports regarding the expression of miRNAs in the spinal cord of rats following SCC and OPC transplantation. The present study aimed to investigate the effects of OPC transplantation on rats with SCC; initially, BBB score and paw withdrawal threshold were tested following OPC transplantation into the spinal cord of rats with SCC. Subsequently, in order to detect the differentially expressed miRNAs in the spinal cord of rats in the OPC group compared with in the Medium group, a miRNA assay was conducted. Finally, qPCR was performed to verify the significantly altered miRNAs.
Firstly, following identification of OPCs, the cells were transplanted into rats that were subjected to SCC for 7 days. The BBB score demonstrated that OPC transplantation was able to improve the motor recovery of rats with SCC. A previous study demonstrated that OPC transplantation improved forelimb motor function following cervical SCI (25). In addition, animals that were subjected to SCC and hESC-derived OPCs transplantation at 7 days performed significantly better on the BBB locomotor test compared with those transplanted with human fibroblasts or DMEM (22). Furthermore, CNTF-OPC-grafted animals exhibited significantly increased BBB locomotor scores compared with those receiving DMEM, enhanced green fluorescent protein (EGFP)-fibroblasts or CNTF-fibroblasts at 3–7 weeks following injury; the BBB scores of the EGFP-OPC-grafted animals were also significantly higher than the other three groups at 5–7 weeks following injury (26). In the present study, paw withdrawal testing indicated that OPC transplantation may relieve SCC-induced mechanical allodynia. It has previously been reported that transplanted hESC-derived OPCs promoted repair of the sensory pathways of rats with SCC (46). The results of the present study were concordant with those of the previous aforementioned studies.
The results of a miRNA assay detected 40 upregulated miRNAs and 5 downregulated miRNAs in the spinal cord of rats from the OPC group compared with in the Medium group, according to the following criteria: Fold change >2 and P<0.05. Bioinformatics analysis indicated that positive regulation of transcription from RNA polymerase II promoter was the most significantly enriched GO term, and metabolic pathways was the most significantly enriched KEGG pathway, according to the number of target genes of differentially expressed miRNAs enriched in them. A previous study detected 97 differentially expressed miRNAs in the adult rat spinal cord following traumatic SCI through miRNA analysis, and the potential targets for these SCI-altered miRNAs included genes associated with inflammation, oxidation and apoptosis, which are known to serve important roles in the pathogenesis of SCI (37). Furthermore, microarray comparisons of the injury sites of spinal cords from contused and sham rats, harvested 4 and 14 days following SCC, demonstrated that 32 miRNAs, including miR-124, miR-129, and miR-1, were significantly downregulated. In addition, a bioinformatics analysis of validated miRNA-targeted genes indicated that miRNA dysregulation may explain the apoptotic susceptibility and aberrant cell cycle associated with a loss of neuronal identity, which underlies the pathogenesis of secondary SCC (38). Furthermore, microarray data has previously revealed that the induction of a specific miRNA expression pattern following moderate SCC is characterized by a marked increase in the number of downregulated miRNAs, particularly at 7 days after injury, and a bioinformatics analysis indicated that alterations in miRNA expression may affect key processes in SCI physiopathology, including inflammation and apoptosis (31). To the best of our knowledge, the present study is the first to report that dysregulated miRNA expression is present in the spinal cord of SCC rats transplanted with OPCs compared with in SCC rats treated with medium.
The present study also demonstrated that the most highly upregulated (miR-375-3p and miR-1-3p) and downregulated (miR-363-3p, miR-449a-5p and miR-3074) spinal cord miRNAs identified in the miRNA assay were consistently altered in a qPCR analysis. Furthermore, functional analysis demonstrated that miR-375 and miR-1 may primarily act to inhibit cell proliferation and apoptosis through the regulation of transcription and translation, whereas miR-363, miR-449a and miR-3074 may primarily act to inhibit cell proliferation and neuronal differentiation through the regulation of transcription. A previous study demonstrated that overexpression of miR-375 significantly inhibited gastric cancer cell proliferation by targeting Janus kinase 2 in vitro and in vivo (47). In addition, by regulating the expression of B-cell lymphoma (Bcl)-2, Bcl-2-associated X protein and miR-375 in vitro and in vivo, formononetin was able to induce apoptosis of U2OS human osteosarcoma cells (48). miR-375 could also affect the proliferation and apoptosis of human papilloma virus 16-positive human cervical cancer cells by targeting insulin-like growth factor 1 receptor (49). In addition, it has been reported that miR-1 may control cell proliferation and cell cycle in human osteosarcoma through modulation of MET protein expression (50). miR-1 may also inhibit gastric cancer cell proliferation by targeting MET (51). Furthermore, miR-1 has been reported to regulate heat shock protein 60 expression, thus contributing to glucose-mediated apoptosis in cardiomyocytes (52). Overexpression of miR-1 may also induce apoptosis in mesothelioma cell lines (53). miR-1 has also been revealed to be involved in the protective effects of hydrogen sulfide against cardiomyocyte apoptosis via ischemia/reperfusion (54). Introduction of miR-363 mimics has been reported to result in inhibition of cell proliferation in renal cell carcinoma (55). Conversely, miR-363 may promote proliferation of human gastric cancer cells via targeting FBW7 ubiquitin ligase expression (56). With regards to miR-449a, it has been reported to regulate proliferation in response to cisplatin by targeting cyclin D1 and Bcl-2 in SGC7901 cells (57). In addition, overexpression of miR-449a significantly inhibits the proliferation of HEC-1B endometrial cancer cells (58). miR-449a acts as a tumor suppressor by reducing the proliferation of human glioblastoma cell lines and glioblastoma stem cells (59). miR-449a may also inhibit liver cancer cell proliferation by suppressing POU class 2 homeobox 1 and calpain 6 (60). miRNAs have various target genes and serve roles in several diseases by regulating the associated target genes, thus, some miRNAs have exhibited opposing effects in different studies. OPC transplantation may alter miRNA expression following SCC in rats, and may be useful for treating SCC. To the best of our knowledge, the present study is the first to identify OPC transplantation-responsive spinal cord miRNAs in rats subjected to SCC, which may suggest potential functional roles for these miRNAs in transplantation of OPCs in rats with SCC.
In conclusion, the present study demonstrated that OPC transplantation could improve motor recovery and relieve mechanical allodynia of rats with SCC. After verifying the results of a miRNA assay, the most highly upregulated (miR-375-3p and miR-1-3p) and downregulated (miR-363-3p, miR-449a-5p and miR-3074) spinal cord miRNAs were detected in SCC rats transplanted with OPCs. These results suggested that OPC transplantation may promote functional recovery of rats with SCC, which may be associated with various miRNAs in the spinal cord, particularly miR-375-3p, miR-1-3p, miR-363-3p, miR-449a-5p and miR-3074.
Acknowledgements
The authors of the present study would like to thank Professor Qing-Jie Xia (West China Hospital, Sichuan University, China) for suggestions on the paper and Professor Su Liu (Johns Hopkins University, Baltimore, MD, USA) for providing the viruses.
References
- 1.Hulsebosch CE. Recent advances in pathophysiology and treatment of spinal cord injury. Adv Physiol Educ. 2002;26:238–255. doi: 10.1152/advan.00039.2002. [DOI] [PubMed] [Google Scholar]
- 2.Penas C, Verdu E, Asensio-Pinilla E, Guzmán-Lenis MS, Herrando-Grabulosa M, Navarro X, Casas C. Valproate reduces CHOP levels and preserves oligodendrocytes and axons after spinal cord injury. Neuroscience. 2011;178:33–44. doi: 10.1016/j.neuroscience.2011.01.012. [DOI] [PubMed] [Google Scholar]
- 3.Beaumont E, Whitaker CM, Burke DA, Hetman M, Onifer SM. Effects of rolipram on adult rat oligodendrocytes and functional recovery after contusive cervical spinal cord injury. Neuroscience. 2009;163:985–990. doi: 10.1016/j.neuroscience.2009.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee BB, Cripps RA, Fitzharris M, Wing PC. The global map for traumatic spinal cord injury epidemiology: Update 2011, global incidence rate. Spinal Cord. 2014;52:110–116. doi: 10.1038/sc.2012.158. [DOI] [PubMed] [Google Scholar]
- 5.Emery E, Aldana P, Bunge MB, Puckett W, Srinivasan A, Keane RW, Bethea J, Levi AD. Apoptosis after traumatic human spinal cord injury. J Neurosurg. 1998;89:911–920. doi: 10.3171/jns.1998.89.6.0911. [DOI] [PubMed] [Google Scholar]
- 6.Bunge RP, Puckett WR, Becerra JL, Marcillo A, Quencer RM. Observations on the pathology of human spinal cord injury. A review and classification of 22 new cases with details from a case of chronic cord compression with extensive focal demyelination. Adv Neurol. 1993;59:75–89. [PubMed] [Google Scholar]
- 7.Kakulas BA. A review of the neuropathology of human spinal cord injury with emphasis on special features. J Spinal Cord Med. 1999;22:119–124. doi: 10.1080/10790268.1999.11719557. [DOI] [PubMed] [Google Scholar]
- 8.Buss A, Brook GA, Kakulas B, Martin D, Franzen R, Schoenen J, Noth J, Schmitt AB. Gradual loss of myelin and formation of an astrocytic scar during Wallerian degeneration in the human spinal cord. Brain. 2004;127:34–44. doi: 10.1093/brain/awh001. [DOI] [PubMed] [Google Scholar]
- 9.Guest JD, Hiester ED, Bunge RP. Demyelination and Schwann cell responses adjacent to injury epicenter cavities following chronic human spinal cord injury. Exp Neurol. 2005;192:384–393. doi: 10.1016/j.expneurol.2004.11.033. [DOI] [PubMed] [Google Scholar]
- 10.Crowe MJ, Bresnahan JC, Shuman SL, Masters JN, Beattie MS. Apoptosis and delayed degeneration after spinal cord injury in rats and monkeys. Nat Med. 1997;3:73–76. doi: 10.1038/nm0197-73. [DOI] [PubMed] [Google Scholar]
- 11.Li GL, Farooque M, Holtz A, Olsson Y. Apoptosis of oligodendrocytes occurs for long distances away from the primary injury after compression trauma to rat spinal cord. Acta Neuropathol. 1999;98:473–480. doi: 10.1007/s004010051112. [DOI] [PubMed] [Google Scholar]
- 12.Shuman SL, Bresnahan JC, Beattie MS. Apoptosis of microglia and oligodendrocytes after spinal cord contusion in rats. J Neurosci Res. 1997;50:798–808. doi: 10.1002/(SICI)1097-4547(19971201)50:5<798::AID-JNR16>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 13.McTigue DM, Wei P, Stokes BT. Proliferation of NG2-positive cells and altered oligodendrocyte numbers in the contused rat spinal cord. J Neurosci. 2001;21:3392–3400. doi: 10.1523/JNEUROSCI.21-10-03392.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McEwen ML, Springer JE. A mapping study of caspase-3 activation following acute spinal cord contusion in rats. J Histochem Cytochem. 2005;53:809–819. doi: 10.1369/jhc.4A6467.2005. [DOI] [PubMed] [Google Scholar]
- 15.Casha S, Yu WR, Fehlings MG. Oligodendroglial apoptosis occurs along degenerating axons and is associated with FAS and p75 expression following spinal cord injury in the rat. Neuroscience. 2001;103:203–218. doi: 10.1016/S0306-4522(00)00538-8. [DOI] [PubMed] [Google Scholar]
- 16.Grossman SD, Rosenberg LJ, Wrathall JR. Temporal-spatial pattern of acute neuronal and glial loss after spinal cord contusion. Exp Neurol. 2001;168:273–282. doi: 10.1006/exnr.2001.7628. [DOI] [PubMed] [Google Scholar]
- 17.Hernández J, Torres-Espin A, Navarro X. Adult stem cell transplants for spinal cord injury repair: Current state in preclinical research. Curr Stem Cell Res Ther. 2011;6:273–287. doi: 10.2174/157488811796575323. [DOI] [PubMed] [Google Scholar]
- 18.Grabel L. Prospects for pluripotent stem cell therapies: Into the clinic and back to the bench. J Cell Biochem. 2012;113:381–387. doi: 10.1002/jcb.23364. [DOI] [PubMed] [Google Scholar]
- 19.Führmann T, Tam RY, Ballarin B, Coles B, Donaghue Elliott I, van der Kooy D, Nagy A, Tator CH, Morshead CM, Shoichet MS. Injectable hydrogel promotes early survival of induced pluripotent stem cell-derived oligodendrocytes and attenuates longterm teratoma formation in a spinal cord injury model. Biomaterials. 2016;83:23–36. doi: 10.1016/j.biomaterials.2015.12.032. [DOI] [PubMed] [Google Scholar]
- 20.McKerracher L, Winton MJ. Nogo on the go. Neuron. 2002;36:345–348. doi: 10.1016/S0896-6273(02)01018-8. [DOI] [PubMed] [Google Scholar]
- 21.Domeniconi M, Filbin MT. Overcoming inhibitors in myelin to promote axonal regeneration. J Neurol Sci. 2005;233:43–47. doi: 10.1016/j.jns.2005.03.023. [DOI] [PubMed] [Google Scholar]
- 22.Faulkner J, Keirstead HS. Human embryonic stem cell-derived oligodendrocyte progenitors for the treatment of spinal cord injury. Transpl Immunol. 2005;15:131–142. doi: 10.1016/j.trim.2005.09.007. [DOI] [PubMed] [Google Scholar]
- 23.Kuypers NJ, Bankston AN, Howard RM, Beare JE, Whittemore SR. Remyelinating oligodendrocyte precursor cell miRNAs from the Sfmbt2 cluster promote cell cycle arrest and differentiation. J Neurosci. 2016;36:1698–1710. doi: 10.1523/JNEUROSCI.1240-15.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang HL, Jiang ZS, Wang FW. Analysis of gene expression profiles associated with functional recovery after spinal cord injury caused by sema4D knockdown in oligodendrocytes. Cell Biochem Biophys. 2014;69:655–661. doi: 10.1007/s12013-014-9848-0. [DOI] [PubMed] [Google Scholar]
- 25.Sharp J, Frame J, Siegenthaler M, Nistor G, Keirstead HS. Human embryonic stem cell-derived oligodendrocyte progenitor cell transplants improve recovery after cervical spinal cord injury. Stem Cells. 2010;28:152–163. doi: 10.1002/stem.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cao Q, He Q, Wang Y, Cheng X, Howard RM, Zhang Y, DeVries WH, Shields CB, Magnuson DS, Xu XM, et al. Transplantation of ciliary neurotrophic factor-expressing adult oligodendrocyte precursor cells promotes remyelination and functional recovery after spinal cord injury. J Neurosci. 2010;30:2989–3001. doi: 10.1523/JNEUROSCI.3174-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bartel DP. MicroRNAs: Target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 29.Zamore PD, Haley B. Ribo-gnome: The big world of small RNAs. Science. 2005;309:1519–1524. doi: 10.1126/science.1111444. [DOI] [PubMed] [Google Scholar]
- 30.Liu Z, Sall A, Yang D. MicroRNA: An emerging therapeutic target and intervention tool. Int J Mol Sci. 2008;9:978–999. doi: 10.3390/ijms9060978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yunta M, Nieto-Diaz M, Esteban FJ, Caballero-López M, Navarro-Ruiz R, Reigada D, Pita-Thomas DW, del Águila A, Muñoz-Galdeano T, Maza RM. MicroRNA dysregulation in the spinal cord following traumatic injury. PLoS One. 2012;7:e34534. doi: 10.1371/journal.pone.0034534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 33.Krichevsky AM. MicroRNA profiling: From dark matter to white matter, or identifying new players in neurobiology. ScientificWorldJournal. 2007;7:155–166. doi: 10.1100/tsw.2007.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kosik KS. The neuronal microRNA system. Nat Rev Neurosci. 2006;7:911–920. doi: 10.1038/nrn2037. [DOI] [PubMed] [Google Scholar]
- 35.Bak M, Silahtaroglu A, Møller M, Christensen M, Rath MF, Skryabin B, Tommerup N, Kauppinen S. MicroRNA expression in the adult mouse central nervous system. RNA. 2008;14:432–444. doi: 10.1261/rna.783108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miska EA, Alvarez-Saavedra E, Townsend M, Yoshii A, Sestan N, Rakic P, Constantine-Paton M, Horvitz HR. Microarray analysis of microRNA expression in the developing mammalian brain. Genome Biol. 2004;5:R68. doi: 10.1186/gb-2004-5-9-r68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu NK, Wang XF, Lu QB, Xu XM. Altered microRNA expression following traumatic spinal cord injury. Exp Neurol. 2009;219:424–429. doi: 10.1016/j.expneurol.2009.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Strickland ER, Hook MA, Balaraman S, Huie JR, Grau JW, Miranda RC. MicroRNA dysregulation following spinal cord contusion: Implications for neural plasticity and repair. Neuroscience. 2011;186:146–160. doi: 10.1016/j.neuroscience.2011.03.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lü HZ, Wang YX, Li Y, Fu SL, Hang Q, Lu PH. Proliferation and differentiation of oligodendrocyte progenitor cells induced from rat embryonic neural precursor cells followed by flow cytometry. Cytometry A. 2008;73:754–760. doi: 10.1002/cyto.a.20577. [DOI] [PubMed] [Google Scholar]
- 40.Basso DM, Beattie MS, Bresnahan JC. A sensitive and reliable locomotor rating scale for open field testing in rats. J Neurotrauma. 1995;12:1–21. doi: 10.1089/neu.1995.12.1. [DOI] [PubMed] [Google Scholar]
- 41.Liu R, Zhao W, Zhao Q, Liu SJ, Liu J, He M, Xu Y, Wang W, Liu W, Xia QJ, et al. Endoplasmic reticulum protein 29 protects cortical neurons from apoptosis and promoting corticospinal tract regeneration to improve neural behavior via caspase and Erk signal in rats with spinal cord transection. Mol Neurobiol. 2014;50:1035–1048. doi: 10.1007/s12035-014-8681-1. [DOI] [PubMed] [Google Scholar]
- 42.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 43.Alvarez-Garcia I, Miska EA. MicroRNA functions in animal development and human disease. Development. 2005;132:4653–4662. doi: 10.1242/dev.02073. [DOI] [PubMed] [Google Scholar]
- 44.Ozdemir M, Attar A, Kuzu I. Regenerative treatment in spinal cord injury. Curr Stem Cell Res Ther. 2012;7:364–369. doi: 10.2174/157488812802481481. [DOI] [PubMed] [Google Scholar]
- 45.Zhang HL, Wang J, Tang L. Sema4D knockdown in oligodendrocytes promotes functional recovery after spinal cord injury. Cell Biochem Biophys. 2014;68:489–496. doi: 10.1007/s12013-013-9727-0. [DOI] [PubMed] [Google Scholar]
- 46.All AH, Bazley FA, Gupta S, Pashai N, Hu C, Pourmorteza A, Kerr C. Human embryonic stem cell-derived oligodendrocyte progenitors aid in functional recovery of sensory pathways following contusive spinal cord injury. PLoS One. 2012;7:e47645. doi: 10.1371/journal.pone.0047645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ding L, Xu Y, Zhang W, Deng Y, Si M, Du Y, Yao H, Liu X, Ke Y, Si J, Zhou T. MiR-375 frequently downregulated in gastric cancer inhibits cell proliferation by targeting JAK2. Cell Res. 2010;20:784–793. doi: 10.1038/cr.2010.79. [DOI] [PubMed] [Google Scholar]
- 48.Hu W, Xiao Z. Formononetin induces apoptosis of human osteosarcoma cell line U2OS by regulating the expression of Bcl-2, Bax and MiR-375 in vitro and in vivo. Cell Physiol Biochem. 2015;37:933–939. doi: 10.1159/000430220. [DOI] [PubMed] [Google Scholar]
- 49.Yu X, Zhao W, Yang X, Wang Z, Hao M. miR-375 affects the proliferation, invasion, and apoptosis of HPV16-positive human cervical cancer cells by targeting IGF-1R. Int J Gynecol Cancer. 2016;26:851–858. doi: 10.1097/IGC.0000000000000711. [DOI] [PubMed] [Google Scholar]
- 50.Novello C, Pazzaglia L, Cingolani C, Conti A, Quattrini I, Manara MC, Tognon M, Picci P, Benassi MS. miRNA expression profile in human osteosarcoma: Role of miR-1 and miR-133b in proliferation and cell cycle control. Int J Oncol. 2013;42:667–675. doi: 10.3892/ijo.2012.1717. [DOI] [PubMed] [Google Scholar]
- 51.Han C, Zhou Y, An Q, Li F, Li D, Zhang X, Yu Z, Zheng L, Duan Z, Kan Q. MicroRNA-1 (miR-1) inhibits gastric cancer cell proliferation and migration by targeting MET. Tumour Biol. 2015;36:6715–6723. doi: 10.1007/s13277-015-3358-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shan ZX, Lin QX, Deng CY, Zhu JN, Mai LP, Liu JL, Fu YH, Liu XY, Li YX, Zhang YY, et al. miR-1/miR-206 regulate Hsp60 expression contributing to glucose-mediated apoptosis in cardiomyocytes. FEBS Lett. 2010;584:3592–3600. doi: 10.1016/j.febslet.2010.07.027. [DOI] [PubMed] [Google Scholar]
- 53.Xu Y, Zheng M, Merritt RE, Shrager JB, Wakelee H, Kratzke RA, Hoang CD. miR-1 induces growth arrest and apoptosis in malignant mesothelioma. Chest. 2013;144:1632–1643. doi: 10.1378/chest.12-2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kang B, Hong J, Xiao J, Zhu X, Ni X, Zhang Y, He B, Wang Z. Involvement of miR-1 in the protective effect of hydrogen sulfide against cardiomyocyte apoptosis induced by ischemia/reperfusion. Mol Biol Rep. 2014;41:6845–6853. doi: 10.1007/s11033-014-3570-2. [DOI] [PubMed] [Google Scholar]
- 55.Li Y, Chen D, Li Y, Jin L, Liu J, Su Z, Qi Z, Shi M, Jiang Z, Ni L, et al. Oncogenic cAMP responsive element binding protein 1 is overexpressed upon loss of tumor suppressive miR-10b-5p and miR-363-3p in renal cancer. Oncol Rep. 2016;35:1967–1978. doi: 10.3892/or.2016.4579. [DOI] [PubMed] [Google Scholar]
- 56.Zhang PF, Sheng LL, Wang G, Tian M, Zhu LY, Zhang R, Zhang J, Zhu JS. miR-363 promotes proliferation and chemo-resistance of human gastric cancer via targeting of FBW7 ubiquitin ligase expression. Oncotarget. 2016;7:35284–35292. doi: 10.18632/oncotarget.9169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hu J, Fang Y, Cao Y, Qin R, Chen Q. miR-449a Regulates proliferation and chemosensitivity to cisplatin by targeting cyclin D1 and BCL2 in SGC7901 cells. Dig Dis Sci. 2014;59:336–345. doi: 10.1007/s10620-013-2923-3. [DOI] [PubMed] [Google Scholar]
- 58.Ye W, Xue J, Zhang Q, Li F, Zhang W, Chen H, Huang Y, Zheng F. MiR-449a functions as a tumor suppressor in endometrial cancer by targeting CDC25A. Oncol Rep. 2014;32:1193–1199. doi: 10.3892/or.2014.3303. [DOI] [PubMed] [Google Scholar]
- 59.Yao Y, Ma J, Xue Y, Wang P, Li Z, Li Z, Hu Y, Shang X, Liu Y. MiR-449a exerts tumor-suppressive functions in human glioblastoma by targeting Myc-associated zinc-finger protein. Mol Oncol. 2015;9:640–656. doi: 10.1016/j.molonc.2014.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liu Y, Wang Y, Sun X, Mei C, Wang L, Li Z, Zha X. miR-449a promotes liver cancer cell apoptosis by downregulation of Calpain 6 and POU2F1. Oncotarget. 2016;7:13491–13501. doi: 10.18632/oncotarget.4821. [DOI] [PMC free article] [PubMed] [Google Scholar]