Abstract
Given the emerging role of microRNAs (miRs) in cancer progression, the present study investigated the role and underlying mechanism of miR-103 in colorectal cancer (CRC). Reverse transcription-quantitative polymerase chain reaction was conducted to quantify the expression levels of miR-103 in clinical specimens and cell lines. The role of miR-103 in CRC was examined using MTT, colony formation and transwell assays. In addition, a luciferase reporter assay was used to confirm an associated between the 3′ untranslated region of zonula occuldens-1 (ZO-1) and miR-103. The results demonstrated that miR-103 was upregulated in CRC. Overexpression of miR-103 promoted CRC cell proliferation and migration in vitro, whereas downregulation of miR-103 inhibited cell proliferation and migration. ZO-1 was identified as a direct target of miR-103, revealing its expression to be inversely correlated with miR-103 expression in CRC samples. In conclusion, the present study revealed that miR-103 has strong tumor-promoting effects via of targeting ZO-1 in CRC and has potential development of miRNA-based targeted approaches for the treatment of CRC.
Keywords: colorectal cancer, microRNA-103, zonula occuldens-1
Introduction
Colorectal cancer (CRC) is one of the most prevalent cancers worldwide, and the fifth leading cause of cancer-associated mortality in China (1). The primary cause of death in patients with CRC is tumor cell invasion and metastasis. Despite efforts to prevent CRC, its incidence is still increasing (2). Thus, elucidating the molecular mechanisms underlying the development of CRC may contribute to the development of therapies against CRC.
MicroRNAs (miRNAs or miRs) are a class of small non-coding RNAs that repress protein translation through complimentary base pairing with the 3′-untranslated region (3′-UTR) of mRNAs, resulting in either mRNA degradation or inhibition of their post-transcriptional translation (3). Emerging evidence has suggested that the dysregulation of miRNAs is involved in a variety of diseases, including human cancers (4–7). Dysfunction of miRNA is associated with CRC tumorigenesis and progression (4). Thus, investigating tumor-specific miRNAs and their targets is critical in understanding their role in cancer tumorigenesis and to identify new molecular markers for the diagnosis and treatment of CRC.
miR-103 is a member of the miR-103/107 family, located on human chromosome 5 (8). A previous study has demonstrated that miR-103 expression is significantly lower in patients with heart failure compared with healthy volunteers and may therefore be used as a diagnostic predictor (9). Previous reports revealed that miR-103 serves a role in the regulation of endometrial cancer cells, in post-transcriptionally reducing the expression of the tumor suppressor, tissue inhibitor of metalloproteinase 3 (TIMP3), and stimulating growth and invasion (10). miR-103 demonstrates an inverse association with DNA-binding protein inhibitor ID-2, a repressor of nervous system cancers, during neuroblastoma cell differentiation induced by retinoic acid (11). A previous study revealed that miR-103 may be a potential minimally-invasive biomarker for the diagnosis of mesothelioma (12). In CRC, high expression levels of miR-103 are associated with metastatic potential of CRC cell lines and predict poor prognosis (13). In addition, certain reports indicate that miR-103/107 promotes CRC metastasis through targeting tumor suppressors death-associated protein kinase-1 (DAPK) and Krueppel-like factor 4 (KLF4) (14).
In the present study, the role of miR-103 in CRC cell proliferation and migration was investigated and the molecular mechanism underlying its effects was partially elucidated. In addition, based on bioinformatic analyses, zonula occuldens-1 (ZO-1) was identified as a potential and functional target for miR-103. Therefore, the present study may provide the first evidence of the regulatory mechanisms of miR-103 and ZO-1 and their roles in CRC carcinogenesis and metastasis, thus providing candidate targets for CRC treatment.
Materials and methods
Tissue samples and cell lines
A total of 26 pairs of human CRC and adjacent non-tumor tissues were collected between 2014 and 2016 at Affiliated Hospital of Nantong University (Jiangsu, China). Tissue samples were immediately snap-frozen in liquid nitrogen. Both tumor tissues and adjacent non-tumor tissues were histologically examined. All human materials were obtained with written informed consent, and the present study was approved by the Institute Research Ethics Committee at the Affiliated Hospital of Nantong University.
Five human CRC cell lines (HCT-116, HCT-8, HT-29, SW480 and SW620) and normal colon cell line FHC were purchased from the Cell Bank of Chinese Academy of Sciences (Shanghai, China). All cell lines were maintained in Dulbecco's modified Eagle's medium (DMEM; Gibco, Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented with 10% fetal bovine serum (FBS). All cells were maintained in a humidified incubator at 37°C and 5% CO2.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted from cells or frozen tissues with TRIzol (Invitrogen; Thermo Fisher Scientific, Inc.) and cDNA was synthesized with a TaqMan MicroRNA Reverse Transcription kit (Applied Biosystems; Thermo Fisher Scientific, Inc.) and PrimeScript RT Master Mix (Takara Biotechnology Co., Ltd., Dalian, China) according to the manufacturer's protocol. The sequences of the primers used for PCR amplification were as follows: ZO-1, 5′-CCCTCAAGGAGCCATTC-3′ (forward) and 5′-CAGTTTGCTCCAACGAGA-3′ (reverse); β-actin, 5′-TAGTTGCGTTACACCCTTTCTTG-3′ (forward) and 5′-GCTGTCACCTTCACCGTTCC-3′ (reverse); miR-103, 5′-AGCAGCATTGTACAGGGCTATGA-3′ (forward) and 5′-TGGTGTCGTGGAGTCG-3′ (reverse); U6, 5′-CTCGCTTCGGCAGCACA-3′ (forward) and 5′-AACGCTTCACGAATTTGCGT-3′ (reverse). qPCR was performed using TaqMan 2X Universal PCR Master Mix (Applied Biosystems; Thermo Fisher Scientific, Inc.) using the following cycling conditions: Preliminary denaturation at 96°C for 2 min, followed by 40 cycles of denaturation at 96°C for 15 sec, annealing at 60°C for 1 min and elongation at 60°C for 1 min. The relative expression levels of each gene were calculated and normalized using the 2−ΔΔCq method relative to U6 or β-actin (15). All the reactions were run in triplicate.
Western blot analysis
Cells and tissue extracts were prepared using CelLytic™ MT mammalian tissue lysis reagent (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) according to the manufacturer's protocol and protein concentrations were detected using a bicinchoninic acid assay. Equal amounts of total protein (20 µg) were separated by 10% SDS-PAGE and transferred to polyvinylidene difluoride membranes. After blocking in 5% non-fat milk for 2 h at room temperature, membranes were incubated with primary antibodies (ZO-1 antibody, cat no. ab59720; Abcam, Cambridge, MA, USA) overnight at 4°C followed by incubation with horse radish peroxidase-conjugated secondary antibodies (1:10,000; cat nos. 31430 and 31460; Pierce; Thermo Fisher Scientific, Inc.) for 2 h at room temperature on a shaker. The bands were visualized using Western Lightning Enhanced Chemiluminescence Pro substrate with horseradish peroxidase and then exposed to medical X-ray films (Kodak, Rochester, NY, USA). The intensities of bands were measured using ImageJ software (National Institutes of Health, Bethesda, MD, USA). β-actin was used as a loading control.
Transfection
miR-103 mimic, miR-103 inhibitor and the corresponding mimic/inhibitor negative control (NC) oligos were purchased from GenePharm, Inc. (Sunnyvale, CA, USA; http://www.genepharma.com/productview.asp?id=7&parentid=15&sortname=miRNA). Cells were transfected with the miR-103 mimic or miR-103 inhibitor, miR-103 mimic or inhibitor NCs using Lipofectamine 2000 reagent (Invitrogen; Thermo Fisher Scientific, Inc.), following the manufacturer's protocol. After 48 h of transfection at 37°C, cells were collected for further analysis.
Bioinformatics analysis
The analysis of predicted miR-103 targets was performed using three well-known microRNA target prediction programs, the algorithms TargetScan (http://targetscan.org/), PicTar (http://pictar.mdc-berlin.de/), and miRanda (http://www.microrna.org/microrna/home.do/) website tools.
Luciferase reporter assay
For the luciferase reporter assay, the pGL3 plasmid encoding a luciferase reporter gene was purchased from Promega Corporation (Madison, WI, USA). Recombinant plasmid of pGL3-ZO-1-3′-UTR (wild-type; wt; UGUUACAUUUUUAAGUGCUGCA or pGL3-ZO-1-3′-UTR-mutant (UGUUACAUUAUAAUGACCACGA) was constructed in-house. SW620 cells (1–2×105 cells/well) were seeded in a 24-well plate and cotransfected with 40 nM of either miR-103 or miRNA control, 20 ng of either pGL3-ZO-1-3′-UTR-wt or pGL3-ZO-1-3′-UTR-mutant, and 2 ng pRL-TK (Promega Corporation) using Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.). The pRL-TK vector was used as a normalization control. After transfection for 48 h at 37°C, cells were harvested and assayed with a Dual-Luciferase Reporter Assay system (Promega Corporation) according to the manufacturer's protocol.
Cell viability assay
MTT assays were performed to analyze cellular proliferation. Cells were seeded at a density of 2×103 cells/well in 96-well plates. After 1, 2, 3, 4 or 5 days incubation, 20 ml MTT (5 mg/ml) was added and incubated for 4 h at 37°C in a 5% CO2 incubator. The supernatant was removed and 150 ml dimethyl sulfoxide was added. Cell viability was then determined using a microplate reader (Model 550; Bio-Rad Laboratories, Inc., Hercules, CA, USA) at a wavelength of 490 nm.
Colony formation assay
A total of 1×103 cells (transfected with miRNA mimic or inhibitor) were placed in each well of 6-well plate and maintained in media containing 10% FBS for 10 days. Colonies were fixed with methanol and stained with 0.1% crystal violet in 20% methanol for 15 min. Colonies were counted using an inverted microscope, and three fields of view were counted.
Transwell assay
Cell migration was evaluated using 24-well uncoated Transwell cell culture chambers (Corning Incorporated, Corning, NY, USA). Briefly, 1×105 transfected cells (transfected with miRNA mimic or inhibitor) were re-suspended in 200 µl serum-free medium and placed in the upper chamber with the lower chamber containing 600 µl DMEM and 10% FBS. After 24 h, cells in the lower chamber were fixed with 4% paraformaldehyde in PBS for 10 min at room temperature and stained with 0.1% crystal violet for 30 min at room temperature. The migrated cells were counted and captured at ×200 magnification in 5 random fields of view with an inverted microscope (Olympus Corporation, Tokyo, Japan) and the mean ± standard deviation was calculated accordingly.
Statistical analysis
All data were expressed as the mean ± standard deviation. A Student's t-test (two-tailed) was used to compare two groups, and a Chi-squared test was used for comparison of the rates of two groups. A one-way analysis of variance and Tukey's multiple comparison post hoc test were used to determine differences between multiple groups. Correlation analysis was performed with Spearman's rank correlation analysis. P<0.05 was considered to indicate a statistically significant difference.
Results
miR-103 is upregulated in colorectal cancer tissues
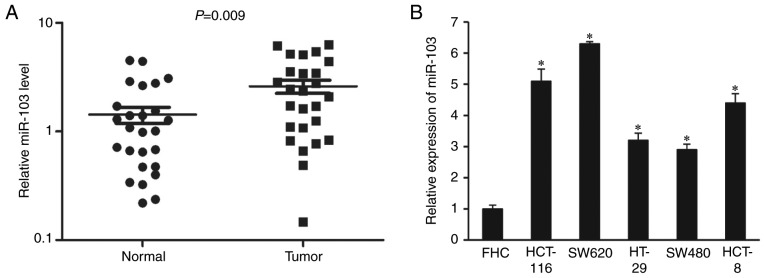
To explore the role of miR-103 in CRC carcinogenesis, the expression levels of miR-103 in 26-CRC tissues and their matched adjacent non-tumor tissues were determined using RT-qPCR. miR-103 was significantly upregulated in tumor tissues compared with adjacent non-tumor tissues (Fig. 1A; P=0.009). Consistent with this, the expression levels of miR-103 was significantly upregulated in all five CRC cell lines compared with the normal colonic cell line (Fig. 1B; P<0.001 vs. FHC). Taken together, these results suggested that miR-103 was upregulated in CRC tissues and that miR-103 overexpression may contribute to tumor development and progression.
Figure 1.
miR-103 is upregulated in colorectal cancer. Reverse transcription-quantitative polymerase chain reaction analysis of miR-103 expression levels in tissues and cell lines. (A) Relative miR-103 expression levels in 26 paired colorectal cancer tissues and adjacent non-tumor tissues. (B) Relative miR-103 expression levels in five colorectal cancer cell lines (HCT116, SW620, HT29, SW480 and HCT-8) and a human colon epithelial cell line (FHC). U6 was used as an internal control. The data represents the mean ± standard deviation. *P<0.001 vs. FHC. miR-103, microRNA-103.
miR-103 promotes the proliferation and migration of colorectal cancer cells
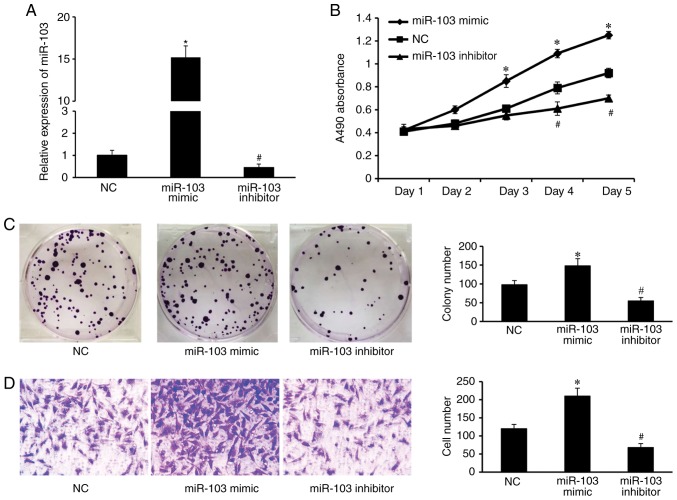
To further assess the role of miR-103 on CRC, cell proliferation was measured using MTT and colony formation assays. Following transfection of SW620 cells with the miR-103 mimic, inhibitor or NC, RT-qPCR analysis revealed high expression levels of miR-103 in the miR-103 mimic transfected group (P<0.001 vs. NC group), and low levels in the miR-103 inhibitor group (Fig. 2A; P=0.02 vs. NC group). MTT assays demonstrated that overexpression of miR-103 promoted cell proliferation, whereas downregulation of miR-103 expression inhibited cell proliferation compared with NC (Fig. 2B; P<0.05). Colony formation assays revealed that SW620 cells transfected with the miR-103 mimic exhibited significantly greater number of colonies compared with the NC group (Fig. 2C; P=0.008). Conversely, SW620 cells transfected with the miR-103 inhibitor significantly suppressed cell colony formation (Fig. 2C; P=0.02). Transwell assays revealed that SW620 cells transfected with the miR-103 mimic exhibited significantly greater numbers of cells compared with the NC group (Fig. 2D; P≤0.001). Conversely, SW620 cells transfected with the miR-103 inhibitor had significantly reduced cell numbers compared with the NC (Fig. 2D; P=0.004).
Figure 2.
miR-103 affects cell proliferation and migration in vitro. (A) Reverse transcription-quantitative polymerase chain reaction analysis of miR-103 expression in SW620 cells transfected with NC oligonucleotide, miR-103 mimic or miR-103 inhibitor (*P<0.001 vs. NC; #P=0.02 vs. NC). (B) Growth rates of the transfected SW620 cells and control cells in vitro. Cell viability was evaluated with the MTT assay using absorbance readings at 490 nm at the indicated times. The values shown are the mean of three experiments. (C) Colony numbers of SW620 cells transfected with miR-103 mimic, miR-103 inhibitor and NC via the colony formation assay (*P=0.008 vs. NC; #P=0.02 vs. NC). (D) Transwell assay (original magnification, ×200) demonstrated the migration ability of SW620 cells, and the results showed that the ability of migration was significantly inhibited in miR-103 inhibitor-transfected cells and the ability of migration was significantly increased in cells transfected with the miR-103 mimic (*P<0.001 vs. NC; #P=0.004 vs. NC). The values shown are the mean of three independent experiments, and represent the mean ± standard deviation. NC, negative control; miR-103, microRNA-103.
miR-103 directly targets the 3′UTR of ZO-1
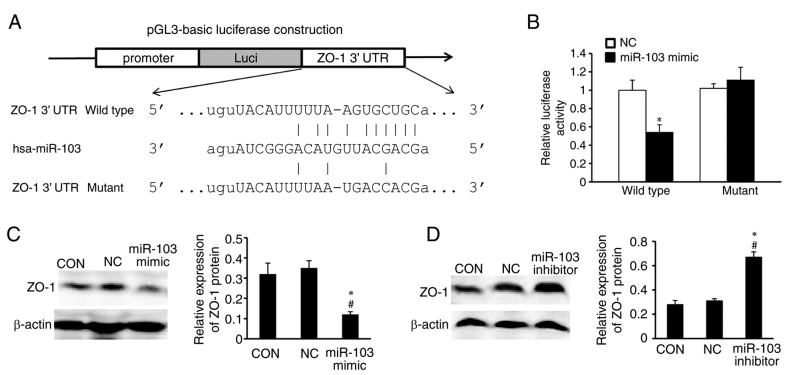
To explore the underlying molecular mechanism of miR-103 in CRC, three available databases (TargetScan, PicTar and miRanda) were utilized to search for predicted direct target genes of miR-103. The predicted targets were arranged according to the binding probability score. The targets with high score and shared by the three databases were chosen. Among the numerous targets, ZO-1 was chosen for further analysis. miR-103 has conserved binding sites in the 3′UTR of ZO-1 in humans (Fig. 3A). To elucidate whether miR-103 interacts with the 3′-UTR of ZO-1, luciferase reporter assays were performed. Plasmids containing the wt and mutant fragments of the 3′-UTR were co-transfected with the miR-103 mimic or NC, respectively. The luciferase activity of the reporter containing the wt 3′ UTR of ZO-1 was decreased in cells transfected with miR-103 mimic, whereas the activity of the mutant reporter was not significantly altered, compared with the NC (Fig. 3B).
Figure 3.
ZO-1 is a direct target of miR-103 in colorectal cancer. (A) The miR-103 wild-type binding sequence or its mutated form was inserted into the C-terminal of the luciferase gene to generate the pGL3-ZO-1-3′ UTR or pGL3-ZO-1-mut-3′ UTR plasmids, respectively. (B) miR-103 targeted the wild type but not the mutant 3′ UTR of ZO-1. The data represent the mean ± standard deviation. *P<0.05 vs. NC. (C) Western blotting and densitometric analysis revealed that miR-103 overexpression inhibited the protein expression levels of ZO-1 in SW620 cells. SW620 cells were transfected with the miR-103 mimic, mimic control or NC. β-actin served as the loading control. The data represent the mean ± standard deviation. *P<0.05 vs. NC; #P<0.05 vs. CON. (D) Western blotting and densitometric analysis demonstrated that knockdown of miR-103 upregulated the protein expression levels of ZO-1 in SW620 cells. SW620 cells were transfected with the miR-103 inhibitor, inhibitor control or NC. The data represent the mean ± standard deviation. *P<0.05 vs. NC; #P<0.05 vs. CON. ZO-1, zonula occuldens-1; miR-103, microRNA-103; 3′UTR, 3′-untranslated region; NC, transfection negative control; CON, untransfected control.
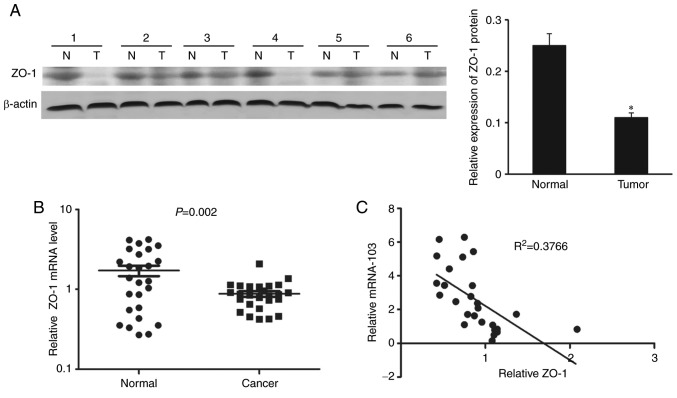
ZO-1 expression was further detected in stably transfected SW620 cells using western blotting. Results revealed that the protein expression levels of ZO-1 were reduced in cells transfected with the miR-103 mimic compared with the NC (Fig. 3C), whereas inhibition of miR-103 resulted in upregulated levels of ZO-1 compared with the NC (Fig. 3D). ZO-1 protein expression levels were also characterized in CRC and normal adjacent tissues (Fig. 4A). A significant inverse correlation was demonstrated between the levels of miR-103 and ZO-1 mRNA expression in cancer patient tissues (r2=0.3766, P<0.001; Fig. 4B and C).
Figure 4.
Expression of ZO-1 in CRC tissues. (A) Western blotting and densitometric analysis of ZO-1 in 26 pairs of CRC tissues and corresponding adjacent non-tumorous tissues, with representative images shown of 6 tissue pairs. β-actin served as a loading control. Relative ZO-1 protein expression level was reduced in 18 of 26 (69.2%) CRC tissues compared with the corresponding adjacent non-tumorous tissues (*P=0.020). (B) Reverse transcription-quantitative polymerase chain reaction of the relative expression of ZO-1 in the 26 CRC and normal adjacent tissues. The data represents the mean ± standard deviation. (C) Spearman's rank correlation analysis demonstrated an inverse correlation between miR-103 and ZO-1 expression in colorectal cancer tissues. The miR-103 expression level was normalized to the U6, and ZO-1 expression levels were normalized to the β-actin. ZO-1, zonula occuldens-1; CRC, colorectal cancer; N, non-tumorous tissues; C, CRC tissues; miR-103, microRNA-103.
Discussion
Numerous studies have demonstrated aberrant expression of miR-103 in different human cancers (16–18). It is reported that miR-103 post-transcriptionally downregulates expression levels of the tumor suppressor gene, TIMP3, and promotes growth and invasion of endometrial cancer cell lines (10). Also, it has been reported that miR-103 is expressed in colorectal cancer as an oncogenic miRNA by targeting DAPK, KLF4 and circadian clock gene period 3 (13,18). Certain reports have revealed that miR-103 is significantly downregulated in heart failure (HF) and may be used as a marker for diagnosis of HF (9). In addition, observations have demonstrated that miR-103 is upregulated in obese mice and silencing of miR-103 leads to improved glucose homeostasis and insulin sensitivity (19). In the present study, miR-103 was upregulated in CRC tissues and significantly promoted CRC cell proliferation and migration in vitro. Conversely, downregulation of miR-103 via transfection of an miR-103 inhibitor inhibited CRC cell proliferation and migration. Collectively, these findings suggested that miR-103 serves an important role in CRC carcinogenesis. Furthermore, ZO-1 was identified as a direct functional target of miR-103.
ZO-1 is a 220 kDa membrane scaffold protein that is a member of the membrane associated guanylate kinase protein family and a key component of junctional complexes that regulate tight junction formation (20,21). Additionally, the initial phosphorylation of ZO-1 seemed to be concomitant with the decrease in the signal intensity of ZO-1 in the region of cell-cell junctions, which suggested that the phosphorylation of ZO-1 causes degradation of the ZO-1 protein (22). Hyperphosphorylation of ZO-1 usually coincides with its departure from tight junctions into the cytoplasm and with increased permeability (23,24), which leads to increased motility and contributes to oncogenic behavior. Furthermore, several studies have indicated that downregulation of ZO-1 is involved in tumor development and progression, and that ZO-1 is considered to be a tumor suppressor (25–27). Decreased expression of ZO-1 is reported to be associated with invasion of breast cancer (20,26), gastrointestinal stromal tumor (27), hepatocellular carcinoma (28) and lung cancer (29). In the present study, based on bioinformatic analyses, ZO-1 was identified as a novel, direct target of miR-103, as confirmed by a luciferase reporter assay. This result was supported by the observation that miR-103 overexpression impaired ZO-1 expression whereas miR-103 knockdown increased ZO-1 protein expression in CRC cells. In addition, the expression levels of miR-103 were inversely correlated with ZO-1 expression in CRC tissues. In summary, the present study demonstrated that miR-103 significantly promoted CRC cell proliferation and migration and may directly target the 3′UTR of ZO-1. The newly identified miR-103/ZO-1 axis may help to further elucidate the complex underlying molecular mechanisms that regulate metastasis and progression of CRC, and has the potential to be a prognostic marker or therapeutic target for patients with CRC. However, the specific molecular mechanisms underlying the miR-103/ZO-1 axis regulation of colon cancer cell proliferation and migration remain unclear, as are the mechanisms by which miR-103 is upregulated in colon cancer, and therefore, further studies are required.
Acknowledgements
The present study was supported by National Natural Science Foundation of China (grant no. 81541046), Jiangsu Post-doctoral Program (grant no. 1402201C), Jiangsu 333 Talent Project (grant no. BRA2015490), Six Talent Peaks Program of Jiangsu Province (grant no. WSW-051 and WS-064), the Social Development Foundation of Nantong City (MS32016017) and the 226 Talent Training Program of Nantong City (The Fourth Batch).
References
- 1.Zhao P, Dai M, Chen W, Li N. Cancer trends in China. Jpn J Clin Oncol. 2010;40:281–285. doi: 10.1093/jjco/hyp187. [DOI] [PubMed] [Google Scholar]
- 2.Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN2008. Int J Cancer. 2010;127:2893–2917. doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- 3.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 4.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 5.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 6.Lee YS, Dutta A. The tumor suppressor microRNA let-7 represses the HMGA2 oncogene. Genes Dev. 2007;21:1025–1030. doi: 10.1101/gad.1540407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bonci D, Coppola V, Musumeci M, Addario A, Giuffrida R, Memeo L, D'Urso L, Pagliuca A, Biffoni M, Labbaye C, et al. The miR-15a-miR-16-1 cluster controls prostate cancer by targeting multiple oncogenic activities. Nat Med. 2008;14:1271–1277. doi: 10.1038/nm.1880. [DOI] [PubMed] [Google Scholar]
- 8.Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ellis KL, Cameron VA, Troughton RW, Frampton CM, Ellmers LJ, Mark RA. Circulating microRNAs as candidate markers to distinguish heart failure in breathless patients. Eur J Heart Fail. 2013;15:1138–1147. doi: 10.1093/eurjhf/hft078. [DOI] [PubMed] [Google Scholar]
- 10.Yu D, Zhou H, Xun Q, Xu X, Ling J, Hu Y. microRNA-103 regulates the growth and invasion of endometrial cancer cells through the downregulation of tissue inhibitor of metalloproteinase 3. Oncol Lett. 2012;3:1221–1226. doi: 10.3892/ol.2012.638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Annibali D, Gioia U, Savino M, Laneve P, Caffarelli E, Nasi S. A new module in neural differentiation control: Two microRNAs upregulated by retinoic acid, miR-9 and −103, target the differentiation inhibitor ID2. PLoS One. 2012;7:e40269. doi: 10.1371/journal.pone.0040269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weber DG, Johnen G, Bryk O, Jöckel KH, Brüning T. Identification of miRNA-103 in the cellular fraction of human peripheral blood as a potential biomarker for malignant mesothelioma-a pilot study. PLoS One. 2012;7:e30221. doi: 10.1371/journal.pone.0030221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hong Z, Feng Z, Sai Z, Tao S. PER3, a novel target of miR-103, plays a suppressive role in colorectal cancer in vitro. BMB Rep. 2014;47:500–505. doi: 10.5483/BMBRep.2014.47.9.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mazzoccoli G, Panza A, Valvano MR, Palumbo O, Carella M, Pazienza V, Biscaglia G, Tavano F, Di Sebastiano P, Andriulli A, Piepoli A. Clock gene expression levels and relationship with clinical and pathological features in colorectal cancer patients. Chronobiol Int. 2011;28:841–851. doi: 10.3109/07420528.2011.615182. [DOI] [PubMed] [Google Scholar]
- 15.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 16.Zhang Y, Qu X, Li C, Fan Y, Che X, Wang X, Cai Y, Hu X, Liu Y. miR-103/107 modulates multidrug resistance in human gastric carcinoma by downregulating Cav-1. Tumour Biol. 2015;36:2277–2285. doi: 10.1007/s13277-014-2835-7. [DOI] [PubMed] [Google Scholar]
- 17.Brewster BL, Rossiello F, French JD, Edwards SL, Wong M, Wronski A, Whiley P, Waddell N, Chen X, Bove B, et al. Identification of fifteen novel germline variants in the BRCA1 3′UTR reveals a variant in a breast cancer case that introduces a functional miR-103 target site. Hum Mutat. 2012;33:1665–1675. doi: 10.1002/humu.22159. [DOI] [PubMed] [Google Scholar]
- 18.Chen HY, Lin YM, Chung HC, Lang YD, Lin CJ, Huang J, Wang WC, Lin FM, Chen Z, Huang HD, et al. miR-103/107 promote metastasis of colorectal cancer by targeting the metastasis suppressors DAPK and KLF4. Cancer Res. 2012;72:3631–3641. doi: 10.1158/0008-5472.CAN-12-0667. [DOI] [PubMed] [Google Scholar]
- 19.Trajkovski M, Hausser J, Soutschek J, Bhat B, Akin A, Zavolan M, Heim MH, Stoffel M. MicroRNAs 103 and 107 regulate insulin sensitivity. Nature. 2011;474:649–653. doi: 10.1038/nature10112. [DOI] [PubMed] [Google Scholar]
- 20.Chai Z, Goodenough DA, Paul DL. Cx50 requires an intact PDZ-binding motif and ZO-1 for the formation of functional intercellular channels. Mol Biol Cell. 2011;22:4503–4512. doi: 10.1091/mbc.E11-05-0438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Giri S, Poindexter KM, Sundar SN, Firestone GL. Arecoline induced disruption of expression and localization of the tight junctional protein ZO-1 is dependent on the HER 2 expression in human endometrial Ishikawa cells. BMC Cell Biol. 2010;11:53. doi: 10.1186/1471-2121-11-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kawauchiya T, Takumi R, Kudo Y, Takamori A, Sasagawa T, Takahashi K, Kikuchi H. Correlation between the destruction of tight junction by patulin treatment and increase of phosphorylation of ZO-1 in Caco-2 human colon cancer cells. Toxicol Lett. 2011;205:196–202. doi: 10.1016/j.toxlet.2011.06.006. [DOI] [PubMed] [Google Scholar]
- 23.Van Itallie CM, Balda MS, Anderson JM. Epidermal growth factor induces tyrosine phosphorylation and reorganization of the tight junction protein ZO-1 in A431 cells. J Cell Sci. 1995;108:1735–1742. doi: 10.1242/jcs.108.4.1735. [DOI] [PubMed] [Google Scholar]
- 24.Antonetti DA, Barber AJ, Hollinger LA, Wolpert EB, Gardner TW. Vascular endothelial growth factor induces rapid phosphorylation of tight junction proteins occludin and zonula occluden 1. A potential mechanism for vascular permeability in diabetic retinopathy and tumors. J Biol Chem. 1999;274:23463–23467. doi: 10.1074/jbc.274.33.23463. [DOI] [PubMed] [Google Scholar]
- 25.Németh Z, Szász AM, Somorácz A, Tátrai P, Németh J, Gyorffy H, Szíjártó A, Kupcsulik P, Kiss A, Schaff Z. Zonula occludens-1, occludin and E-cadherin protein expression in biliary tract cancers. Pathol Oncol Res. 2009;15:533–539. doi: 10.1007/s12253-009-9150-4. [DOI] [PubMed] [Google Scholar]
- 26.Martin TA, Watkins G, Mansel RE, Jiang WG. Loss of tight junction plaque molecules in breast cancer tissues is associated with a poor prognosis in patients with breast cancer. Eur J Cancer. 2004;40:2717–2725. doi: 10.1016/j.ejca.2004.08.008. [DOI] [PubMed] [Google Scholar]
- 27.Zhu H, Lu J, Wang X, Zhang H, Tang X, Zhu J, Mao Y. Upregulated ZO-1 correlates with favorable survival of gastrointestinal stromal tumor. Med Oncol. 2013;30:631. doi: 10.1007/s12032-013-0631-7. [DOI] [PubMed] [Google Scholar]
- 28.Orbán E, Szabó E, Lotz G, Kupcsulik P, Páska C, Schaff Z, Kiss A. Different expression of occludin and ZO-1 in primary and metastatic liver tumors. Pathol Oncol Res. 2008;14:299–306. doi: 10.1007/s12253-008-9031-2. [DOI] [PubMed] [Google Scholar]
- 29.Ni S, Xu L, Huang J, Feng J, Zhu H, Wang G, Wang X. Increased ZO-1 expression predicts valuable prognosis in non-small cell lung cancer. Int J Clin Exp Pathol. 2013;6:2887–2895. [PMC free article] [PubMed] [Google Scholar]