Abstract
Curcumin is a well-known phenolic substance and has many pharmacological effects associated with metabolism. However, the exact molecular mechanisms underlying this process have yet to be determined. The Notch pathway is a signal transduction pathway involved in energy metabolism. The present study aimed to investigate the effects of curcumin administration on glucose-lipid metabolism in rats subjected to a high fat diet, and investigate changes in Notch-1 signaling. Sprague-Dawley rats (n=40) were randomly divided into four groups (10 rats/group): Control diet group, high fat diet group, high fat diet plus curcumin low dose group and high fat diet plus curcumin high dose group. Following 8 weeks of treatment with curcumin (100 mg/kg in the low dose group and 200 mg/kg in the high dose group), serum metabolic markers and hepatic gene expression patterns were investigated. No differences in body weight following 8 weeks of curcumin administration (P>0.05) were observed; however, curcumin treatment did reduce visceral fat levels (peri-epididymal and peri-renal), and decreased cholesterol, triglyceride and low-density lipoprotein levels in serum compared with the high fat diet rats that did not receive curcumin (P<0.05, P<0.01). An oral glucose tolerance test and an intraperitoneal insulin tolerance test revealed that insulin resistance was reduced (P<0.05 or P<0.01) and tissue section analysis revealed that hepatosteatosis was attenuated following treatment with curcumin. Furthermore, the protein expression of Notch-1 and its downstream target Hes-1 were suppressed. These effects were also in parallel with an upregulation of fatty acid oxidation-associated gene expression, including peroxisome proliferator-activated receptor (PPAR)-α, carnitine palmitoyltransferase 1 and PPAR-γ (P<0.05). In addition, curcumin administration led to a downregulation in the expression of lipogenic genes, including sterol regulatory element-binding protein, fatty acid synthase and acetyl-CoA carboxylase (P<0.05). The expression of inflammation-associated genes, including nuclear factor-κB, tumor necrosis factor-α and prostaglandin-endoperoxide synthase 2 were also suppressed. The results of the present study suggest that the hepatic Notch-1 pathway can be suppressed via curcumin treatment, which may ameliorate fatty liver and insulin resistance in rats subjected to a high fat diet.
Keywords: curcumin, fatty liver, insulin resistance, Notch-1 signaling
Introduction
Non-alcoholic fatty liver disease (NAFLD) is a prevalent chronic liver disease, which threatens the health of both adults and children. Furthermore, NAFLD is also regarded as a risk factor for metabolism syndromes and is strongly associated with type 2 diabetes mellitus, obesity and insulin resistance (1). However, the pathophysiology of NAFLD remains unclear, and many strategies, such as diet control, weight loss and exercise are difficult to maintain for a long time, and available pharmacotherapy are still deficient (2). Thus, the development of novel therapeutics and the identification of novel therapeutic targets are required.
Notch signaling is a conserved signaling pathway involved in cell commitment and development (3). Previous research has demonstrated that Notch-1 can act as an oncogene and is upregulated in cases of hepatocellular carcinoma (4). Recently, Notch signaling has been recognized as a key regulator of energy homeostasis, which is strongly associated with glycometabolism and lipid accumulation (5). In addition, Notch signaling amplifies signal transmissions responsible for fat production in cases of obesity and diabetes. Furthermore, Notch-1 activation enhances lipogenesis in the liver, resulting in NAFLD, and suppression of Notch-1 signaling in the liver reduces hepatosteatosis (6). Thus, the Notch signaling pathway may be a potential therapeutic target for the treatment of NAFLD and insulin resistance.
Curcumin is the active ingredient of Curcuma longa, which has various pharmacological activities, including anti-inflammatory, anti-oxidant and anti-cancer activities (7). Numerous studies have demonstrated that administration of curcumin suppresses tumor growth via inhibition of the Notch signaling pathway in hepatic cancers (8–11), thus indicating the potential of using curcumin to target the Notch signaling pathway. Furthermore, curcumin has also been demonstrated to reduce serum lipids and glucose levels in animal models (12), however, the underlying molecular mechanism responsible for said effects remains unclear.
It has previously been revealed that curcumin downregulates Notch signaling in hepatocellular carcinoma (13), and that this cancer type can also develop as a result of hepatosteatosis and hepatocirrhosis. Considering previous findings, the present study aimed to investigate whether curcumin reduces the development of NAFLD, and whether this is associated with inhibition of the Notch signaling pathway. In the present study, rats were fed a high-fat diet and were used to investigate the effects of curcumin on hepatic steatosis and insulin resistance. Furthermore, Notch signaling and associated metabolic targets were investigated to determine the potential mechanisms involved.
Materials and methods
Chemicals and reagents
Curcumin was purchased from Sigma-Aldrich; Merck KGaA (Darmstadt, Germany), dissolved in dimethyl sulfoxide, and then olive oil was added to give a final concentration of 40 mg/ml. Antibodies were purchased from Cell Signaling Technology, Inc. (Danvers, MA, USA). Fluorescein isothiocyanate-conjugated goat anti-rat antibody (1:3,000; cat. no. A0258) and immunoglobulin G were purchased from Beyotime Institute of Biotechnology (Haimen, China). Primers and other chemical reagents were purchased from Sangon Biotech Co., Ltd. (Shanghai, China).
Animals and treatment
Male Sprague Dawley rats (n=40; 6-week-old) weighing ~200 g were obtained from the Shanghai Center of Experimental Animals (Shanghai, China) and were housed in a standard environment (20±2°C; 50±5% humidity; 12 h light/dark cycle, diet and filtered water ad libitum). Following acclimatization for 1 week, 10 rats were weaned to a normal chow diet (7% fat, 64.7% carbohydrate and 18.7% protein), and the other 30 began a high-fat diet (HFD; 43.6% fat, 24.5% carbohydrate and 14.7% protein). The 30 rats were randomly allocated into three groups (10 per group): HFD group, the low dose group (HFD + curcumin 100 mg/kg/day), and the high dose group (HFD + curcumin 200 mg/kg/day). Rats were given daily intraperitoneal injections of either curcumin or vehicle (mixture of dimethyl sulfoxide and olive oil, at a ratio of 1:20) daily for 8 weeks. Food consumption and body weight were measured weekly. The ethics committee of the First Affiliated Hospital of Xiamen University (Xiamen, China) granted ethical approval for the care and use of animals in the present study.
Systemic glucose tolerance and insulin sensitivity test [oral glucose tolerance test (OGTT) and intra-peritoneal insulin tolerance test (IPITT)]
In preparation for the performance of glucose tolerance tests, rats were fasted for 12 h and then administered an intragastric injection of glucose at a dose of 2 g/kg/body weight (bw). Blood glucose was then measured at 0, 30, 60, 90, 120 and 180 min time intervals using a One Touch Ultra blood glucose meter (LifeScan, Inc., San Jose, CA, USA). In order to perform insulin sensitivity tests, rats were administered an intraperitoneal injection of insulin (0.75 U/kg/bw), which was then followed by 12 h of fasting. Blood glucose levels were then measured at the same time intervals as detailed with regards to the OGTT. The areas under the glucose curves were evaluated according to the trapezoidal rule (14).
Biochemical analyses
Blood samples were collected from the rats and serum was immediately centrifuged at 1,600 × g for 10 min at 4°C. Serum levels of cholesterol (CHO), triglycerides (TG), high-density lipoprotein (HDL) and low-density lipoprotein-c (LDL) were analyzed using an automatic chemistry analyzer (Hitachi Ltd., Tokyo, Japan).
Hepatic TG measurement
In order to quantitatively assess hepatic TG content, a Triglyceride assay kit (Applygen Technologies, Inc., Beijing, China) was used. Briefly, 50 mg liver samples were homogenized and extracted using a mixture of lipid extract (chloroform/methanol, 2:1). Samples were then centrifuged and the organic solvent layer was collected. Hepatic TG concentrations were measured using the enzymatic hydrolysis method, according to the manufacturer's protocol.
Hematoxylin and eosin (H&E) staining
H&E staining was used to assess hepatic steatosis. Briefly, liver tissues were fixed in 4% paraformaldehyde at 4°C for 12 h and embedded in paraffin, prepared into 5-µm thick sections, and stained with hematoxylin for 8 min, wash in running tap water for 60 min, counterstain with eosin for 60 sec at room temperature. Images were scored under a light microscope (magnification, ×200; Nikon Corporation, Tokyo, Japan) according to the following criteria (15): Score 1 for 0–25% steatosis, score 2 for 26–50% steatosis, score 3 for 51–75% and score 4 for over 76% steatosis.
RNA isolation and reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total mRNA was isolated from the liver using TRIzol Reagent (Qiagen China Co., Ltd., Shanghai, China). RT was performed using the BeyoRT™ II First Strand cDNA Synthesis Kit (Beyotime Institute of Biotechnology) and then qPCR was performed using the Light-Cycler 480 System (Roche Diagnostics, Basel, Switzerland) with SYBR Green reagent (Takara Biotechnology, Co., Ltd., Dalian, China). The amplification reactions were according to the following thermal cycling conditions: 95°C for 10 min, 40 cycles of 95°C for 15 sec and 60°C for 30 sec. The quantification was expressed as the ratio of target genes to GAPDH mRNA using the 2−ΔΔCq method (16). RT-qPCR was performed using the primers in Table I.
Table I.
Sequences of primers designed for reverse transcription-quantitative polymerase chain reaction.
| Gene | Forward primer | Reverse primer |
|---|---|---|
| CPT-1 | GCTTTGAAGTTTTTGGTGAAATTGA | GCTATGGTTTCATCACCTACCGT |
| PPAR-γ | AAAGAAGCCGACACTAAACC | CTTCCATTACGGAGAGATCC |
| PPAR-α | TGTCACACAATGCAATCCGTTT | TTCAGGTAGGCTTCGTGGATTC |
| SREBP-1 | GGAGCCATGGATTGCACATT | CCTGTCTCACCCCCAGCATA |
| FAS | CACAGCATTCAGTCCTATCCACAGA | CACAGCCAACCAGATGCTTCA |
| ACC | CAATCCTCGGCACATGGAGA | GCTCAGCCAACCGGATGTAGA |
| NF-κB | TCTGGCGCAGAAGTTAGG | CCAGAGACCTCATAGTTGT |
| TNF-α | TACAGGCTTGTCACTCGA ATT | GATGGTGGGTATGGGTCAGAA GGA |
| COX-2 | ACTTGCTCACTTTGTTGAGTCATTC | TTTGATTAGTACTGTAGGGGTTAATG |
| GAPDH | ACAGCAACAGGGTGGTGGAC | TTTGAGGGTGCAGCGAACTT |
CPT-1, carnitine palmitoyltransferase 1; PPAR-γ, peroxisome proliferator-activated receptor-γ; PPAR-α, peroxisome proliferator-activated receptor-α; SREBP-1c, sterol regulatory element-binding protein; FAS, fatty acid synthase; ACC, acetyl-CoA carboxylase; NF-κB, nuclear factor-κB; TNF-α, tumor necrosis factor-α; COX-2, prostaglandin-endoperoxide synthase 2.
Protein extraction and western blot analysis
The protein in the liver samples was extracted and homogenized using a radioimmunoprecipitation assay lysis buffer with + phenylmethyl sulfonyl fluoride. Proteins (100 µg) were separated by 10% SDS-PAGE and transferred to polyvinylidene fluoride membranes (Roche Diagnostics), blocked using 10% skimmed milk (Guangming Group, Shanghai, China). Membranes were incubated overnight at 4°C with primary antibodies against Notch-1 (rabbit polyclonal; 1:1,000; cat. no. D6F11; Cell Signaling Technology, Inc., Danvers, MA, USA), HES1 (rabbit polyclonal; 1:800; cat. no. D6P2U; Cell Signaling Technology, Inc.) and GAPDH (rabbit polyclonal; 1:1000; cat. no. no. 5174; Cell Signaling Technology, Inc.). The blots were then incubated at 37°C for 30 min with a horseradish peroxidase-conjugated goat anti rabbit immunoglobulin G secondary antibody (1:2,000; cat. no. A0208; Beyotime Institute of Biotechnology). A UVP Bioimaging system (BioSpectrum 410; UVP Inc., Upland, CA, USA) and Gel-Pro-Analyzer software version 5.0 (Media Cybernetics, Inc., Bethesda, MD, USA) were used for detection and analysis.
Statistical analysis
Quantitative data were expressed as the mean ± standard deviation. Statistical significance among the groups was determined using one-way analysis of variance followed by Tukey's multiple comparison test. P<0.05 was considered to indicate a statistically significant difference.
Results
Administration of curcumin decreases visceral fat and liver weight
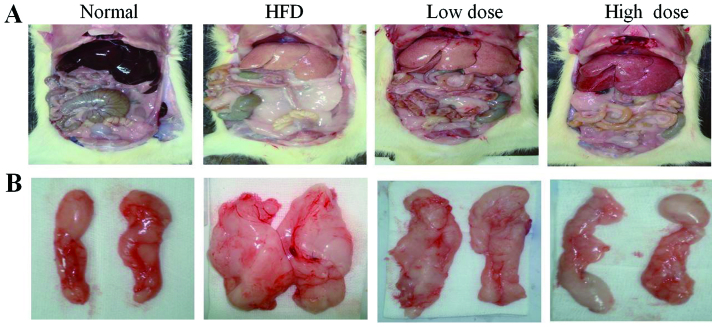
With regards to liver morphology, the liver color was grayer in the HFD group and less gray after curcumin treatment (Fig. 1A). The visceral fat was reduced in both the low dose and the high dose curcumin-treated groups (Fig. 1B), and the liver weight was also reduced following curcumin treatment in a dose-dependent manner (Table II; P<0.05). However, curcumin treatment did not reduce the total body weight of the rats compared with the HFD group (Table II). In addition, the food consumption of the rats did not change following treatment with curcumin (data not shown). These results demonstrate that curcumin treatment decreases abdominal obesity, but not via reduction of food intake.
Figure 1.
Morphological changes of visceral and peri-epididymal fat in sham and HFD rats following 8 weeks of treatment with low and high dose curcumin. (A) Liver and visceral fat, and (B) peri-epididymal fat. HFD, high fat diet.
Table II.
Body weight of different groups at the conclusion of the study.
| Parameter | Control | HFD | Low dose curcumin | High dose curcumin |
|---|---|---|---|---|
| Body weight (g) | 612.1±27.35 | 695.1±67.30a | 674.0±56.59 | 665.8±61.29 |
| Peri-epididymal fat (g) | 11.1±3.13 | 18.5±6.23b | 13.8±5.74c | 11.8±4.15d |
| Peri-renal fat (g) | 11.2±3.37 | 28.0±3.41a | 22.5±4.39c | 19.7±5.82c |
| Liver weight (g) | 17.1±0.61 | 30.4±3.57b | 26.4±5.41 | 23.4±5.74c |
P<0.01
P<0.05 vs. control group
P<0.05
P<0.01 vs. HFD group. HFD, high fat diet.
Curcumin improves glucose tolerance and increases insulin sensitivity
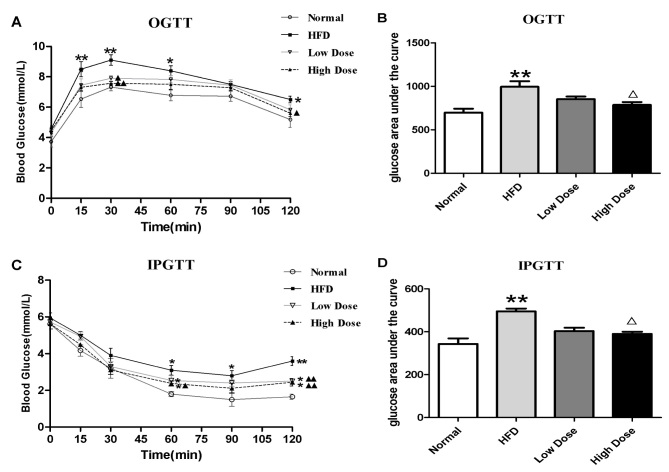
OGTT and IPGTT were performed in order to investigate the effects of curcumin on glucose homeostasis and insulin sensitivity. As revealed by Fig. 2A, the HFD group experienced a significant reduction in levels of glucose tolerance compared with the normal control group. Curcumin treatment significantly reduced blood glucose levels at 30 and 120 min compared with the HFD group (P<0.05 and P<0.01). The area under the curve also showed that glucose concentrations were lower in both the curcumin low dose (non-significantly) and high dose groups. (compared to the HFD groups (P<0.01) (Fig. 2B). In comparison with the control group, HFD led to insulin resistance, which manifested as a blunted response in the IPGTT (Fig. 2C). Furthermore, administration of curcumin significantly decreased the blood glucose concentration at 60, 90 and 120 min compared with the HFD group (P<0.05 and P<0.01; Fig. 2C). The area under the curve showed that insulin caused a significant reduction in glucose in curcumin high dose groups (P<0.01 vs. the HFD group) (Fig. 2D). These results suggest that curcumin treatment is effective in increasing glucose tolerance and insulin sensitivity in HFD-induced obesity in rats.
Figure 2.
Effect of curcumin on OGTT and IPGTT in HFD rats. Results are presented as (A) OGTT and (B) OGTT glucose area under the curve, (C) IPGTT and (D) IPGTT glucose area under the curve. *P<0.01, **P<0.01 vs. normal group; ∆P<0.05, ∆∆P<0.01 vs. HFD group. OGTT, oral glucose tolerance test; IPGTT, intra-peritoneal insulin tolerance test; HFD, high fat diet.
Curcumin decreases the concentration of blood lipids
As presented in Table III, the concentrations of serum CHO, TG and LDL were significantly decreased in the curcumin-treated groups compared with HFD rats (P<0.05 and P<0.01). Total HDL did not change significantly among the groups.
Table III.
Characteristics of serum chemistry.
| Group | CHO (mmol/l) | TG (mmol/l) | LDL (mmol/l) | HDL (mmol/l) |
|---|---|---|---|---|
| Normal | 0.53±0.04 | 2.23±0.72 | 0.45±0.18 | 0.47±0.12 |
| HFD | 1.45±0.32b | 4.93±1.17a | 1.77±0.21a | 0.27±0.04b |
| Low dose | 1.24±0.44 | 3.14±1.10c | 1.46±0.29c | 0.25±0.06 |
| High dose | 1.12±0.26c | 2.41±0.72d | 1.28±0.13d | 0.28±0.06 |
P<0.05
P<0.01 vs. control group
P<0.05
P<0.01 vs. HFD group. HFD, high fat diet; CHO, cholesterol; TG, triglyceride; LDL, low-density lipoprotein; HDL, high-density lipoprotein.
Curcumin reduces hepatosteatosis
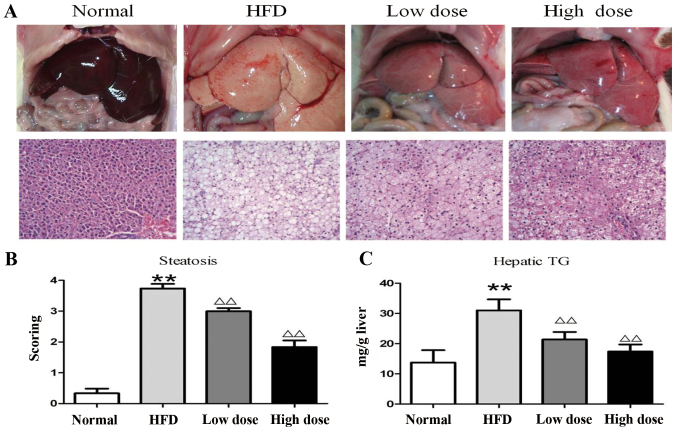
H&E staining was performed to determine the effect of curcumin administration on hepatosteatosis. HFD-treated rats had significantly increased TG levels compared with normal diet-treated rats. However, treatment with curcumin, both at low and high doses, reduced both TG levels and liver steatosis, which was demonstrated by the decreased visualization of lipid droplets following H&E staining of liver tissue sections (P<0.01; Fig. 3A and C). Under higher magnification, large cytoplasmic lipid droplets were observed throughout the sections (>90%) in the HFD rat liver, but only ~75% fatty infiltration in the low dose and 55% in the high dose curcumin groups was observed (Fig. 3B).
Figure 3.
Analysis of liver steatosis following curcumin treatment in HFD rats. (A) Liver morphological appearance (hematoxylin and eosin staining, ×200). (B) Semi-quantitative scoring of steatosis in histological samples. (C) Hepatic tissue TG levels. Data are presented as the mean ± standard deviation. **P<0.01 vs. control group; ∆∆P<0.01 vs. HFD group. HFD, high fat diet; TG, triglyceride.
Curcumin regulates the expression of genes involved in hepatic lipid metabolism and inflammation
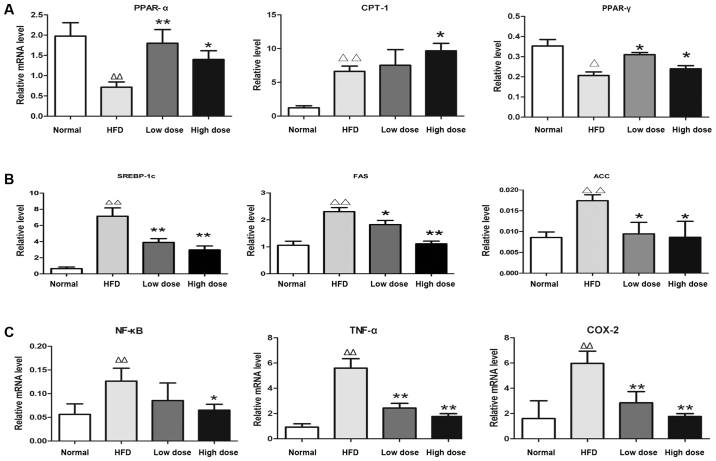
In order to investigate the underlying mechanism of curcumin in NAFLD, the expression of genes associated with hepatic lipid metabolism, including those involved in fatty acid oxidation [peroxisome proliferator-activated receptor-γ (PPAR-γ), peroxisome proliferator-activated receptor-α (PPAR-α) and carnitine palmitoyltransferase 1 (CPT-1)], lipogenesis [sterol regulatory element-binding protein (SREBP-1c), acetyl-CoA carboxylase (ACC) and fatty acid synthase (FASN)] and inflammation [nuclear factor-κB (NF-κB), tumor necrosis factor-α (TNF-α) and prostaglandin-endoperoxide synthase 2 (COX-2)] were investigated using RT-qPCR. It was revealed that curcumin treatment enhanced mRNA expression of PPAR-α, CPT-1 and PPAR-γ (Fig. 4A), whilst suppressing mRNA expression of SREBP-1c, ACC and FASN in the liver compared with HFD rats (Fig. 4B). Thus, this suggested that curcumin administration improves hepatic lipid metabolism in rats via promotion of fat degradation and suppression of lipogenesis. Furthermore, the expression levels of inflammation-associated genes NF-κB, TNF-α and COX-2 were reduced by curcumin treatment compared with the HFD rats (Fig. 4C), therefore demonstrating the anti-inflammatory effect of curcumin on rats.
Figure 4.
Effects of curcumin on mRNA expression in the livers of HFD rats. The results are presented as (A) lipolysis-associated genes, (B) lipogenesis-associated genes and (C) inflammation-associated genes. *P<0.01, **P<0.01 compared with the control group; ∆P<0.05, ∆∆P<0.01 compared with the HFD group. HFD, high fat diet; PPAR-α, peroxisome proliferator-activated receptor-α; CPT-1, carnitine palmitoyltransferase 1; PPAR-γ, peroxisome proliferator-activated receptor-γ; SREBP-1c, sterol regulatory element-binding protein; FAS, fatty acid synthase; ACC, acetyl-CoA carboxylase; NF-κB, nuclear factor-κB; TNF-α, tumor necrosis factor-α; COX-2, prostaglandin-endoperoxide synthase 2.
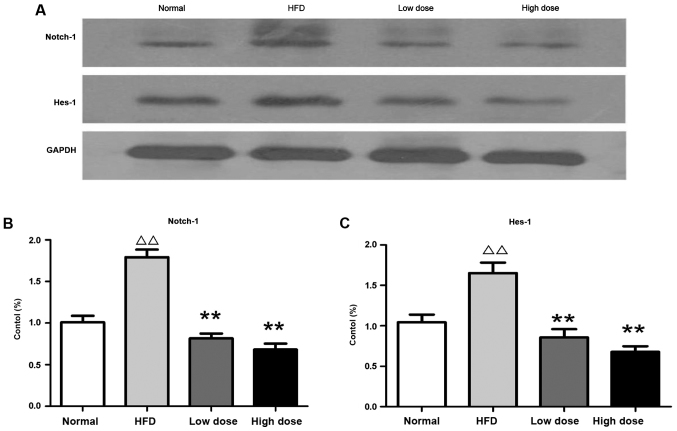
Curcumin reduces hepatic Notch signaling protein expression
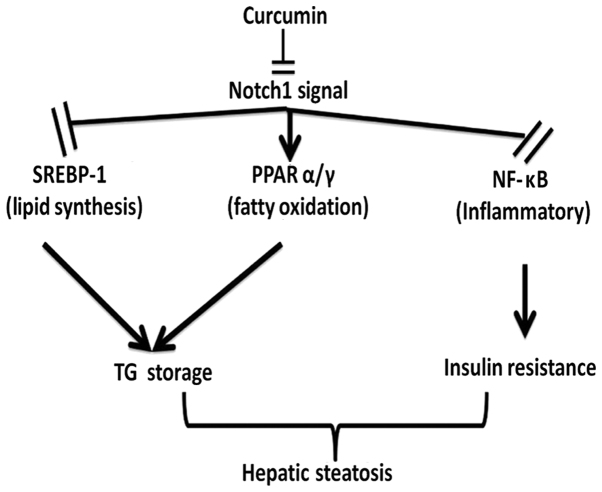
The protein expression of Notch-1 and Hes-1 (which are the primary Notch-1 target genes that regulate cell fate decision) were investigated using western blotting. It was revealed that Notch-1 and Hes-1 were upregulated in the liver of HFD rats compared with the normal controls, whereas curcumin treatment reversed this effect in a dose-dependent manner (Fig. 5). Therefore, the results suggest that curcumin may ameliorate fatty liver and insulin resistance via downregulation of Notch signaling (Fig. 6).
Figure 5.
Effects of curcumin on protein expression in the liver of HFD rats. (A) Expression of Notch-1 and Hes-1 was detected using western blotting. (B) Relative densitometry analysis of Notch-1 expression (expressed as a percentage). (C) Relative densitometry analysis of Hes-1 (expressed as a percentage). ∆∆P<0.05 vs. control group; **P<0.01 vs. HFD group. HFD, high fat diet; HES-1, transcription factor HES-1.
Figure 6.
Potential mechanisms for curcumin-induced suppression of Notch-1 signaling and amelioration of fatty liver. SREBP-1, sterol regulatory element binding proten; PPAR, peroxisome proliferator-activated receptor; NF-κB, nuclear factor κB; TG, triglyceride.
Discussion
Several important results were demonstrated in the present study: Curcumin suppresses serum CHO and TG levels, reduces visceral fat accumulation and abdominal obesity, increases insulin sensitivity and attenuates HFD-induced hepatic steatosis.
NAFLD is always coupled with insulin resistance and other metabolic abnormalities (17), and liver fat synthesis and lipolysis imbalance are associated with these pathological processes. Various transcription factors are also involved in NAFLD pathogenesis, including SREBP-1c, FAS, ACC and PPARs. SREBP-1c is a key pathogenic factor in NAFLD, and is responsible for the suppression of ACC and FAS expression, which are involved in fat synthesis and TG aggregation (18). The PPAR family of nuclear receptors is composed of three family members: PPAR-α, PPAR-β and PPAR-γ. The activation of PPARs results in the proliferation of peroxisomes and subsequent upregulation of genes, such as CPT-1, leptin and the insulin receptor; and thereby attenuates adipogenesis and insulin resistance (19).
In order to determine the therapeutic mechanism of curcumin with regards to the amelioration of metabolism disturbance and NAFLD, the expression levels of lipogenic and lipolytic genes were investigated. The results of these analyses revealed that curcumin upregulated the expression of PPAR-α, CPT-1 and PPAR-γ lipolytic genes (P<0.05 or P<0.01); and downregulated the expression of SREBP-1c, FAS and ACC lipogenic genes in HFD rats (P<0.05). The expression levels of inflammation-associated genes (NF-κB, COX-2 and TNF-α) were also downregulated compared with HFD rats following administration of curcumin. Therefore, the results of the present study suggest that curcumin suppresses the synthesis of fatty acids, enhances lipolysis and decreases inflammation, which all contribute to amelioration of fatty liver and insulin resistance.
Notch signaling is an important transduction pathway for cell fate decisions. Cell surface ligands bind to neighboring receptors, which result in the formation of active notch intracellular domains (NICD) (20). NICDs translocate from the membrane to the nucleus, and then interact with the recombining binding protein suppressor of hairless DNA-binding protein, in order to promote the formation of an active complex. This formation leads to the transcription of Hes-1 and hairy/enhancer-of-split related with YRPW motif protein 2, which are Notch signaling pathway targets that mediate transcriptional control of downstream genes (21).
The Notch pathway is always involved in the pathological process underlying fatty liver, and the major Notch signaling proteins, Notch-1 and Hes-1, are commonly implicated. Notch-1 expression has previously been reported to be upregulated in obese mice (22). Previous studies have revealed that hepatic Notch signaling is upregulated and positively correlates with insulin resistance and fatty liver in humans and other mammals (23). Furthermore, it has also been demonstrated that Notch gain-of-function causes fatty liver, and is correlated with an increasing rate of SREBP-1c-mediated lipogenesis (24). In addition, pharmacological inhibition of the Notch pathway via a γ-secretase inhibitor has been revealed to reduce obesity, increase glucose tolerance and improve insulin sensitivity in diet-induced obesity in mice (25). Furthermore, silencing of Notch genes via RNA interference was demonstrated to inhibit the fatty degeneration of liver cells (24). In addition, it has previously been revealed that Notch-1 can inhibit adipogenic differentiation via suppression of PPAR-γ and PPAR-α expression levels (26). The small interfering RNA-mediated suppression of Hes-1 expression can also inhibit adipogenesis (27). The results of these studies suggest that Notch signaling boosts lipogenesis and inhibits lipolysis via regulation of the expression of SREBP-1 and PPAR-α/γ transcription factors.
In the present study, an increased level of Notch-1 in fatty liver of rats was identified, coupled with the upregulation of Hes-1 expression, and downregulation of PPAR-α/γ expression. By contrast, expression of the SREBP-1, FAS and ACC lipolysis genes were upregulated in the fatty livers of rats. The results of the present study suggest that administration of curcumin inhibits Notch-1 signaling, downregulates SREBP-1 expression, and suppresses the expression of FAS and ACC lipogenesis genes in the liver. Furthermore, the present study also demonstrated that curcumin promotes expression of the PPAR-α/γ and CPT-1 lipolysis genes in rats, which reduces the development of hyperglycemia and fatty liver.
Fatty liver is occasionally accompanied by inflammation (28). Pro-inflammatory cytokines, such as TNF-α, can directly increase inflammation in liver tissues and impair insulin sensitivity. NF-κB is heavily involved in the regulation of inflammatory-associated gene expression in mammals. Activation of NF-κB can enhance the expression of pro-inflammatory cytokines, which are major factors in HFD-induced liver damage (29). High expression of Notch-1 upregulates NF-κB transcriptional activity; whereas downregulation of Notch-1 suppresses NF-κB expression (30). The present study demonstrated that curcumin can attenuate liver injury, inhibit NF-κB activation and suppress downstream activation of COX-2 and TNF-α.
With regards to the aforementioned results, the current study has revealed that curcumin can downregulate Notch signaling and differentially regulate SREBP-1, PPAR-α/γ and NF-κB transcription factors; which may contribute to the therapeutic amelioration of fatty liver and insulin resistance produced by curcumin. The results of the present study therefore suggest that the Notch-1 pathway may represent an attractive therapeutic target for the treatment of insulin resistance and fatty liver induced by a HFD.
Although the underlying mechanisms of the curcumin-induced effects in fatty liver and insulin resistance are complex, the present study, to the best of our knowledge, is the first to demonstrate that the Notch signaling pathway may be involved in the pathogenesis of fatty liver and insulin resistance, and thus represent a potential novel therapeutic target. Further research is required to validate the therapeutic potential of the Notch signaling pathway as a target for the treatment of NAFLD and insulin resistance.
Acknowledgements
The present study was supported by the Science Foundation of Shanghai (grant no. 15401932900), the Natural Science Foundation of Xiamen City (grant no. 3502Z20154011), and the National Natural Science Foundation of China (grant no. 81503579).
References
- 1.Chartoumpekis DV, Palliyaguru DL, Wakabayashi N, Khoo NK, Schoiswohl G, O'Doherty RM, Kensler TW. Notch intracellular domain overexpression in adipocytes confers lipodystrophy in mice. Mol Metab. 2015;4:543–550. doi: 10.1016/j.molmet.2015.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carr RM, Oranu A, Khungar V. Nonalcoholic fatty liver disease: Pathophysiology and Management. Gastroenterol Clin North Am. 2016;45:639–652. doi: 10.1016/j.gtc.2016.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schwanbeck R. The role of epigenetic mechanisms in Notch signaling during development. J Cell Physiol. 2015;230:969–981. doi: 10.1002/jcp.24851. [DOI] [PubMed] [Google Scholar]
- 4.Lu J, Xia Y, Chen K, Zheng Y, Wang J, Lu W, Yin Q, Wang F, Zhou Y, Guo C. Oncogenic role of the Notch pathway in primary liver cancer. Oncol Lett. 2016;12:4278. doi: 10.3892/ol.2016.4609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gridley T, Kajimura S. Lightening up a notch: Notch regulation of energy metabolism. Nat Med. 2014;20:811–812. doi: 10.1038/nm.3650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim GW, Lin JE, Blomain ES, Waldman SA. Antiobesity pharmacotherapy: New drugs and emerging targets. Clin Pharmacol Ther. 2014;95:53–66. doi: 10.1038/clpt.2013.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kuo JJ, Chang HH, Tsai TH, Lee TY. Positive effect of curcumin on inflammation and mitochondrial dysfunction in obese mice with liver steatosis. Int J Mol Med. 2012;30:673–679. doi: 10.3892/ijmm.2012.1049. [DOI] [PubMed] [Google Scholar]
- 8.Sha J, Li J, Wang W, Pan L, Cheng J, Li L, Zhao H, Lin W. Curcumin induces G0/G1 arrest and apoptosis in hormone independent prostate cancer DU-145 cells by down regulating Notch signaling. Biomed Pharmacother. 2016;84:177–184. doi: 10.1016/j.biopha.2016.09.037. [DOI] [PubMed] [Google Scholar]
- 9.Hackler L, Jr, Ózsvári B, Gyuris M, Sipos P, Fábián G, Molnár E, Marton A, Faragó N, Mihály J, Nagy LI, et al. The Curcumin Analog C-150, Influencing NF-κB, UPR and Akt/notch pathways has potent anticancer activity in vitro and in vivo. PLoS One. 2016;11:e0149832. doi: 10.1371/journal.pone.0149832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang Z, Zhang Y, Banerjee S, Li Y, Sarkar FH. Notch-1 down-regulation by curcumin is associated with the inhibition of cell growth and the induction of apoptosis in pancreatic cancer cells. Cancer. 2006;106:2503–2513. doi: 10.1002/cncr.21904. [DOI] [PubMed] [Google Scholar]
- 11.Yang Y, Duan W, Liang Z, Yi W, Yan J, Wang N, Li Y, Chen W, Yu S, Jin Z, Yi D. Curcumin attenuates endothelial cell oxidative stress injury through Notch signaling inhibition. Cell Signal. 2013;25:615–629. doi: 10.1016/j.cellsig.2012.11.025. [DOI] [PubMed] [Google Scholar]
- 12.Leclercq IA, Farrell GC, Sempoux C, dela Peña A, Horsmans Y. Curcumin inhibits NF-kappaB activation and reduces the severity of experimental steatohepatitis in mice. J Hepatol. 2004;41:926–934. doi: 10.1016/j.jhep.2004.08.010. [DOI] [PubMed] [Google Scholar]
- 13.Liu ZC, Yang ZX, Zhou JS, Zhang HT, Huang QK, Dang LL, Liu GX, Tao KS. Curcumin regulates hepatoma cell proliferation and apoptosis through the Notch signaling pathway. Int J Clin Exp Med. 2014;7:714–718. [PMC free article] [PubMed] [Google Scholar]
- 14.Feng XT, Tang SY, Jiang YX, Zhao W. Anti-diabetic effects of zhuoduqing formula, a chines herbal decoction, on a rat model of type 2 diabetes. Afr J Tradit Complement Altern Med. 2017;14:42–50. doi: 10.21010/ajtcam.v14i3.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ha SK, Chae C. Inducible nitric oxide distribution in the fatty liver of a mouse with high fat diet-induced obesity. Exp Anim. 2010;59:595–604. doi: 10.1538/expanim.59.595. [DOI] [PubMed] [Google Scholar]
- 16.Livak KJ1, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 17.Lallukka S, Yki-Järvinen H. Non-alcoholic fatty liver disease and risk of type 2 diabetes. Best Pract Res Clin Endocrinol Metab. 2016;30:385–395. doi: 10.1016/j.beem.2016.06.006. [DOI] [PubMed] [Google Scholar]
- 18.Kohjima M, Higuchi N, Kato M, Kotoh K, Yoshimoto T, Fujino T, Yada M, Yada R, Harada N, Enjoji M, et al. SREBP-1c, regulated by the insulin and AMPK signaling pathways, plays a role in nonalcoholic fatty liver disease. Int J Mol Med. 2008;21:507–511. [PubMed] [Google Scholar]
- 19.Gross B, Pawlak M, Lefebvre P, Staels B. PPARs in obesity-induced T2DM, dyslipidaemia and NAFLD. Nat Rev Endocrinol. 2016;13:36–49. doi: 10.1038/nrendo.2016.135. [DOI] [PubMed] [Google Scholar]
- 20.Brzozowa-Zasada M, Piecuch A, Dittfeld A, Mielańczyk Ł, Michalski M, Wyrobiec G, Harabin-Słowińska M, Kurek J, Wojnicz R. Notch signalling pathway as an oncogenic factor involved in cancer development. Contemp Oncol (Pozn) 2016;20:267–272. doi: 10.5114/wo.2016.61845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cai Z, Zhao B, Deng Y, Shangguan S, Zhou F, Zhou W, Li X, Li Y, Chen G. Notch signaling in cerebrovascular diseases (Review) Mol Med Rep. 2016;14:2883–2898. doi: 10.3892/mmr.2016.5641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Valenti L, Mendoza RM, Rametta R, Maggioni M, Kitajewski C, Shawber CJ, Pajvani UB. Hepatic notch signaling correlates with insulin resistance and nonalcoholic fatty liver disease. Diabetes. 2013;62:4052–4062. doi: 10.2337/db13-0769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lai PY, Tsai CB, Tseng MJ. Active form Notch4 promotes the proliferation and differentiation of 3T3-L1 preadipocytes. Biochem Biophys Res Commun. 2013;430:1132–1139. doi: 10.1016/j.bbrc.2012.12.024. [DOI] [PubMed] [Google Scholar]
- 24.Pajvani UB, Shawber CJ, Samuel VT, Birkenfeld AL, Shulman GI, Kitajewski J, Accili D. Inhibition of Notch signaling ameliorates insulin resistance in a FoxO1-dependent manner. Nat Med. 2011;17:961–967. doi: 10.1038/nm.2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pajvani UB, Qiang L, Kangsamaksin T, Kitajewski J, Ginsberg HN, Accili D. Inhibition of Notch uncouples Akt activation from hepatic lipid accumulation by decreasing mTorc1 stability. Nat Med. 2013;19:1054–1060. doi: 10.1038/nm.3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ba K, Yang X, Wu L, Wei X, Fu N, Fu Y, Cai X, Yao Y, Ge Y, Lin Y. Jagged-1-mediated activation of notch signalling induces adipogenesis of adipose-derived stem cells. Cell Prolif. 2012;45:538–544. doi: 10.1111/j.1365-2184.2012.00850.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lei T, Bi Y, Gao MJ, Gao SM, Zhou LL, Zheng HL, Chen XD. HES1 inhibits adipogenesis of porcine mesenchymal stem cells via transcriptional repression of FAD24. Domest Anim Endocrinol. 2013;45:28–32. doi: 10.1016/j.domaniend.2013.03.003. [DOI] [PubMed] [Google Scholar]
- 28.Musso G, Cassader M, Cohney S, De Michieli F, Pinach S, Saba F, Gambino R. Fatty liver and chronic kidney disease: Novel mechanistic insights and therapeutic opportunities. Diabetes Care. 2016;39:1830–1845. doi: 10.2337/dc15-1182. [DOI] [PubMed] [Google Scholar]
- 29.Yao L, Kan EM, Kaur C, Dheen ST, Hao A, Lu J, Ling EA. Notch-1 signaling regulates microglia activation via NF-κB pathway after hypoxic exposure in vivo and in vitro. PLoS One. 2013;8:e78439. doi: 10.1371/journal.pone.0078439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu Y, Su C, Shan Y, Yang S, Ma G. Targeting Notch1 inhibits invasion and angiogenesis of human breast cancer cells via inhibition Nuclear Factor-κB signaling. Am J Transl Res. 2016;8:2681–2692. [PMC free article] [PubMed] [Google Scholar]