Introduction
Stroke is a leading cause of death and disability across the globe, affecting 15 million people each year1. Stoke represents an archetypical common complex disease with both genetic and environmental determinants2, 3 playing a role in its occurrence. The proportion of stroke risk that can be attributed to genetic variation has been estimated to be 30%4–6. While this estimate provides an indication of the overall importance of genetic variation in stroke, the key to developing new treatment strategies is to identify the specific genetic variants (mutations) that modify an individual’s risk of stroke. Genetic association studies seek to identify these variants and link them to specific genes, which, in turn, point to specific cellular processes to become therapeutic targets for drug development. In addition, newly discovered genetic risk loci can be utilized to improve existing phenotyping systems, enhance prediction tools aimed to identify high risk patients, and aid in establishing causality for associations involving non-genetic exposures.
Successfully identifying the range of genetic variants that cause stroke and leveraging these discoveries to reduce the suffering caused by this condition requires overcoming several key challenges. First, stroke is the final result of multiple different pathological processes and must therefore be accurately subtyped in order to identify underlying biology. Second, because large numbers of cases and controls are required to identify the culprit genetic variants, tens (even hundreds) of thousands of cases must be studied, requiring the collaboration of multiple centers, many of which utilize different ascertainment methods and criteria. Third, because genetic variation differs across the globe, representative populations from all ethnicities must be studied. Finally, all these data must be shared rapidly and widely in order to ensure the most rapid progress in research and enable investigators with the brightest ideas to utilize these data provided by patients to facilitate stroke research. The Cerebrovascular Disease Knowledge Portal (CDKP) has been created by the International Stroke Genetics Consortium to overcome these challenges and enable data sharing as freely and rapidly as possible.
The evolution of the revolution: open access to data from high throughput technologies
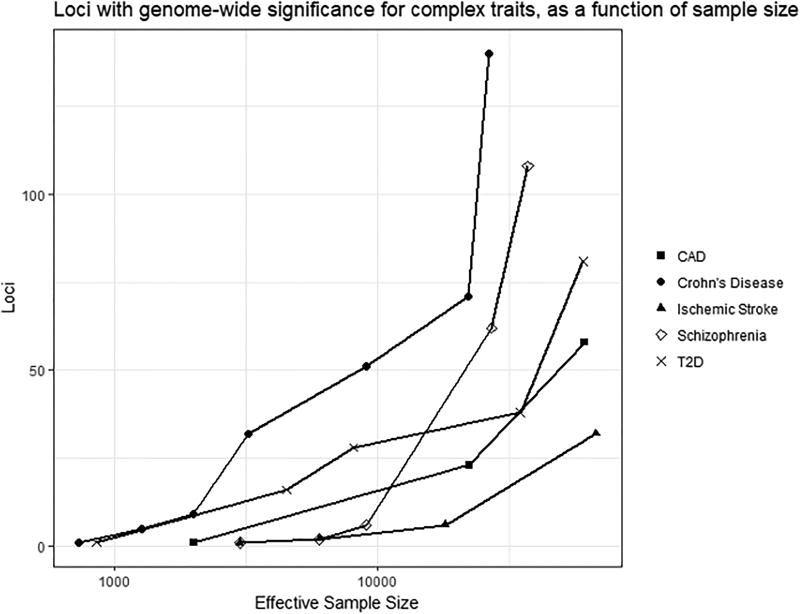
In the past decade, the introduction of high throughput genotyping technologies revolutionized the way we think about, and work with, genomic data. Genome-wide genotyping arrays, whole exome sequencing, and whole genome sequencing allow us collect massive amounts of common and rare genetic data from the entire genome, and agnostically test these variants for association with human disease. These new technologies have been potentiated by the creation of large collaboration networks that allow the assembly of sample sizes unimaginable not long ago. Importantly, the experience accumulated with other complex diseases indicates that the field of stroke genomics is now approaching a breakthrough point. Concretely, as the overall sample size for stroke approximates the order of 100,000 cases, and sequencing technologies are applied to existing samples, it is expected that the discovery pace for new susceptibility loci will accelerate exponentially (Figure 1)7–11. This discovery power will be especially helpful in successfully identifying low frequency and rare mutations as well as hard-to-detect risk alleles for specific stroke subtypes and ethnic groups12. Formed in 2007, the International Stroke Genetics Consortium (ISGC) was created to bring together clinicians, investigators, geneticists and statisticians to assemble the sample sizes and expertise necessary to understand the genetic underpinnings of cerebrovascular disease and leverage these discoveries to help patients13. Through its launch of collaborative studies including METASTROKE, the NINDS-SiGN Consortium and MEGASTROKE, and its partnership with the CHARGE consortium14, 15, the ISGC has been responsible for most of the confirmed stroke risk loci discovered in the era of GWAS16.
Figure 1.
Number of genome-wide significant loci for different complex traits.
Open data access is an opportunity to accelerate scientific discoveries in stroke. The CDKP is a comprehensive web-based resource that enables access and exploration of genetic and phenotypic data related to cerebrovascular diseases (available at www.cerebrovascularportal.org). The mission of the CDKP is to potentiate stroke research by democratizing access to high-quality genetic, phenotypic and imaging data on large numbers of stroke patients. To accomplish this mission, the CDKP collects, securely stores, harmonizes, displays and shares stroke genomic data with investigators from around the world. The CDKP houses two substantially different types of data: summary level data and individual level data. Summary level data (summary statistics) consist of full sets of association results by Single Nucleotide Polymorphism (SNP) that form the basis of published genetic association studies; they generally cannot be used to identify individual subjects. Individual level data consist of complete datasets containing phenotypic and genotypic information by subject. Subjects must give broad consent to allow for the sharing of their individual level data, in part because of the risk of identifiability.
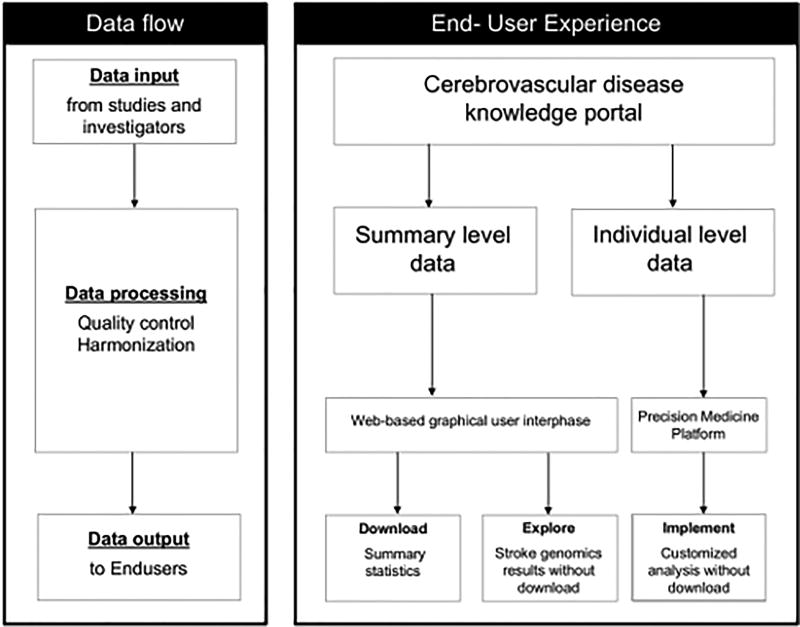
These two types of data are accessed through different paths (Figure 2). Summary level are accessed through the CDKP web-based platform itself, while the CDKP will direct the user to the American Heart Association Precision Medicine Platform (or the PMP, www.precision.heart.org) to access individual level data. Within the PMP, individual-level data is hosted in a secure environment that allows investigators to implement complex genetic analyses without downloading the data. These analyses are run in a cloud computing-based workspace powered by Amazon Web Services. This workspace is simple to use and was specifically designed to guide and teach first time users on how to migrate to a cloud-based ecosystem. New data and tools are constantly being incorporated to both the CDKP and the PMP.
Figure 2.
Data access through the CDKP. Data flow describes the generalized path that all data take through the CDKP. The End-User Experience details how a user interacts with the resources. Summary level data can be access through the CDKP’s web based GUI designed to run queries and allow for download of data while Individual level data can be accessed through the PMP to run custom analysis.
Here, we describe the design and available features of this new resource, emphasizing opportunities and challenges that may arise in the near future.
Design and development of the CDKP
The creation of the CDKP, funded by the National Institute of Neurological Disorders and Stroke, has been a joint effort of the ISGC, the Broad Institute, and the American Heart Association Institute for Precision Cardiovascular Medicine. Given its extensive experience with similar projects, the Broad Institute hosts the data available through the platform and developed the informatic framework and analytical tools utilized by the CDKP. A dedicated steering committee, consisting of ISGC members who have contributed data to the portal, oversees the CDKP. An Operations Committee, consisting of ISGC members and Broad Institute staff, is responsible for the everyday operations of the CDKP. In addition, the ISGC has created a Data Access Committee that is responsible for ensuring access to all data. Any investigator wishing to contribute data to the CDKP can contact the Operations Committee at cerebrovascular.disease.portalgmail.com. Several different types of data can be deposited in the CDKP, including summary statistics, raw individual level genetic data, and post processing (e.g, post quality control and imputation) individual level genetic data. Furthermore, as the CDKP evolves, there will be opportunities to deposit a broad range of phenotypic data, including imaging. Prior to transfer to the CDKP, proper IRB and legal approval for each dataset is required, including assurance from the originating institution that all subjects are consented for this type of data sharing. Datasets transferred to the portal are harmonized following a standardized pipeline that evaluates the presence of important data points (such as a unique identifier for each subject) and identifies systematic errors. Phenotype-genotype harmonization takes place at the Broad Institute, which serves as the Data Coordinating Center. Individual level data is subsequently transferred to the secure working space of the PMP.
Specific tools available through the Cerebrovascular Disease Knowledge Portal
Within the CDKP and PMP, genomic data can be accessed through three main tools: (1) a graphical user interphase (GUI) that allows quick-and-easy exploration of both individual and summary level data contained in the portal; (2) a repository of full sets of summary level data produced by landmark published studies in the field; and (3) a repository of individual level data that investigators can analyze directly in a secure workspace (Figure 2).
Web-based graphical user interphase
The GUI offers users a wide menu of integrated tools for data visualization and analysis, as well as the possibility to implement on-the-fly descriptive and association analyses focused on specific items of interest: base pair positions, genomic regions, genes, or SNPs, to name a few possibilities. Users can also explore descriptive and association results from GWAS of other related (hypercholesterolemia, hypertension, diabetes, coronary artery disease) and unrelated traits (psychiatric conditions). Figures, tables and results generated with this tool can be easily exported and downloaded. This functionality does not require a formal request, although users are required to register to utilize the portal.
Summary level data (summary statistics)
The CDKP contains full sets of summary statistics from landmark published studies. Because all association results are provided without filtering for specific p-value thresholds, several million results are available for each specific study. These summary level data can be accessed through a single mouse click, without submitting a formal application. The rationale for this strategy is to encourage and accelerate preliminary analyses to evaluate different working hypotheses. By providing summary level results from published studies only, users can easily identify the methods and study populations that were used to generate the summary statistics. While the majority of currently available stroke data comes from subjects of European descent, data from populations of African-American, African, and Asian descent are scheduled to be added in the coming year. Each set of summary statistics has an accompanying document that describes the rules and limitations of use, including: (1) data are provided as-is, meaning each user is responsible for the design, implementation and interpretation of any analyses based on this summary level data; (2) commercial use of these data is prohibited (including deposition of these data into commercial databases); (3) cohorts of stroke cases and matched controls are usually used by more than one study; consequently, is the user’s responsibility to check for possible overlaps when using summary statistics from different studies; and (4) scientific communications based on CDKP data must acknowledge the use of the portal. Currently, the CDKP contains full sets of results from 4 landmark papers (Table 1), and this list will rapidly grow in coming months.
Table 1.
Summary level data (summary statistics) contained in the ISGC CDKP.
| Reference Number |
Author | Year | Phenotype | Discovered Loci | Title |
|---|---|---|---|---|---|
| PMC3980413 | Woo, et al. | 2014 | Intracerebral Hemorrhage | TRHDE PMF1/SLC25A44 | Meta-analysis of Genome-wide Association Studies Identifies 1q22 as a Susceptibility Locus for Intracerebral Hemorrhage |
| PMC4818561 | Malik, et al. | 2016 | Ischemic Stroke | ABO HDAC9 PITX2 ZFHX3 | Low-frequency and common genetic variation in ischemic stroke: The METASTROKE collaboration |
| PMC3490334 | Traylor, et al. | 2012 | Ischemic Stroke | HDAC9 PITX2 ZFHX3 | Genetic risk factors for ischaemic stroke and its subtypes (the METASTROKE collaboration): a meta-analysis of genome-wide association studies. |
| PMC4912948 | SiGN, et al. | 2015 | Ischemic Stroke | TSPAN2 HDAC9 PITX2 ZFHX3 ALDH2 | Loci associated with ischaemic stroke and its subtypes (SiGN): a genome-wide association study. |
Individual level data
Through the PMP, the CDKP also allows users to search, discover and analyze individual level data directly (Table 2). For administrative, legal and security reasons, this option requires submission of a research plan as well as approval by the CDKP Data Acces Committee. Once access to the data is granted, users are able to analyze data in the secure cloud-based computational workspace provided by the PMP. This secure workspace provides users with access to dataset(s) for which authorization has been granted; while results can be easily downloaded, the individual level data never leaves the PMP (e.g., cannot be downloaded). The PMP workspace offers powerful cloud-based cluster computing capabilities through a script driven environment that utilizes Jupyter Notebooks (www.jupyter.org), an open-source web application that allows users to create and share script documents to run different kernels – or computer languages - in the cloud (https://precision.heart.org/ICH_GWAS_2014_notebook.html). Commands contained in these scripts are submitted to the cloud computing system, where they are executed in parallel utilizing the appropriate kernels.
Table 2.
Individual Level data available through the CDKP.
| Cohort | Site | PI | Upload Date |
Cases | Controls | Phenotypes | Platform used for genotyping |
|---|---|---|---|---|---|---|---|
| ICH GWAS I | Boston, MA, USA | Jonathan Rosand | 2016 | 316 | 457 |
|
Illumina HumanHap610-Quad |
| ICH GWAS I | Cincinnati, OH, USA | Daniel Woo | 2016 | 797 | 539 |
|
Affymetrix 6.0 |
| GERFHS III | Cincinnati, OH, USA | Daniel Woo | 2016 | 628 | 856 |
|
Affymetrix 6.0 |
| VISP | Winston-Salem, North Carolina, USA | Bradford Worrall | 2017 | 2100 | 0 |
|
Illumina HumanOmni1-Quad_v1-0_B BeadChip |
Interaction with existing data resources
The CDKP seeks to complement other related platforms, like the Database of Genotypes and Phenotypes (dbGaP) and the European Genome-phenome Archive (EGA).In contrast to these repositories, the CDKP specializes in capturing genomic data related to cerebrovascular diseases and is not limited to a specific country or continent. In addition, the CDKP has been specifically designed to provide fast and easy access to stroke genomic information, without specific requirements (other than registering a name and email address) to browse results and download summary statistics, and with a brief and efficient application process to access individual level data. The primary advantage in using the CDKP is that data have already been harmonized and cerebrovascular disease specific phenotypes have been gathered. This allows the CDKP data to be immediately ready for analysis upon access. Further, cerebrovascular disease is not one single condition, but a collection of unique sub-types, each with a distinct biology and genetic risk. Therefore, to properly study cerebrovascular disease, more samples than usual and especial attention to phenotyping are needed to account for aforementioned biological heterogeneity observed across the different cerebrovascular disease subtypes.
Problems and Solutions
The creation of international collaborative data platforms is not straightforward. A number of challenges arose during the design, development and deployment of the CDKP. One important barrier encountered was the variability in approval processes and required assurances, especially when comparing institutions located in different countries. The differences in governance structure and IRBs that exist internationally are something that must be dealt with on a case by case and country by country basis.
Administrative and legal barriers exist for both summary and individual level data. For summary level data, the main barrier is that some studies explicitly prohibit the use of results by commercial users. This limitation is clearly explained in the README document that accompanies each downloadable dataset that describes the conditions of use for each set of summary statistics available on the portal. For individual level data, the main barriers are: (1) the possibility that users may try to use genetic data to identify study subjects; and (2) that users may use the data for different purposes than those described in their application for access. The CDKP addresses both risks by providing access to individual level data through the PMP, a dedicated cloud-based workspace that allows the implementation of sophisticated analyses without downloading data (e.g., individual level data never leaves the workspace) and offers the possibility to fully monitor the work undertaken by each user.
Another identified challenge is how to integrate datasets with overlapping individuals, an issue with which the field continues to struggle17, 18. This challenge is faced when working with multiple consortia, which often contain overlapping subsets of participating subjects. A number of analytical tools have been developed to handle this problem when individual level data are available.18 In the cases of summary statistics, when overlap between two datasets is unknown, linkage disequilibrium score regression can be implemented to estimate the redundancy across multiple groups, although this approach can be inaccurate when working with small sample sizes. We envision that the widespread availability of summary and individual level data related to stroke genomics will propel research on this and other technical and analytical problems. Figure 3 displays the overlap across summary level data currently available in the CDKP.
Figure 3.
Subject overlap across the datasets within the CDKP.
Future of the CDKP
Two windows exist for individuals interested in search and discoverability as well as analysis of stroke data: the CDKP (www.cerebrovascularportal.org) and the PMP (www.precision.heart.org). Each window provides complementary services that will accelerate stroke research. We anticipate the addition of multiple new analytical and data resources. Ten additional landmark papers have already been identified by the CDKP steering committee and conversations are ongoing to deposit their summary results in the portal. In addition, as the ISGC’s official data sharing portal, the CDKP, will contain, moving forward, summary and individual level data from new studies conducted by the consortium. As an important example, the ISGC will share, immediately after publication, the full set of association results of the MEGASTROKE Collaboration, the largest genetic study of stroke conducted to date including more than 65,000 cases and 450,000 controls.
An important priority in coming months will be to expand the CDKP and PMP to include new types of phenotypic data. Most of the summary and individual level genetic data currently contained by the portal pertains to stroke risk: either summary statistics from case/control analyses or full datasets of matched cases and controls. Plans are underway to move beyond risk analyses to incorporate information related to clinical status during admission, neuroimaging biomarkers and functional outcome, extending the range of scientific questions to be tackled within the CDKP’s framework. In addition to clinical and radiological data points, we will also endeavor to enrich the portal with other omics data, including transcriptomics, metabolomics and proteomics.
The guiding principle for the future development of the CDKP and PMP is to meet the needs and interests of the portal’s users. For this reason, we will deploy several tools aimed at establishing a fluent communication with those utilizing the resource, and will strive to integrate the users’ feedback to make decisions on future steps.
Acknowledgments
We would like to recognize all members of the technical teams at the Broad Institute and American Heart Association Institute for Cardiovascular Precision Medicine.
Sources of Funding
The CKDP was launched with funding from the National Institute of Neurological Disorders and Stroke (R24 NS092983) to Dr. Rosand. Dr. Rosand is supported by the National Institute of Neurologic Disorders and Stroke (NS073344, NS093870). Dr. Falcone is supported by a Yale Pepper Scholar Award (P30AG021342) and the Neurocritical Care Society Research Fellowship. Dr. Malik is supported by the European Union Horizon2020 project CoSTREAM (grant agreement No 667375).
Footnotes
Conflicts of Interest / Disclosures
None.
References
- 1.The top 10 causes of death. World Health Organization; Jan, 2017. [Accessed March 15, 2017]. http://www.who.int/mediacentre/factsheets/fs310/en/ [Google Scholar]
- 2.Hankey GJ. Stroke. The Lancet. 2017;389:641–654. doi: 10.1016/S0140-6736(16)30962-X. [DOI] [PubMed] [Google Scholar]
- 3.Benjamin EJ, Blaha MJ, Chiuve SE, Cushman M, Das SR, Deo R, et al. Heart disease and stroke statistics—2017 update: A report from the american heart association. Circulation. 2017;135:e146–e603. doi: 10.1161/CIR.0000000000000485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bevan S, Traylor M, Adib-Samii P, Malik R, Paul NL, Jackson C, et al. Genetic heritability of ischemic stroke and the contribution of previously reported candidate gene and genomewide associations. Stroke. 2012;43:3161–3167. doi: 10.1161/STROKEAHA.112.665760. [DOI] [PubMed] [Google Scholar]
- 5.Traylor M, Rutten-Jacobs LC, Holliday EG, Malik R, Sudlow C, Rothwell PM, et al. Differences in common genetic predisposition to ischemic stroke by age and sex. Stroke. 2015;46:3042–3047. doi: 10.1161/STROKEAHA.115.009816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bluher A, Devan W, Holliday E, Nalls M, Parolo S, Bione S, et al. Heritability of young-and old-onset ischaemic stroke. European journal of neurology. 2015;22:1488–1491. doi: 10.1111/ene.12827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.(SiGN) NSGN, Consortium ISG. Loci associated with ischaemic stroke and its subtypes (sign): A genome-wide association study. The Lancet Neurology. 2016;15:174–184. doi: 10.1016/S1474-4422(15)00338-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jostins L, Ripke S, Weersma RK, Duerr RH, McGovern DP, Hui KY, et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491:119–124. doi: 10.1038/nature11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu JZ, Anderson CA. Genetic studies of crohn's disease: Past, present and future. Best Practice & Research. Clinical Gastroenterology. 2014;28:373–386. doi: 10.1016/j.bpg.2014.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schizophrenia Working Group of the Psychiatric Genomics C. Ripke S, Neale BM, Corvin A, Walters JTR, Farh K-H, et al. Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511:421–427. doi: 10.1038/nature13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wood AR, Esko T, Yang J, Vedantam S, Pers TH, Gustafsson S, et al. Defining the role of common variation in the genomic and biological architecture of adult human height. Nature genetics. 2014;46:1173–1186. doi: 10.1038/ng.3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Malik R, Traylor M, Pulit SL, Bevan S, Hopewell JC, Holliday EG, et al. Low-frequency and common genetic variation in ischemic stroke the metastroke collaboration. Neurology. 2016;86:1217–1226. doi: 10.1212/WNL.0000000000002528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Anderson CD, Boncoraglio G, Falcone G. Proceedings of the 19th and 20th international stroke genetics consortium workshops. Neurology Genetics. 2017;3:S1. [Google Scholar]
- 14.Traylor M, Farrall M, Holliday EG, Sudlow C, Hopewell JC, Cheng Y-C, et al. Genetic risk factors for ischaemic stroke and its subtypes (the metastroke collaboration): A meta-analysis of genome-wide association studies. The Lancet Neurology. 2012;11:951–962. doi: 10.1016/S1474-4422(12)70234-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chauhan G, Arnold CR, Chu AY, Fornage M, Reyahi A, Bis JC, et al. Identification of additional risk loci for stroke and small vessel disease: A meta-analysis of genome-wide association studies. The Lancet Neurology. 2016;15:695–707. doi: 10.1016/S1474-4422(16)00102-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Falcone GJ, Malik R, Dichgans M, Rosand J. Current concepts and clinical applications of stroke genetics. The Lancet Neurology. 2014;13:405–418. doi: 10.1016/S1474-4422(14)70029-8. [DOI] [PubMed] [Google Scholar]
- 17.Pasaniuc B, Price AL. Dissecting the genetics of complex traits using summary association statistics. Nature reviews. Genetics. 2017;18:117–127. doi: 10.1038/nrg.2016.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lin D-Y, Sullivan PF. Meta-analysis of genome-wide association studies with overlapping subjects. The American Journal of Human Genetics. 2009;85:862–872. doi: 10.1016/j.ajhg.2009.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]