Abstract
Background
Disease recurs rapidly in the majority of pancreatic cancer patients undergoing curative resection. In previous small studies, high expression of mismatch-repair protein MLH1 in pancreatic cancers was associated with better outcomes. We sought to validate the association between MLH1 expression and survival amongst subjects with resected pancreatic cancer receiving adjuvant chemoradiation.
Methods
Samples were obtained from the NRG Oncology RTOG 9704 prospective randomized trial (NCT00003216), that compared two adjuvant protocols in subjects with resected pancreatic cancers. Tissue microarrays (TMA) were prepared from formalin-fixed, paraffin embedded resected tumor tissue. MLH1 expression was quantified using fluorescence immunohistochemistry (IHC) and AQUAnalysis®. MLH1 expression levels were dichotomized above and below the median expression value.
Results
Immunohistochemistry staining was successfully performed on 117 patients for MLH1 (60 and 57 from the 2 arms). Characteristics from those with tissue were similar to the trial population as a whole. At time of analysis 84% of the participants had died, with a median survival of 17 months. Elevated MLH1 expression levels in tumor nuclei were significantly correlated with longer DFS and OS in each arm individually, and both arms combined. Two-year overall survival was 16% in those with low MLH1 expression levels and 53% for those with high levels, p<0.0001 (both arms combined). This association remained true on multivariate analysis allowing for nodal status (HR=0.41, CI 0.27–0.63, p<0.0001).
Conclusions
Within our sample, expression of MLH1 correlates with long-term survival. Further studies should assess whether MLH1 expression predicts which patients with localized pancreatic cancer benefit most from aggressive multi-modality treatment.
Keywords: Pancreatic Neoplasms, MutL Protein Homolog 1(MLH1), Chemotherapy, Adjuvant, Radiotherapy, Adjuvant, Clinical Trial Phase III, Biomarkers, Tumor
Introduction
Over 48,000 people are diagnosed with pancreatic cancer each year in the United States; overall five-year survival is seven percent.1 At diagnosis approximately 20% have resectable disease, yet even amongst these patients median survival is only 15 months due to the early development of regional and metastatic disease.2 There is an unmet need for identifying upfront the subset of patients that demonstrate prolonged survival and derive benefit from such an aggressive approach.
Adjuvant chemotherapy is the standard of care for resected pancreatic cancer.3 Additional adjuvant options include radiation therapy to the tumor bed and regional lymphatic drainage.4 NRG Oncology RTOG 9704 was a randomized phase III clinical trial comparing two different adjuvant protocols in patients with grossly resected pancreatic adenocarcinoma.5 All patients received concurrent 5FU-radiation (RT) therapy; in addition the control arm received 5FU prior to- and following RT, whereas the experimental arm received gemcitabine. Five-year overall survival (OS) was 18% and 22% in the 5FU and gemcitabine arms, respectively (not significant).6 On multivariate analysis, patients in the gemcitabine arm with pancreatic head tumors experienced a trend toward improved OS (p = 0.08).
The DNA mismatch repair (MMR) pathway corrects base substitution and insertion-deletion errors.7 MMR dysfunction leads to the cellular acquisition of both point mutations and the instability of long repetitive DNA sequences, i.e., microsatellite instability (MSI). Inherited MMR dysfunction is associated with increased rates of carcinogenesis (hereditary nonpolyposis colon cancer HNPCC or Lynch syndrome). 8 The MMR machinery has an additional function sensing DNA lesions, triggering cell cycle checkpoints and apoptosis.9
MutL homolog 1 (MLH1) is a pivotal member of the MMR pathway, with complete loss of MLH1 being the most common cause of microsatellite instability.10 Germline mutations in MLH1 are rare in pancreatic cancer, with estimated frequency of <1%.11 Likewise, whole-genome sequencing has demonstrated that structural somatic mutations in MLH1 are rare (about 2%), however single allelic loss is seen in about 9% of tumors.12 Whole exome sequencing of 15 pancreatic cancer derived cell lines showed expression of MLH1 protein to be decreased in cells with MLH1 allelic loss.13 Cells haplodeficient for MLH1 had decreased MLH1 expression and an increased number of indel mutations, however, they nonetheless tested negative on a PCR based-MSI assay. Hence it appears that even a relative decrease in MLH1 expression may impair DNA fidelity, without inducing MSI.
The clinical significance of decreased MLH1 expression in the absence of MSI is unknown; one small retrospective series found that elevated MLH1 expression was significantly associated with favorable differentiation, fewer lymph node metastases, and improved overall survival.14 Another study found increased MLH1 expression to be associated with high tumor differentiation, fewer lymph node metastasis, and tumor location.15
In this current study, it was hypothesized that resected pancreatic tumors with decreased MLH1 expression would demonstrate impaired MMR function, and resistance to adjuvant chemoradiation, influencing overall survival.
Methods
Inclusion criteria for NRG Oncology RTOG 9704 included non-metastatic histologically confirmed pancreatic adenocarcinoma that had undergone gross total tumor resection. Patients with poor performance status, inadequate organ function or previous cytotoxic treatments were excluded. Protocol therapy was required to begin 3 to 8 weeks after resection.
Specimen and biomarker analysis
Tissue microarrays (TMAs) with 0.6mm cores were prepared from standard paraffin blocks of tissue removed at time of operation (i.e. prior to delivery of systemic therapy). MLH1 was detected using primary monoclonal antibody (clone EPR3894, Epitomics, Burlingame, CA, USA) and visualized with EnVision+ (K4011, DAKO). Pan-cytokeratin was detected with polyclonal antibody (BP5069, Acris, San Diego, CA, USA) and visualized with Alexa-555 conjugated secondary antibody (A21435, Life Technologies, Burlington, ON, Canada).
Automated image acquisition was performed using the HistoRx PM-2000™ platform (Branford, CT, USA), and digital images were analyzed using the HistoRX AQUAnalysis® program, version 2.3.4.1 as described.16, 17 Briefly, high-resolution digital images were obtained for each TMA core using separate filters to define the nuclear (DAPI), tumor (Cy3), and MLH1 (Cy5) compartments. An analysis algorithm was constructed to generate a tumor-specific mask by thresholding the pan-cytokeratin images, and the tumor nuclear compartment was created by isolating DAPI-positive tumor nuclei within that area. The nuclear AQUA score (nAQUA, Supplemental Figure 1), representing MLH1 expression, is defined as the average Cy5 pixel intensity within the tumor nuclear area for each TMA core.
Unusable areas were removed before each image was processed using optimized threshold values. Images were validated according to the following: 1) >10% of the tissue area is pan-cytokeratin positive, 2) >50% of the image was usable (i.e. no overlapping or out of focus tissue), and 3) the thresholding produced accurate masked areas. An example of the staining for MLH1 is presented in Supplemental Figure 1. Of note, immunohistochemical staining and scoring was performed on blinded non-annotated specimens.
Statistical Methods
MLH1 expression data was forwarded to RTOG central office where the samples were matched with clinical outcomes, and survival analysis performed. Failure for overall survival (OS) was defined as death due to any cause and was measured from date of randomization to date of death or last follow-up for censored patients. Failure for disease-free survival (DFS) was defined as local, regional or distant relapse, appearance of a second primary lesion or death due to any cause and was measured from date of randomization to date of first failure or last follow-up for censored patients. The following baseline characteristics were dichotomized: pathological t-stage (T1, T2 vs. T3, T4) and AJCC stage (I, II vs. III, IV). Race was categorized as White vs. African American/other. Statistical comparisons to assess potential associations between baseline characteristics and MLH1 grouping were carried out using the chi-square or Fisher’s exact test. OS and DFS were estimated univariately with the Kaplan-Meier method and MLH1 grouping were compared using the log-rank test. Reported median survival and DFS times are the times (in years) at which 50% of the patients had failed.
Antibody staining for MLH1 expression was analyzed as a continuous variable and as a categorical variable using its median for all patients as a cut point (< median vs. ≥ median). Univariate and multivariate Cox proportional hazards models were used to determine if there are any associations between MLH1 expression with OS and DFS. The first level in the MLH1 grouping (< median) was used as the reference level. A hazard ratio (HR) > 1 indicates an increased risk of failure for ≥ median relative to < median. For the multivariate analysis, only the MLH1 grouping was forced into the models and a stepwise selection procedure was used to choose other variables using α=0.05 level as the entry and exit criteria for the model building. The following variables were assessed in the models along with MLH1 expression: treatment (RX) arm, age, gender, race, tumor location, nodal status, largest tumor dimension, and surgical margin status. All analyses were performed on all patients and then within each treatment arm.
To adjust for the multiple comparisons in this exploratory analysis, a two-sided p-value < 0.01 was considered to be statistically significant. A p-value ≥ 0.01 and < 0.05 was considered as showing a trend towards statistical significance. All analyses were performed using SAS/STAT® software (Version 9.2 SAS Institute Inc., Cary, NC, USA).
Results
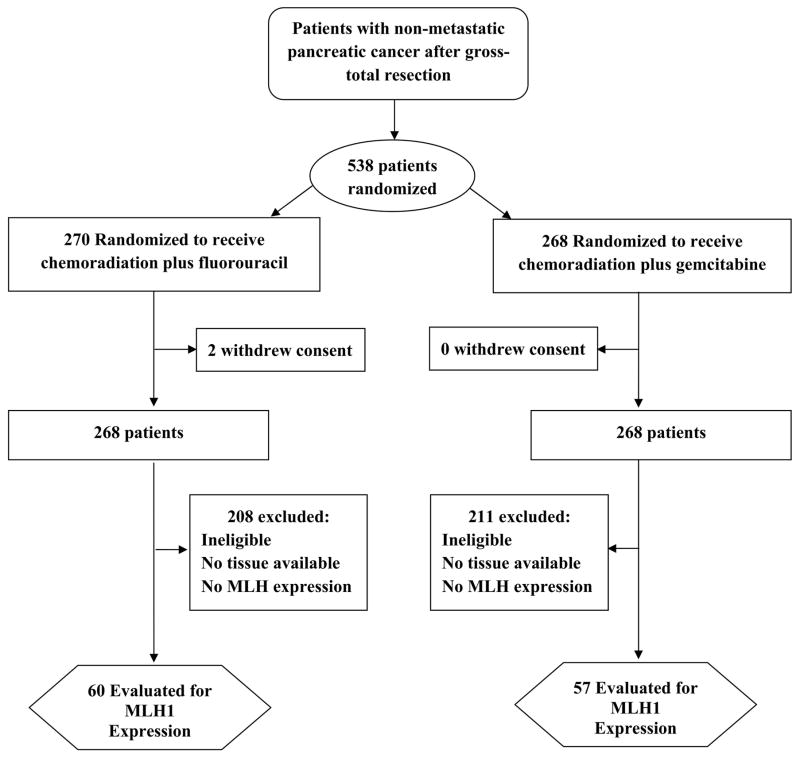
A total of 538 patients were enrolled onto the trial, tissue was available for 220 patients, of whom MLH1 expression was quantifiable in 131 patients. MLH1 expression could not be quantified on 74 patients due to the low quantity / quality of tumor in core, and on 15 patients due to variability with IHC staining for PCK and DAPI). Fourteen patients were excluded from this analysis because they did not meet eligibility requirements for NRG Oncology RTOG 9704. Hence, there were 117 eligible and analyzable patients. Figure 1 shows the breakdown of the 117 eligible and analyzable patients by treatment arm. Table 1 shows baseline characteristics, there are no statistically significant associations seen between pretreatment characteristics and MLH1 nuclear expression, nor were there significant differences in OS or DFS between those who were analyzable and those who were not analyzable (Supplemental Table 1). Supplemental table 2 shows the follow-up and outcomes summary for the 117 patients by treatment arm and for the entire study. Median follow-up for surviving patients was 7 years in each arm (min-max: 2–9 years). For the 117 patients, the median expression for MLH1 in tumor nuclei was 3636.1 (min-max: 408.0–6321.0).
Figure 1.
The flow of patients through the study
Table 1.
Baseline Characteristics of Patients, overall study population and by MLH1 Nuclear expression including patients from Both Arms of study
| Entire population for whom MLH1 was evaluated (n=117) | < median (n=58) | ≥ median (n=59) | |||||
|---|---|---|---|---|---|---|---|
| Age | |||||||
| Median | 63 | 63 | 63 | ||||
| Min-Max | 35–80 | 37–80 | 35–80 | ||||
| n | % | n | % | n | % | p-value† | |
|
|
|||||||
| Gender | 0.31 | ||||||
| Male | 62 | 53.0 | 28 | 48.3 | 34 | 57.6 | |
| Female | 55 | 47.0 | 30 | 51.7 | 25 | 42.4 | |
| Race | 0.21 | ||||||
| White | 105 | 89.7 | 50 | 86.2 | 55 | 93.2 | |
| African-American/Other | 12 | 10.3 | 8 | 13.8 | 4 | 6.8 | |
| Primary Tumor Location | 0.48 | ||||||
| Head | 98 | 83.8 | 50 | 86.2 | 48 | 81.4 | |
| Everything Else | 19 | 16.2 | 8 | 13.8 | 11 | 18.6 | |
| KPS | 0.65 | ||||||
| 60,70,80 | 42 | 35.9 | 22 | 37.9 | 20 | 33.9 | |
| 90,100 | 75 | 64.1 | 36 | 62.1 | 39 | 66.1 | |
| T-stage (surgical) | 0.96 | ||||||
| T1,T2 | 28 | 23.9 | 14 | 24.1 | 14 | 23.7 | |
| T3,T4 | 89 | 76.1 | 44 | 75.9 | 45 | 76.3 | |
| N-stage (surgical) | 0.51 | ||||||
| N0 | 37 | 31.6 | 20 | 34.5 | 17 | 28.8 | |
| N1 | 80 | 68.4 | 38 | 65.5 | 42 | 71.2 | |
| AJCC Stage (5th Edition) | 0.64 | ||||||
| I,II | 36 | 30.8 | 19 | 32.8 | 17 | 28.8 | |
| III,IV | 81 | 69.2 | 39 | 67.2 | 42 | 71.2 | |
| Largest tumor dimension of primary | 0.52 | ||||||
| <3 cm | 47 | 40.2 | 25 | 43.1 | 22 | 37.3 | |
| ≥ 3cm | 70 | 59.8 | 33 | 56.9 | 37 | 62.7 | |
| Surgical Margins | 0.12 | ||||||
| Complete resection/negative margins | 47 | 40.2 | 21 | 36.2 | 26 | 44.1 | |
| Complete resection/positive margins | 40 | 34.2 | 25 | 43.1 | 15 | 25.4 | |
| Complete resection/unknown margins | 30 | 25.6 | 12 | 20.7 | 18 | 30.5 | |
| RX | 0.23 | ||||||
| RT + 5-FU | 60 | 51.3 | 33 | 56.9 | 27 | 45.8 | |
| RT + Gemcitabine | 57 | 48.7 | 25 | 43.1 | 32 | 54.2 | |
p-value from Chi-square or Fisher’s exact test
Abbreviations: KPS= Karnofsky Performance Status; RX= treatment; RT= radiation therapy; 5-FU=5-Fluorouracil; AJCC= American Joint Committee on Cancer
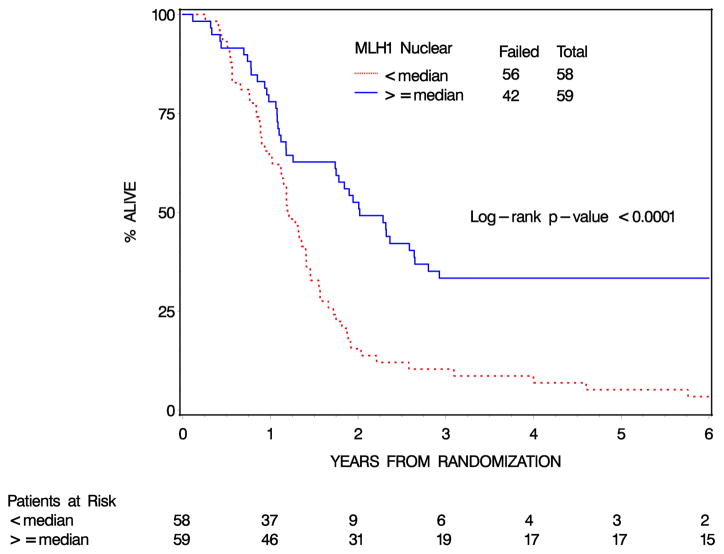
Increased expression of MLH1 in tumor nuclei was associated with both a longer OS and DFS in both the 5FU (OS, 9% vs 33% four-year survival p< 0.005) and gemcitabine (OS, 8% vs 33% four-year survival p< 0.005) treatment arms. Since MLH1 expression was similarly predictive in the two arms, and overall outcomes in the two arms were very similar, the two arms were combined in subsequent analyses. The 4-year OS rates for MLH1 Nuclear expression < median vs. MLH1 Nuclear expression ≥ median were 9% (95% confidence interval [CI]: 3%–18%) and 33% (95% CI: 22%–45%), respectively [log rank p-value<0.0001 (Table 2, Figure 2)]. On univariate Cox analysis, patients with MLH1 Nuclear expression ≥ median have a 57% decrease in the risk of dying than those with MLH1 Nuclear expression < median (HR=0.43, 95% CI: [0.28, 0.64], p-value<0.0001). Similarly, the 4-year DFS rates for MLH1 Nuclear expression < median vs. MLH1 Nuclear expression ≥ median were 2% (95% CI: 0%–1%) and 18% (95% CI: 9%–29%), respectively [log rank p-value=0.0002 (Supplemental Figure 2)]. Patients with high MLH1 expressing tumor have a 51% decrease in the risk of failing than those with low MLH1 Nuclear expression (HR=0.49, 95% CI: [0.33, 0.72], p-value=0.0003). When MLH1 expression in tumor nuclei is analyzed as a continuous variable, a 500-unit increase in MLH1 Nuclear expression corresponds to an 11%/10% reduction in the risk of dying/failing (OS: HR=0.89, 95% CI: [0.81, 0.98], p=0.013 and DFS: HR=0.90, 95% CI: [0.83, 0.98], p=0.017).
Table 2.
Overall and disease-free survival by Nuclear MLH1 Expression (n=117)
| OS | DFS | |||
|---|---|---|---|---|
| 2y-% (95% CI) | 4y-% (95% CI) | 2y-% (95% CI) | 4y-% (95% CI) | |
| <median | 16 (8,26) | 9 (3,18) | 10 (4,20) | 2 (0,8) |
| ≥median | 53 (39,64) | 33 (22,45) | 37 (25,49) | 18 (9,29) |
| p-value† | <0.0001 | 0.0002 | ||
| HR‡ | 0.43 (0.28, 0.64) | 0.49 (0.33, 0.72) | ||
Abbreviations: OS=overall survival; DFS= disease-free survival; HR= hazard ratio; CI= confidence interval
From the log-rank test
HR < 1 indicates a decreased risk of failure for MLH1 expression ≥ median relative to MLH1 expression < median.
Figure 2.
Overall Survival by nuclear MLH1 expression: All Patients (n=117)
The multivariate Cox proportional hazards models for OS are shown in Table 3. After adjusting for nodal status, a 500-unit increase in MLH1 Nuclear expression is associated with a 12% decrease in the risk of dying (HR=0.88, 95% C.I. = [0.80, 0.96], p=0.0045). Likewise, after adjusting for nodal status, patients with MLH1 Nuclear expression ≥ median are associated with a 59% reduction in the risk of dying compared to patients with MLH1 Nuclear expression < median (HR=0.41, 95% C.I. = [0.27, 0.63], p<0.0001). In the multivariate Cox proportional hazards models for DFS, only MLH1 Nuclear expression was statistically significantly associated with DFS; no other variables were added to the models (results not shown).
Table 3.
OS by Nuclear MLH1 expression, Multivariate Cox Proportional Hazards Model (n=117)
| Variables | Comparison | Adjusted HR | (95% C.I.) | p-value† |
|---|---|---|---|---|
| MLH1 Nuclear | Continuous (unit increase = 500) | 0.88 | (0.80, 0.96) | 0.0045 |
|
| ||||
| Nodal Status | Negative | 1.00 | -- | -- |
| Positive | 1.74 | (1.12, 2.71) | 0.013 | |
|
| ||||
| MLH1 Nuclear | < median | 1.00 | -- | -- |
| ≥ median | 0.41 | (0.27, 0.63) | <0.0001 | |
|
| ||||
| Nodal Status | Negative | 1.00 | -- | -- |
| Positive | 1.68 | (1.09, 2.61) | 0.019 | |
p-value from Chi-square test using the Cox proportional hazards model.
Abbreviations: OS=overall survival; HR= hazard ratio; CI= confidence interval
Discussion
This study has demonstrated that higher MLH1 expression in tumor cell nuclei correlates with improved overall survival in resected pancreatic cancer receiving adjuvant chemo-radiation, with a HR of 0.43 on multivariate analysis. Furthermore, the Kaplan-Meier survival curves start to separate at year 1 and then stay apart (Figure 2) suggesting that MLH1 expression predicts long-term outcomes. These findings are in keeping with the hypothesis that tumors with low MLH1 expression may have impaired MMR function and consequently demonstrate treatment resistance.
Function and expression of MLH1
The MLH1 gene is a pivotal member of the mismatch repair (MMR) pathway. MLH1 presence / function may be assessed on the genetic, protein or functional level. Structural mutations within the MLH1 gene are a rare event in pancreatic cancer (less than 1% of sporadic cancers), although single allelic loss is more frequent.12
Multiple factors impact upon MLH1 expression include copy number, allelic loss,13 promoter hypermethylation,15 histone acetylation,18 and microRNA expression.14 On the molecular level the transcription factor GLI1, associated with the Hedgehog signaling pathway decreases MLH1 expression and consequent MMR activity.19 Additionally, external factors such as hypoxia,20 exposure to chemotherapy21 and epigenetic modifiers20, 22 have been found to influence MLH1 expression levels.
Few investigators have examined the clinical significance of MLH1 expression in pancreatic cancer. In agreement with the current results, two small Chinese studies that noted that increased MLH1 expression, as assessed by manual pathologist grading, was associated with good prognostic factors and possibly survival,14, 15 although another small study challenged these findings.23 These studies are limited by their lack of clinical details and the diverse methods used to quantitate MLH1 expression. SNPs within MMR genes correlate with various outcomes in pancreatic cancer, including the response to chemoradiation and overall survival, 24 however it is unclear how the presence of these SNPs relates to gene expression or MMR function. Conversely, Japanese investigators noted that cancers with a high-frequency of MSI were associated with a better prognosis,25, 26, however the incidence of MSI in these studies was an order of magnitude greater than that noted in North America and Europe, raising questions regarding the generalizability of their findings.
MLH1, Microsatellite instability and response to DNA damaging agents
Microsatellite instability (MSI) is considered a functional demonstration of MMR dysfunction. MSI may be due to germline, somatic or epigenetic silencing of one of several MMR genes, including MLH1, MSH2, MSH6, and PMS2. The incidence of MSI in pancreatic cancer is much higher in Japan (around 15%26) than in North America and Europe (around 1%27). However MSI is an imperfect measure of MMR function: heterozygotic loss of MLH1 is associated with decreased MLH1 expression, and increased frequency of indel DNA mutations but not classic MSI.13
Tumors with dysfunctional DNA repair are generally considered sensitive to DNA damaging agents.28 Although the MMR pathway is concerned with DNA repair, paradoxically MMR dysfunction in the pre-clinical setting has repeatedly been noted to be associated with primary resistance to cytotoxic therapies including methylating agents, 6-thioguanine, cisplatin, carboplatin, temozolomide and etoposide.29, 30 Interestingly, several studies have demonstrated that cells with low expression of MMR proteins may demonstrate therapeutic resistance even in the absence of MSI.30, 31 The role of MMR in determining primary sensitivity to radiation46,47 and nucleoside analogues such as 5FU and gemcitabine32, 33 is more controversial.
An alternative mechanistic explanation for our findings relates to MMR’s role in maintaining genomic integrity. MMR deficient cells demonstrate genomic instability associated with increased rates of sporadic mutations, as demonstrated by increased rates of carcinogenesis but also more rapid development of secondary resistance to therapeutic agents such as cisplatin, topotecan, gemcitabine and etoposide.34 Furthermore, when exposed to mutagenic agents MMR deficient cells generate resistant variants more rapidly than MMR intact cells.35 Wang et al. demonstrated a dose response effect, between the MLH1 expression and the degree of genomic instability (as measured by somatic indels).13 Hence compared to MMR proficient cells, cells with MMR dysfunction may more rapidly develop resistance during adjuvant chemo-radiation, facilitating disease recurrence.
A strength of the current study is that it was based upon a large prospectively gathered multi-center clinical trial, representing a homogenous tumor population, who at the time of tissue collection were treatment naive. Another strength is the use of Histo-RX AQUA technology, which has been shown to be objective, reproducible, and suitable for clinical practice. Blinding of the pathological samples helped avoid bias. The two groups were well balanced (Table 1) and their characteristics similar to patients for whom tissue samples were not available (Supplemental Table 1).
A limitation of the study is that a genomic analysis of the samples was not performed. MSI was assumed to be very low based upon previous studies,27, 36 however the frequency of minor genetic changes (e.g. indels) was not known. Furthermore, it is unknown what factors influenced MLH1 protein expression (e.g. promoter hypermethylation or allelic loss) within this population. Although, it was hypothesized that lower levels of MLH1 expression correlate to dysfunctional MMR, additional mechanistic studies are required. This study was based upon the use of archival specimens derived from a TMA that lacked normal tissue samples, consequently we are unable to assess intra-tumoral heterogeneity of MLH1expression, nor are we able to compare tumor expressions levels with those of normal (e.g. pancreatic) tissues. We plan to perform such comparisons in the future studies.
Even though these current findings are in line with two smaller studies that have correlated MLH1 expression with outcomes in pancreatic cancer,14, 15 ideally these results would be validated in a similar large dataset from an additional prospective clinical trial. These findings raise a number of questions: is MLH1 expression a predictive or prognostic factor? Does MLH1 expression correlate with prognosis in other stages of pancreatic cancer, e.g. metastatic, or post-resection not receiving radiation? Is the profound effect of MLH1 expression on overall survival demonstrated here, the result of MMR dysfunction induced primary or secondary resistance? Based upon the proposed mechanism that low MLH1 expression is associated with rapid development of resistance, one may hypothesize that patients with high MLH1 expression bearing tumors may especial benefit from aggressive adjuvant regimens, such as the chemo-radiation employed in RTOG 9704, conversely, those with low MLH1 expression bearing tumors who have a poor prognosis may be best served by receiving chemotherapy alone. Hence MLH1 should join the list of potential biomarkers (e.g. impaired homologous recombination, molecular subtypes based upon genomic analysis) to be considered for use in a personalized approach to pancreatic cancer treatment.
In conclusion, this study suggests that resected tumors with above-average MLH1 expression have a good prognosis in the context of adjuvant chemo-radiation. Once validated, MLH1 expression may prove to be a useful stratification factor for future trials of DNA damaging agents in pancreatic cancer, and furthermore may potentially predict which patients benefit from an aggressive multi-modality approach.
Supplementary Material
Supplemental Figure 1. Quantitative fluorescence immunohistochemistry and digital image analysis for MLH1.
Supplemental Figure 2. Disease-free survival by nuclear MLH1 expression: All Patients (n=117)
Acknowledgments
Funding
This project was supported by grants U10CA21661 (RTOG-Ops-Stat), U10CA37422 (CCOP), U10CA180822 (NRG Oncology SDMC), U24CA114734 (RTOG Specimen Bank) from the National Cancer Institute (NCI) and Eli Lilly. YRL is supported by the EU FP7 Marie Curie program FP7-MC-CIG 303795, and the Rosetrees Trust.
We gratefully acknowledge the substantial assistance of Stephanie Petrillo in the Tumor/Stromal AQUAnalysis, and Michelle Dean for the Antibody workup and immunohistochemical staining.
Footnotes
Conflicts of Interest
Dr. Magliocco reports travel and related expenses associated with being the Co-chair of pathology for NRG Oncology. The remaining authors report no conflicts of interest.
Protection of human subjects:
The analysis was performed on tissue collected in the setting of a prospective clinical trial sponsored by the NIH. The trial was performed with the approval by each medical center’s institutional review board in accordance with an assurance filed with and approved by the U.S. Department of Health and Human Services, in addition each participant provided informed consent.
Author contributions:
Conceptualization YRL CG CC APD, Methodology and Resources AMM AK JS, Formal Analysis JM KW, Investigation and Resources JM AMM ACK WFR ERM TAD WS JS TG, Writing – Original Draft YRL AMM TG APD JM KW CG, Writing – Review & Editing JS ACK WFR RBM TAD WS CC, Supervision, Project Administration and Funding Acquisition APD KW.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65:5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- 2.Tepper J, Nardi G, Sutt H. Carcinoma of the pancreas: review of MGH experience from 1963 to 1973. Analysis of surgical failure and implications for radiation therapy. Cancer. 1976;37:1519–1524. doi: 10.1002/1097-0142(197603)37:3<1519::aid-cncr2820370340>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- 3.Oettle H, Post S, Neuhaus P, et al. Adjuvant chemotherapy with gemcitabine vs observation in patients undergoing curative-intent resection of pancreatic cancer: a randomized controlled trial. JAMA. 2007;297:267–277. doi: 10.1001/jama.297.3.267. [DOI] [PubMed] [Google Scholar]
- 4.Kalser MH, Ellenberg SS. Pancreatic cancer. Adjuvant combined radiation and chemotherapy following curative resection. Arch Surg. 1985;120:899–903. doi: 10.1001/archsurg.1985.01390320023003. [DOI] [PubMed] [Google Scholar]
- 5.Regine WF, Winter KA, Abrams RA, et al. Fluorouracil vs gemcitabine chemotherapy before and after fluorouracil-based chemoradiation following resection of pancreatic adenocarcinoma: a randomized controlled trial. JAMA. 2008;299:1019–1026. doi: 10.1001/jama.299.9.1019. [DOI] [PubMed] [Google Scholar]
- 6.Regine WF, Winter KA, Abrams R, et al. Fluorouracil-based chemoradiation with either gemcitabine or fluorouracil chemotherapy after resection of pancreatic adenocarcinoma: 5-year analysis of the U.S. Intergroup/RTOG 9704 phase III trial. Ann Surg Oncol. 2011;18:1319–1326. doi: 10.1245/s10434-011-1630-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kunkel TA, Erie DA. DNA mismatch repair. Annu Rev Biochem. 2005;74:681–710. doi: 10.1146/annurev.biochem.74.082803.133243. [DOI] [PubMed] [Google Scholar]
- 8.Fishel R, Lescoe MK, Rao MR, et al. The human mutator gene homolog MSH2 and its association with hereditary nonpolyposis colon cancer. Cell. 1993;75:1027–1038. doi: 10.1016/0092-8674(93)90546-3. [DOI] [PubMed] [Google Scholar]
- 9.Anthoney DA, McIlwrath AJ, Gallagher WM, Edlin AR, Brown R. Microsatellite instability, apoptosis, and loss of p53 function in drug-resistant tumor cells. Cancer Res. 1996;56:1374–1381. [PubMed] [Google Scholar]
- 10.Kuismanen SA, Holmberg MT, Salovaara R, de la Chapelle A, Peltomaki P. Genetic and epigenetic modification of MLH1 accounts for a major share of microsatellite-unstable colorectal cancers. Am J Pathol. 2000;156:1773–1779. doi: 10.1016/S0002-9440(10)65048-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grant RC, Selander I, Connor AA, et al. Prevalence of germline mutations in cancer predisposition genes in patients with pancreatic cancer. Gastroenterology. 2015;148:556–564. doi: 10.1053/j.gastro.2014.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Waddell N, Pajic M, Patch AM, et al. Whole genomes redefine the mutational landscape of pancreatic cancer. Nature. 2015;518:495–501. doi: 10.1038/nature14169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang L, Tsutsumi S, Kawaguchi T, et al. Whole-exome sequencing of human pancreatic cancers and characterization of genomic instability caused by MLH1 haploinsufficiency and complete deficiency. Genome Res. 2012;22:208–219. doi: 10.1101/gr.123109.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu WJ, Zhao YP, Zhang TP, et al. MLH1 as a direct target of MiR-155 and a potential predictor of favorable prognosis in pancreatic cancer. J Gastrointest Surg. 2013;17:1399–1405. doi: 10.1007/s11605-013-2230-5. [DOI] [PubMed] [Google Scholar]
- 15.Li M, Zhao ZW. Clinical implications of mismatched repair gene promoter methylation in pancreatic cancer. Med Oncol. 2012;29:970–976. doi: 10.1007/s12032-011-9968-y. [DOI] [PubMed] [Google Scholar]
- 16.Camp RL, Chung GG, Rimm DL. Automated subcellular localization and quantification of protein expression in tissue microarrays. Nat Med. 2002;8:1323–1327. doi: 10.1038/nm791. [DOI] [PubMed] [Google Scholar]
- 17.Otsuka S, Klimowicz AC, Kopciuk K, et al. CXCR4 overexpression is associated with poor outcome in females diagnosed with stage IV non-small cell lung cancer. J Thorac Oncol. 2011;6:1169–1178. doi: 10.1097/JTO.0b013e3182199a99. [DOI] [PubMed] [Google Scholar]
- 18.Edwards RA, Witherspoon M, Wang K, et al. Epigenetic repression of DNA mismatch repair by inflammation and hypoxia in inflammatory bowel disease-associated colorectal cancer. Cancer Res. 2009;69:6423–6429. doi: 10.1158/0008-5472.CAN-09-1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Inaguma S, Riku M, Hashimoto M, et al. GLI1 interferes with the DNA mismatch repair system in pancreatic cancer through BHLHE41-mediated suppression of MLH1. Cancer Res. 2013;73:7313–7323. doi: 10.1158/0008-5472.CAN-13-2008. [DOI] [PubMed] [Google Scholar]
- 20.Lu Y, Wajapeyee N, Turker MS, Glazer PM. Silencing of the DNA mismatch repair gene MLH1 induced by hypoxic stress in a pathway dependent on the histone demethylase LSD1. Cell Rep. 2014;8:501–513. doi: 10.1016/j.celrep.2014.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mackay HJ, Cameron D, Rahilly M, et al. Reduced MLH1 expression in breast tumors after primary chemotherapy predicts disease-free survival. J Clin Oncol. 2000;18:87–93. doi: 10.1200/JCO.2000.18.1.87. [DOI] [PubMed] [Google Scholar]
- 22.Lawrence Y, Pillar N, Goldstein J, et al. Severe Gastrointestinal Complications of Radiation Therapy in Rectal Cancer: Quantifying the Effect of Age. Int J Radiat Oncol Biol Phys. 2014;1:S391. [Google Scholar]
- 23.Tomaszewska R, Okon K, Stachura J. Expression of the DNA mismatch repair proteins (hMLH1 and hMSH2) in infiltrating pancreatic cancer and its relation to some phenotypic features. Pol J Pathol. 2003;54:31–37. [PubMed] [Google Scholar]
- 24.Dong X, Li Y, Hess KR, Abbruzzese JL, Li D. DNA mismatch repair gene polymorphisms affect survival in pancreatic cancer. Oncologist. 2011;16:61–70. doi: 10.1634/theoncologist.2010-0127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nakata B, Wang YQ, Yashiro M, et al. Prognostic value of microsatellite instability in resectable pancreatic cancer. Clin Cancer Res. 2002;8:2536–2540. [PubMed] [Google Scholar]
- 26.Yamamoto H, Itoh F, Nakamura H, et al. Genetic and Clinical Features of Human Pancreatic Ductal Adenocarcinomas with Widespread Microsatellite Instability. Cancer Research. 2001;61:3139–3144. [PubMed] [Google Scholar]
- 27.Laghi L, Beghelli S, Spinelli A, et al. Irrelevance of microsatellite instability in the epidemiology of sporadic pancreatic ductal adenocarcinoma. PLoS One. 2012;7:e46002. doi: 10.1371/journal.pone.0046002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McLornan DP, List A, Mufti GJ. Applying synthetic lethality for the selective targeting of cancer. N Engl J Med. 2014;371:1725–1735. doi: 10.1056/NEJMra1407390. [DOI] [PubMed] [Google Scholar]
- 29.Aebi S, Fink D, Gordon R, et al. Resistance to cytotoxic drugs in DNA mismatch repair-deficient cells. Clin Cancer Res. 1997;3:1763–1767. [PubMed] [Google Scholar]
- 30.McFaline-Figueroa JL, Braun CJ, Stanciu M, et al. Minor changes in expression of the mismatch repair protein MSH2 exert a major impact on glioblastoma response to temozolomide. Cancer Res. 2015 doi: 10.1158/0008-5472.CAN-14-3616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Claij N, Te Riele H. Methylation tolerance in mismatch repair proficient cells with low MSH2 protein level. Oncogene. 2002;21:2873–2879. doi: 10.1038/sj.onc.1205395. [DOI] [PubMed] [Google Scholar]
- 32.Carethers JM, Chauhan DP, Fink D, et al. Mismatch repair proficiency and in vitro response to 5-fluorouracil. Gastroenterology. 1999;117:123–131. doi: 10.1016/s0016-5085(99)70558-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Koi M, Umar A, Chauhan DP, et al. Human chromosome 3 corrects mismatch repair deficiency and microsatellite instability and reduces N-methyl-N′-nitro-N-nitrosoguanidine tolerance in colon tumor cells with homozygous hMLH1 mutation. Cancer Res. 1994;54:4308–4312. [PubMed] [Google Scholar]
- 34.de las Alas MM, Aebi S, Fink D, Howell SB, Los G. Loss of DNA mismatch repair: effects on the rate of mutation to drug resistance. J Natl Cancer Inst. 1997;89:1537–1541. doi: 10.1093/jnci/89.20.1537. [DOI] [PubMed] [Google Scholar]
- 35.Lin X, Howell SB. Effect of loss of DNA mismatch repair on development of topotecan-, gemcitabine-, and paclitaxel-resistant variants after exposure to cisplatin. Mol Pharmacol. 1999;56:390–395. doi: 10.1124/mol.56.2.390. [DOI] [PubMed] [Google Scholar]
- 36.Ghimenti C, Tannergard P, Wahlberg S, et al. Microsatellite instability and mismatch repair gene inactivation in sporadic pancreatic and colon tumours. Br J Cancer. 1999;80:11–16. doi: 10.1038/sj.bjc.6690314. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1. Quantitative fluorescence immunohistochemistry and digital image analysis for MLH1.
Supplemental Figure 2. Disease-free survival by nuclear MLH1 expression: All Patients (n=117)