Abstract
High-dimension single-cell technology is transforming our ability to study and understand cancer. Numerous studies and reviews have reported advances in technology development. The biological insights gleaned from single-cell technology about cancer biology are less reviewed. Here we focus on research studies that illustrate novel aspects of cancer biology that bulk analysis could not achieve, and discuss the fresh insights gained from the application of single-cell technology across basic and clinical cancer studies.
Keywords: single-cell analysis, tumor heterogeneity, subclone phylogeny, cell identity, tumor ecosystem, correlation analysis
1. Introduction
The concept of analyzing heterogeneous cell populations at single-cell resolution has long held great interest for investigators across diverse fields. Methods that characterize single cells, such as flow cytometry and immunohistochemistry, have been workhorses of biological research for decades. However, what has changed over the last few years is the dramatic increase in the number of diverse approaches that address high-dimensional analysis of single cells. These approaches include single-cell transcriptome and genome sequencing, as well as high throughput qPCR and mass cytometry for multiplex detection of proteins. High dimension also refers to the number of cells being analyzed. Depending on the technique, this can be hundreds to millions of single cells. Thus, high-dimension single-cell analysis involves not only high number of targets, but also a high number of cells.
It is thought that cancer starts with changes in a single cell. Shaped by selective forces exerted by the microenvironment, the immune system, and exposure to a wide variety of environmental insults, additional changes accumulate until a tumor is formed that escapes immune surveillance and grows progressively. Indeed, each malignancy is its own experiment in evolution, leading to heterogeneity among cancer cells within one patient and heterogeneity amongst patients with the same disease. One of the hallmarks of high-dimension single-cell analysis is its unparalleled ability to characterize cell-to-cell heterogeneity. Thus, using high-dimension single-cell analysis to study cancer is a natural fit, explaining why this approach is increasingly adopted by cancer researchers. As summarized in Figure 1 and reviewed below, single-cell technology has transformed our understanding of tumor heterogeneity, including intrinsic and extrinsic factors that could co-drive disease initiation, progression, relapse, and metastasis. With the advancement of technology, it is now feasible to collect genome-wide profiles of DNA, RNA, histone modifications, chromatin accessibility, DNA methylation, nuclear lamina interactions and chromosomal contacts, as well as the protein signatures of single cells. This has prompted many reviews and perspectives on applying single-cell technology to cancer (extensively reviewed in Navin, 2015b [see Figure 1 for timeline] and others: Van Loo and Voet, 2014; Navin, 2015a; Sun et al., 2015; Saadatpour et al., 2015; Wills and Mead, 2015; Mato Prado et al., 2016; Schmidt and Efferth, 2016; Tellez-Gabriel, 2016; Ye et al., 2016; Zhang et al., 2016; Zhu et al., 2017; Müller and Diaz, 2017). Rather than focusing on technology, this review addresses how single-cell analysis improves our understanding of tumor heterogeneity at multiple layers (genetic/epigenetic, transcriptomic, proteomic, multiomic).
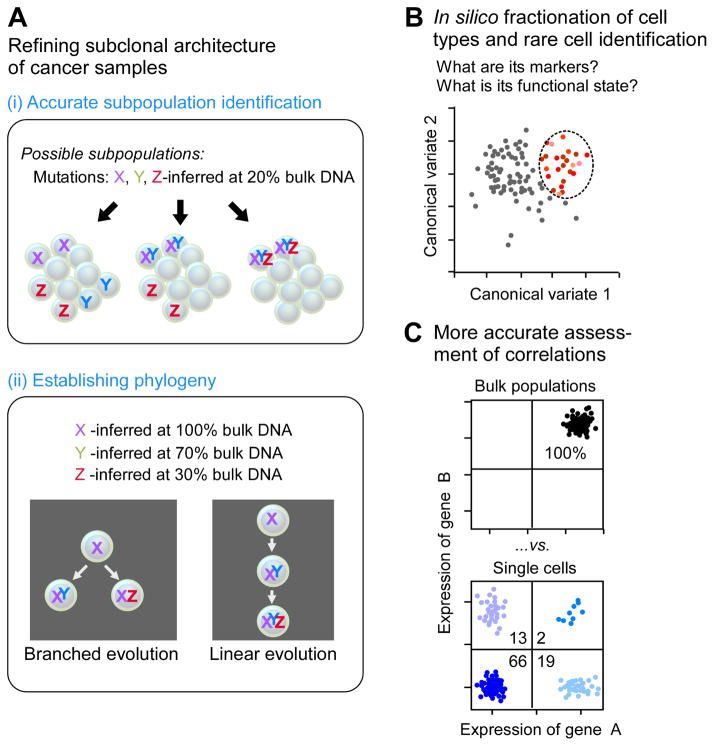
Figure 1.
Key uses of high-dimension single-cell analysis in studying cancer.
2. Overview of single-cell technology
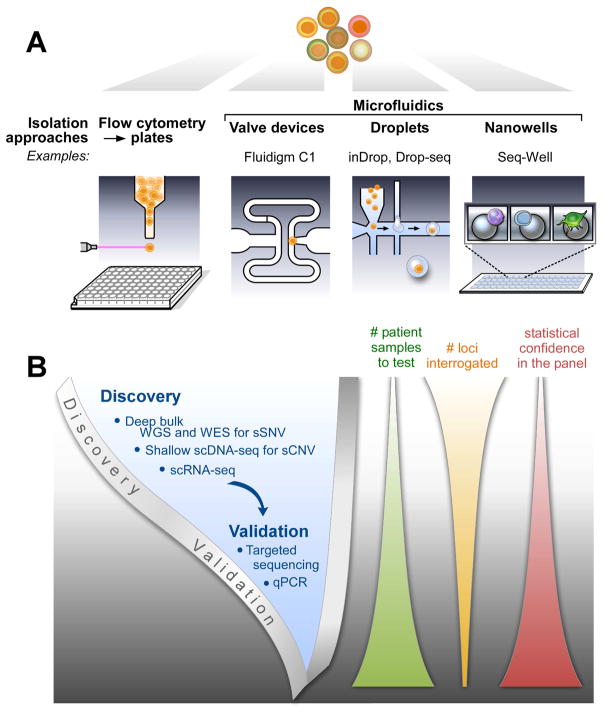
Different aspects of single-cell technology will be briefly summarized. Each section will begin with a list of recent reviews that provide more in-depth descriptions of the topic being discussed. These reviews should be consulted for citations to the primary references. Figure 2 presents the overall workflow for single-cell analysis. After the isolation of single cells, high-dimension technology is applied either to discover heterogeneity (typically in a small set of patient samples) or to validate aspects of heterogeneity (typically in greater numbers of patient samples).
Figure 2. Workflow for high-dimension single-cell experiments.
(A) The predominant methods currently used for isolation of single cells are flow sorting and microfluidics. Examples of microfluidic methods include inDrop (Klein et al., 2015), Drop-seq (Macosko et al., 2015), and Seq-Well (Gierahn et al., 2017). (B) The single-cell analysis of cancer proceeds from discovery of heterogeneity using unbiased, high-dimension methods on a relatively small number of patients to validation in a greater number of patients using targeted techniques.
2.1. Single cell isolation
Most single-cell analysis requires isolation of single cells, previously reviewed by Bheda and Schneider (2014) and Hu et al. (2016). Table 1 in Wang and Navin (2015) effectively summarizes the methods used to isolate both abundant and rare cells. For abundant cells, these methods include serial dilution, mouth pipetting, flow sorting, robotic micromanipulation, and microfluidic platforms. The use of microfluidics is particularly attractive because it reduces the cost and labor required to process hundreds to thousands of single cells. As summarized by Prakadan et al. (2017) the most commonly used microfluidic methods include: (1) valve-based devices, which provide precise control of cells and reagents, and are best suited for implementing complex, integrated workflows; (2) droplets, which provide dramatic advantages in scale and speed, enabling very high throughput (thousands to tens of thousands of cells); and (3) nanowells (devices with nanoliter-sized wells), which provide operational simplicity and lower the barriers to adoption and the development of new protocols. One difficulty often encountered with microfluidics is the disconnect between the availability of microfluidic equipment and the timing/location of sample collection. Flow sorting into conventional microwells (typically 96- or 384-well plates) enables archiving of single-cell lysates, which provides flexibility in the timing and location of sample collection. For example, single-cell lysates from multiple locations can be shipped on dry ice for processing at a central site. In addition, flow sorting enables precise capture of rare populations that are often depleted or lost using other methods. Thus, as shown in Figure 2A, flow sorting and microfluidics are now the predominant methods used for isolating single cells when the cells being analyzed are relatively abundant.
2.2. Genome analysis
Recent reviews of single-cell DNA sequencing (scDNA-seq) include Macaulay and Voet (2014), Wang and Navin (2015), Sun et al. (2015), Szulwach and Livak (2016), and Gawad et al. (2016). The common first step in analyzing the genome of a single cell is whole genome amplification (WGA). Three main types of WGA have been developed (Figure 2 in Gawad et al., 2016): (1) isothermal multiple displacement amplification (MDA); (2) PCR methods such as degenerate oligonucleotide primed PCR (DOP-PCR); and (3) hybrid methods such as PicoPLEX and multiple annealing and looping based amplification cycles (MALBAC) that have a short isothermal amplification step followed by PCR amplification. MDA has greater genomic coverage and a lower error rate, but the other two types of methods have better amplification uniformity. The DNA generated by WGA can be examined by whole-genome sequencing (WGS), whole-exome sequencing (WES), or targeted analysis. The combination of amplification method and mode of analysis depends on the type of variation being evaluated (see Figure 2B). For somatic copy number variation (sCNV), the preferred strategy has been PCR or hybrid amplification because of better uniformity, followed by very shallow WGS. For somatic single nucleotide variation (sSNV), WGS or WES of many single cells is expensive. A more efficient approach that has been commonly adopted is to use deep sequencing of a bulk sample to identify the sSNVs in a given tumor. Then, putative driver mutations or mutations implicated in clonal structure are detected in single cells using MDA because of broader coverage and lower error rate, followed by targeted analysis. The targeted analysis can be performed by sequencing of PCR amplicons or hybrid-selected DNA; or by PCR assays, digital PCR assays, or arrays.
2.3. Transcriptome analysis
Single-cell RNA sequencing (scRNA-seq) has been recently reviewed by Kolodziejczyk et al. (2015), Livak (2016), and Liu and Trapnell (2016). It is important to distinguish the two general methods used in scRNA-seq: whole transcript and end-tagging. Whole transcript analysis provides information about the entire transcript, including splice variants, the presence of mutations, and identification of fusion transcripts that cross translocation or inversion breakpoints. End-tagging methods incorporate cell barcodes during the initial reverse transcriptase step, enabling early pooling of single-cell samples and greatly simplifying the processing of a large number of cells. The highest throughput in terms of number of cells analyzed is achieved using end-tagging methods in droplets or nanowells. End-tagging methods also enable the use of unique molecular identifiers (UMIs; Hug and Schuler, 2003; Kivioja et al., 2011) that improve quantification by reducing the influence of amplification bias on transcript counting. Thus, end-tagging methods are preferred for high-throughput quantification of transcripts while whole transcript methods are chosen when information about splice variants, mutations, and fusion transcripts is important. All scRNA-seq methods, though, still have reduced sensitivity compared to targeted qPCR analysis (Bengtsson et al., 2005; Dalerba et al., 2011; Livak et al., 2013) or in situ hybridization (see 2.6). As depicted in Figure 2B, scRNA-seq is used to discover and cluster the transcriptional profiles of individual cells and qPCR or other methods are used to detect identified signatures in a greater number of patients.
2.4. Epigenome analysis
Aspects of single-cell epigenomic analysis have been reviewed in Bheda and Schneider (2014), Wills et al. (2015), and Clark et al. (2016). Epigenomic analysis methods that have been adapted for single cells include reduced representation bisulfite sequencing (RRBS), whole-genome bisulfite sequencing, chromatin immunoprecipitation followed by sequencing (ChIP-seq), the assay for transposase-accessible chromatin (ATAC-seq), DNase-seq, and the chromatin conformation assay Hi-C. Publications on these single-cell techniques have mostly been proof-of-principle demonstrations and publications documenting meaningful application of epigenomic methods to the study of cancer are just beginning to emerge.
2.5. Proteome analysis
Single-cell proteomic analyses have been reviewed by Heath et al. (2016) and Su et al. (2017) and are quite diverse (summarized in Table 1 in Heath et al., 2016). For the most part, detection of proteins in or from single cells has relied upon antibody recognition. Thus, single-cell proteomics is predominantly a targeted, rather than global, approach. Two approaches that might be considered high dimension are mass cytometry (reviewed by Spitzer and Nolan, 2016) and single-cell barcode chips (SCBCs) for multiplex detection of secreted proteins (Lu et al., 2015).
2.6. Spatial context measurements
To date, most high-dimension single-cell data have been collected from dispersed cancer cells. Spatial context, though, is very important because the microenvironment affects many elements of cancer development. A standard tool for detecting individual RNAs in cells is RNA FISH (fluorescent in situ hybridization). By direct hybridization of probes to RNA, RNA FISH is more sensitive than any method that uses reverse transcriptase to synthesize cDNA and is the gold standard for validating scRNA-seq and single-cell qPCR results. By performing sequential rounds of hybridization with multiplex probes, SeqFISH (Lubeck et al., 2014) and MERFISH (Chen et al., 2015) make RNA FISH into a high-dimension single-cell technology that retains spatial information. For example, with four dyes and eight rounds of hybridization, SeqFISH can cover the entire transcriptome (48 = 65,536). The limitation preventing this whole-transcriptome analysis is the expense of all those fluorescent probes. A related technique is FISSEQ (Lee et al., 2015) that enables in situ sequencing of RNA. Crosetto et al. (2015) have reviewed these and other spatially resolved transcriptomic methods. More recently, Nichterwitz et al. (2016) reported combining laser capture microscopy (LCM) with global transcriptome profiling via Smart-seq2. SWITCH technology (Murray et al., 2015) enables multiple rounds (>20) of antibody labeling to achieve high-dimension proteomic imaging, similar to multiplexed immunohistochemical consecutive staining on single slide (MICSSS) reported by Remark et al. (2016). Bodenmiller (2016) has reviewed other multiplexed epitope-based tissue imaging methods, including mass cytometry imaging and multiplexed ion beam imaging (MIBI). Finally, Cell Painting (Bray et al., 2016) measures approximately 1500 morphological features to generate a rich profile of individual cells. These emerging technologies that combine imaging with high-dimension content are just beginning to be applied to the study of cancer.
2.7. Data analysis
Addressing the computational challenges in analyzing high-dimension single-cell data, especially sequencing data, is beyond the scope of this review. Of note is a recent review by Wagner et al. (2016) on scRNA-seq. Earlier reviews on this topic include Stegle et al. (2015), Bacher and Kendziorski (2016), and Poirion et al. (2016). Analysis of single-cell DNA sequencing data has often used tools developed for bulk analysis. Recent methods that focus on analysis of single-cell data include Ginkgo for sCNV (Garvin et al., 2015) Monovar for sSNV (Zafar et al., 2016) and OncoNEM for reconstructing clonal lineage trees (Ross and Markowetz, 2016).
3. Heterogeneity in cancer
Clinically evident cancer can be conceptualized as emerging from initial malignant transformation and subsequent rounds of evolution in concert with editing due to interactions with the tumor microenvironment (Dunn et al., 2004). Critical questions in the fields of cancer biology and therapeutics include understanding the basis of heterogeneity of cancer patients responding to diverse forms of therapy and dissecting the biologic and genomic features that might be predictive or prognostic of clinical response. Typically, informative specimens collected and characterized from cancer patients include samples collected at initial detection, at time of therapy response, remission, or relapse, and from metastatic lesions.
3.1. Genome heterogeneity
Next-generation sequencing of cancer genomes has not only accelerated the identification of key cancer driving events but also clarified the vast intratumoral genetic heterogeneity present within malignancies. A growing body of literature has pointed to the intratumoral heterogeneity of cancers as the fuel for disease relapse and metastasis, thereby highlighting the importance of understanding clonal structure and tumor evolution in the pathogenesis of disease (Landau et al., 2013; Mroz and Rocco, 2013; Papemmanuil et al., 2013).
3.1.1. Clonal structure and order of genetic alterations
The existing computational approaches for the analysis of intratumoral heterogeneity in bulk samples are based on inference of subclonal structure through analysis of mutant allele frequencies. Precise measurement of the clonal structure, however, requires single-cell analysis because there are certain combinations of mutant allelic frequencies that are impossible to computationally resolve (Paguirigan et al., 2015). DNA analysis at single-cell resolution can resolve the clonal structure (tumor phylogeny), define the order of genetic alterations, and trace dynamic clonal evolution, providing insights to critical steps in oncogenesis.
In the first landmark report of single-cell DNA analysis, Navin et al. (2011) reported WGS analysis of 200 single-cell nuclei (mean coverage 6% of the whole genome) from two breast cancer patients following DOP-PCR amplification and demonstrated the ability of using single-cell DNA characterization to dissect intratumoral heterogeneity. Both samples exhibited a branched mode of copy number evolution, and, in both, metastatic tumors could be genetically linked to subclones from the primary site. Intratumoral heterogeneity in renal cancer, myeloproliferative disorder, and bladder cancer was demonstrated through single-cell WES following MDA amplification (Xu et al., 2012; Hou et al., 2012; Li et al., 2012). Analysis of sSNVs within samples revealed the presence of common founding mutations and several clustered mutations, although clear resolution of subclonal structure and phylogeny was hampered by the high error rates of the method used. This points out the difficulty of using scDNA-seq for de novo discovery of sSNVs. In their study of a colon cancer patient, Yu et al. (2014) addressed this issue by using bulk WES of the tumor to validate their single-cell WES sSNV calls. The subclonal structure observed in their single-cell analysis supports a biclonal origin for this tumor. Using functional validation, they also showed that a mutation in SLC12A5, rarely seen in bulk colon cancer analysis, is a potential cancer driver.
Using MDA amplification followed by low-coverage WGS, Francis et al. (2014) analyzed two primary glioblastomas with focal EGFR amplification present as extrachromosomal amplicons. By performing bulk DNA sequencing on each tumor, they could correct for the errors generated during single-cell MDA. Single-cell resolution enabled deciphering of complex subclonal structures involving differing wild-type EGFR copy numbers, differing variant EGFR copy numbers, different EGFR variant breakpoints, and different breakpoints for other variants. The occurrence of distinct subclones with different breakpoints deleting the same gene is an example of convergent evolution.
Single-cell DNA sequencing of cancer samples has been applied to the question of order of genetic alterations acquired in cancer progression. Wang et al. (2014) analyzed two breast tumors and found that aneuploid rearrangements occurred early in disease history, while point mutations evolved gradually, generating extensive clonal diversity. This study determined sCNVs using DOP-PCR on one set of single nuclei and sSNVs using MDA on a parallel set of single nuclei. Although they reduced the effect of technical errors on sSNV detection by analyzing G2/M nuclei and limiting the time for MDA, they still needed to validate sSNV calls with analysis of bulk samples. In order to address the expense of scDNA-seq, Gao et al. (2016) focused on the detection of sCNVs using DOP-PCR in a highly multiplexed format. They sequenced 1000 cells from 12 triple-negative breast cancer samples and demonstrated that most copy number aberrations were acquired at the earliest stages of tumor evolution in short punctuated bursts, followed by stable clonal expansions that form the tumor mass.
Targeted detection of DNA alterations in single cells has been used to determine the order of genetic events in blood cancer. By multiplex PCR amplification of specific targets in single-cell genomic DNA followed by the use of qPCR assays to detect sCNVs, sSNVs, and a gene fusion, Potter et al. (2013) analyzed three ALL samples (range from 115 to 262 single cells per patient) and deciphered detailed phylogenies showing branched evolution. Using the same method, Papaemmanuil et al. (2014) found that RAG-mediated deletions occur throughout leukemic evolution in ETV6-RUNX1-positive ALL. Gawad et al. (2014) used MDA followed by targeted sequencing of PCR amplicons to analyze 1479 single ALL cells from six patients. Cells were analyzed for sSNVs, large deletions, and IgH sequences. For five of six patients, they resolved branched clonal structures that could not be determined using bulk allele frequency data alone. All three of these studies support a temporal ordering of events in the development of ALL with the ETV6-RUNX1 translocation occurring in utero, followed by preleukemic evolution due to RAG-mediated deletions and accumulation of sSNVs. In a study of CLL, Wang et al. (2017) used MDA followed by targeted sequencing of PCR amplicons to detect sSNVs and sCNVs and showed that four out of five patients had a branched evolutionary structure. Quek et al. (2016) also used targeted scDNA-seq to confirm clonal structures and frequencies in six AML samples.
3.1.2. Chromothripsis
An important aspect of genome heterogeneity is exploring the mechanisms that generate the heterogeneity. Particularly puzzling is the phenomenon of chromothripsis that involves extensive genomic rearrangements and sCNV generation restricted to one or a few chromosomes. Correlation of live-cell tracking with single-cell WGS enabled Zhang et al. (2015) to elucidate the mechanism of chromothripsis. Their analysis showed that genetic rearrangements are restricted to a chromosome or chromosomes mis-segregated into a micronucleus and involved the fragmentation and subsequent reassembly of a single chromatid upon reincorporation of the micronucleus into a daughter nucleus after cell division. Thus, scDNA-seq was instrumental in characterizing a new mutational process fueling cancer evolution, of which chromothripsis is one extreme example.
3.1.3. Understanding cancer biology
3.1.3.1 Tracing disseminated tumor cells DTCs)
Micro-metastases of solid tumors often occur in the bone marrow, generating disseminated tumor cells (DTCs). Detection of DTCs at time of diagnosis is a prognostic marker for poor survival, indicating that DTCs may be a contributing factor to relapse. Demeulemeester et al. (2016) used known surface markers to isolate 56 putative DTCs from the bone marrow of six non-metastatic breast cancer patients. For one of the patients, some of the putative DTCs were isolated three years after diagnosis when a lymph node metastasis was found. These cells were analyzed by MDA followed by low-depth sequencing (1.7× average depth and 23.7% average coverage). After eliminating doublets, 19 of the putative DTCs were morphologically classified as tumor cells. By comparing the sCNVs of single cells to that of the primary tumors, they determined that only 10 of the 19 were true DTCs (53% true positive rate), and these were found in only three of the six patients. In these true DTCs, sSNVs found in the primary tumors were also detected. Many of the isolated cells were aberrant cells of unknown origin that have sCNVs unrelated to the primary tumor and have none of the sSNVs found in the primary tumor. For the three patients with true DTCs, the single-cell data enabled definitive delineation of tumor phylogeny. Interestingly, for the patient with the lymph node metastasis, two of the true DTCs isolated at diagnosis were more closely related to the metastasis than the primary tumor. The results of this study clearly show that all true DTCs were disseminated late in tumor progression. Previous reports of early dissemination were probably due to the confounding effect of the aberrant cells of unknown origin. The targeted scDNA-seq results of Leung et al. (2017) also support a late-dissemination model of metastasis in colorectal cancer.
3.1.3.2 Identifying cancer genes
Sleeping Beauty (SB) insertional mutagenesis has been used for cancer gene discovery in animal models across different types of tumors. By integrating single-cell genome sequencing with a SB-based animal model of myeloid leukemia, Sleeping Beauty capture hybridization sequencing (SBCapSeq), developed by Mann et al. (2016), is a version of targeted scDNA-seq that focuses the sequencing on transposon insertion sites in single tumor cells. The analysis of just 26 cells from one tumor enabled the detection of clonal insertion events not detected by bulk methods and led to the identification of two dominant subclones, each containing a unique pair of interacting trunk drivers. Within each subclone, individual cells had different combinations of additional candidate cancer genes (CCGs) identified from bulk analysis. CCGs in the same cell are potentially genes that cooperated to drive clonal expansion. It is important to note that it is the power of single-cell correlation that enables the clear identification of potentially cooperating genes. This method can be adapted to any transposon-based system and therefore provides a tool to evaluate clonal dynamics and identify potential cooperating cancer genes in model tumors.
3.2. Transcriptome heterogeneity
The emergence of the Human Cell Atlas project (https://www.humancellatlas.org/) shows that scRNA-seq has become the method of choice for discovering cell types and states, and providing an initial characterization. Recent studies have demonstrated the potential for high-content single-cell RNA analysis to revolutionize our understanding of tumor biology ranging from deconvolution of heterogeneous cell populations in the cancer ecosystem, trajectory analysis of cellular state transitions, and dissection of underlying regulatory circuits to the profiling of circulating tumor cells (CTCs) and their relationship to cancer metastasis.
3.2.1. Cell type and developmental stages
A variety of studies have focused on the use of transcriptome analysis of single cells to distinguish subpopulations in solid tumors. An early study by Dalerba et al. (2011) used single-cell qPCR analysis to show that the heterogeneity found in colon cancer cells mirrors the heterogeneity found in normal colon differentiation. This demonstrates that developmental differentiation can be a key source of transcriptional heterogeneity in colon cancer, and perhaps in other cancers as well.
Patel et al. (2014) examined 430 single cells from five glioblastoma patients using SMART-seq. They identified four meta-signatures that characterize individual cells across the tumors: cell cycle, hypoxia, complement/immune response, and oligodendrocyte function. Using a stemness signature derived from bulk RNA analysis, they showed that single tumor cells are continuously distributed along a stemness-differentiation axis, similar to the findings of Dalerba et al. (2011). Based on bulk RNA analysis, each of the tumors in this study could be classifed into one of four glioblastoma subtypes defined by The Cancer Genome Atlas (TCGA): proneural, neural, classical, and mesenchymal. At the single-cell level, though, each tumor was a mixture of these different subtypes. Thus, classification based on population-level data is a simplification of the true transcriptional heterogeneity present in each tumor.
Li et al. (2017) used scRNA-seq to profile 375 cells from the colorectal tumors of 11 patients and 215 cells from the nearby normal mucosa. They devised a new clustering algorithm called reference component analysis (RCA) that projects the single-cell data onto a global reference panel compliled from bulk transcriptome data spanning diverse tissue and cell types. One hallmark of this method is that it claims to normalize for the batch effects inherent in clinical samples. Using RCA, both tumor-derived and normal cells were classifed into seven clusters: epithelial cells, fibroblasts, endothelial cells, B cells, T cells, mast cells and myeloid cells. Further clustering subdivided the normal epithelial cells into nine different types of transit-amplifying (TA) cells, enterocytes, and goblet cells. Tumor epithelial cells were subdivided into stem/TA-like (93% of total), enterocyte 2B–like, and goblet-like cells. By first classifying the cells, analysis of normal/tumor differential expression could be achieved by comparing cells of the same type, for example, normal eptithelial cells to tumor epithelial cells. Such a detailed comparison using bulk analysis is challenging, if not impossible, because it would require the isolation of pure cell types from normal and tumor tissue. Data at single-cell resoluiton enables replacing physical fractionation with in silico fractionation. Strikingly, cancer-associated fibroblasts (CAFs) showed two different expression patterns, and both of these were distinct from fibroblasts found in normal mucosa. Furthermore, analysis of epithelial to mesenchymal transition (EMT) showed that, in these samples, expression of putative EMT transcription factors was limited to CAFs and was not found in tumor epithelial cells.
Two smaller-scale studies have begun applying single-cell transcriptome analysis to ovarian and breast cancer. For high grade serous ovarian cancer (HGSOC), TCGA studies have already defined four molecular subtypes of disease: mesenchymal, immunoreactive, proliferative, and differentiated. In a study by Winterhoff et al. (2017) in which 66 single cells were evaluated using scRNA-seq, clustering and geneset analysis led to a reclassification of disease subtype that was not apparent by bulk analysis. For example, stromal cells, not cancer cells, were enriched for the EMT gene signature, similar to the finding of Li et al. (2017). Furthermore, single cancer epithelial cells classified predominantly as proliferative, rather than mesenchymal as indicated from the bulk analysis. Likewise, Anjanappa et al. (2017) used multiple single-cell qPCR asays to analyze 420 single cells from four breast tumors and 284 adjacent normal cells, and could detect stemness-associated transcripts and the PAM50 geneset (Parker et al., 2009). Their results enabled refined classification of the tumors in terms of basal/luminal characteristics.
3.2.2 Cancer stem cells
The rare nature of putative cancer stem cells has made them challenging to detect and characterize using conventional bulk methods. Multiple emerging studies, however, have demonstrated the ability of single-cell analysis to define these rare populations on the basis of distinct transcriptional signatures, as described below.
First, a series of studies have examined this question in breast cancer. Lawson et al. (2015) used a panel of 116 qPCR assays to analyze single cells from three xenograft models derived from genetically distinct human triple-negative breast cancers. Based on analyzing normal single cells, 49 of the assays constituted a differentiation signature that could distinguish basal/stem, luminal, and luminal progenitor cells. They developed a sorting strategy to robustly isolate human metastatic cells (>99.5% detection rate) and used the qPCR assays to characterize single metastatic cells. Metastases were detected in 77 mice across the three models. Mice were further stratified from low burden to high burden based on the number of metastatic cells detected per animal. Gene expression was compared between early-stage (low burden) and late-stage (high burden) metastatic disease. They analyzed 441 metastatic and 523 primary tumor cells from 12 animals. High-burden metastatic cells were more similar to primary tumor cells than low-burden metastatic cells. Low-burden metastatic cells exhibited a basal/stem-like and quiescent signature compared to high-burden metastatic cells that had a more proliferative signature and a spectrum of luminal-like expression. Transplant experiments indicated that low-burden metastatic cells have tumor-initiating capacity. Across all three models, 1.4% of primary tumor cells cluster with low-burden metastatic cells, thereby providing an estimate of the prevalence of putative cancer stem cells in these breast cancer models. In a separate study using 80 qPCR assays, a stem-like signature could be identified in single cells from four breast cancer cell lines (two ER+ and two ER−) and two primary breast cancer samples collected immediately after surgery (Akrap et al., 2016).
Second, for human oligodendrogliomas, Tirosh et al. (2016b) analyzed 4,347 single cells collected from six patients with untreated grade II disease using Smart-seq2. The tumors had co-deletion of chromosome arms 1p and 19q and mutations in IDH1 or IDH2. Principal component analysis (PCA) showed that oligodendrogliomas are primarily composed of two subpopulations of glial cells that have astrocyte or oligodendrocyte signatures. A stem/progenitor signature was established by profiling normal neural progenitor cells by scRNA-seq. By combining the three PCA-derived signatures, the authors devised scores for stemness/differentiation and for lineage (astrocyte/oligodendrocyte) that were used to assign a cellular state to each tumor cell. By applying this scoring system to the six tumors, the prevalence of putative cancer stem cells was assessed as 5% or less in these samples.
Finally, adult and childhood acute lymphocytic leukemia (ALL) were characterized by Ebinger et al. (2016), focusing on relapse-inducing cells in xenografts. In order to identify the human cells in mice, transplanted tumor cells were transduced with lentivirus to express luceriferase for in vivo imaging, an artificial antigen for cell isolation using magnetic sorting, and a red fluorochrome for cell sorting by flow cytometry. Slowly proliferating cells were identified by retention of the dye carboxyfluorescein diacetate succinimidyl ester (label-retaining cells, LRC). LRC exhibited the adverse characteristics of dormancy, in vivo drug resistance, and leukemia-initating properties upon transplantation. Surprisingly, LRC and non-LRC had similar stem cell frequencies and leukemia-initiating potential, indicating that stemness alone was insufficient to define relapse-inducing cells in ALL, and suggesting that the additional properties of dormancy and drug resistance were possible surrogate attributes critical for aggressive biologic behavior. Supporting this model, LRC and primary minimal residual disease (MRD) cells from patients clustered together using single-cell transcriptome profiling, and both were distinct from tumor cells collected at diagnosis. The LRC properties of dormancy and drug resistance were further associated with their localization to a bone marrow niche close to the endosteum, suggesting that removing MRD cells from their protective niche might sensitize them to treatment.
3.2.3. Tumor ecosystem
In addition to studying heterogeneous subpopulations of tumor cells, single cell approaches are increasingly appreciated as an important methodology to comprehensively deconvolute the cellular components within the microenvironment surrounding tumor cells. One of the first studies to employ such an approach was described by Tirosh et al. (2016a) in a scRNA-seq study of 4645 malignant, immune, and stromal cells from 18 metastatic and one primary melanoma tumors. Malignant and normal cell subpopulations were identified based on the presence of tumor-associated chromosomal deletions or by distinct lineage-specific expressed genes, respectively. Tumor cells exhibited two transcriptional cell states, MITF-high and AXL-high. Analysis of biopsies and cell lines treated with RAF and MEK inhibitors indicated the AXL-high program is associated with drug resistance after MAP kinase-targeted treatment. The precision of single-cell profiling enabled identification of robust signatures for five nonmalignant cell types found in the tumors: T cells, B cells, macrophages, endothelial cells, and CAFs. These signatures were applied to bulk data of 471 tumors from TCGA to infer the relative abundance of each cell type, segregating the tumors into 10 microenvironment clusters. It was observed that abundance of CAFs was associated with expression of the AXL-rich program and a set of CAF-expressed genes correlated strongly with T cell infiltration. Examination of T-cell expression patterns led to identification of a 28-gene core exhaustion signature. Further studies will see if this exhaustion signature can be used to predict response to immunotherapy.
Zheng et al. (2017) focused on tumor infiltrating lymphocytes in their scRNA-seq analysis of 5,063 T cells isolated from tumor, nearby normal tissue, and peripheral blood from six patients with liver cancer. Transcriptional signatures distinguished 11 T cell subsets, six CD4 and five CD8. They investigated relationships among these subgroups using T cell receptor (TCR) reconstruction (Stubbington et al., 2016) to determine clonality and using Monocle 2 (Trapnell et al., 2014) to discern developmental trajectories. Tumor tissue was enriched for regulatory T cells and exhausted CD8 cells, and both showed increased frequencies of clonal TCRs compared to blood and normal liver tissue. Both trajectory analysis and TCR clonality indicated that exhausted CD8 cells likely evolved from other types of CD8 T cells in the tumor. On the other hand, regulatory T cells did not seem to derive from conventional CD4 cells in the tumor or by expansion of regulatory T cells from adjacent normal tissue. This study analyzing just T cells emphasizes the interest in characterizing immune cells in the tumor microenvironment, an effort prompted by breakthroughs in immunotherapy. Thus, many of the scRNA-seq studies in this review include some analysis of tumor-associated immune cells. Furthermore, the proteomic studies of Chevrier et al. (2017) and Lavin et al. (2017) showcase the power of using mass cytometry to characterize the immune response to cancer and therapy (see 3.5).
3.2.4. Transcriptome variability
Nguyen et al. (2016) screened 29,390 cells from 200 clonal subpopulations of two breast cancer cell lines using cell-size imaging and identified sub-lines with highly variable (HV) or lowly variable (LV) cell morphology. Compared to LV, HV subpopulations exhibit increased metastatic capacity and resistance to chemotherapies when inoculated into the arterial circulation of mice. Profiling by scRNA-seq showed that HV and LV subpopulations did not differ significantly in total transcript abundance per cell nor could they be classified separately using linear or non-linear clustering. However, HV subpopulations do have greater variability as assessed by coefficient of variation (CV) per individual transcript. Using pathway analysis, spliceosome machinery and myeloid cell differentiation were identified as the only two gene sets that exhibited significantly higher variability in HV subpopulations derived from both parental cell lines. LV subpopulations engineered to have low expression of the splicing factor SNRNP40 showed increased metastatic capacity. This study revealed transcriptome variablility as a mechansim by which cancer subpopulations can enhance fitness and implicated changes in the splicing machinery as a possible contributor to this variability.
3.3. Genotype/phenotype correlation
Single-cell studies have clearly established the heterogeneity of cancer genomes and transcriptomes, but have raised the question of how these two types of heterogeneity are related. Understanding how heterogeneity affects function and tumor evolution requires correlating genotype with phenotype. Achieving the integration of genotype and functional information at the single-cell level dramatically increases the power of correlation analysis (Wills et al., 2013).
3.3.1. Multiomics
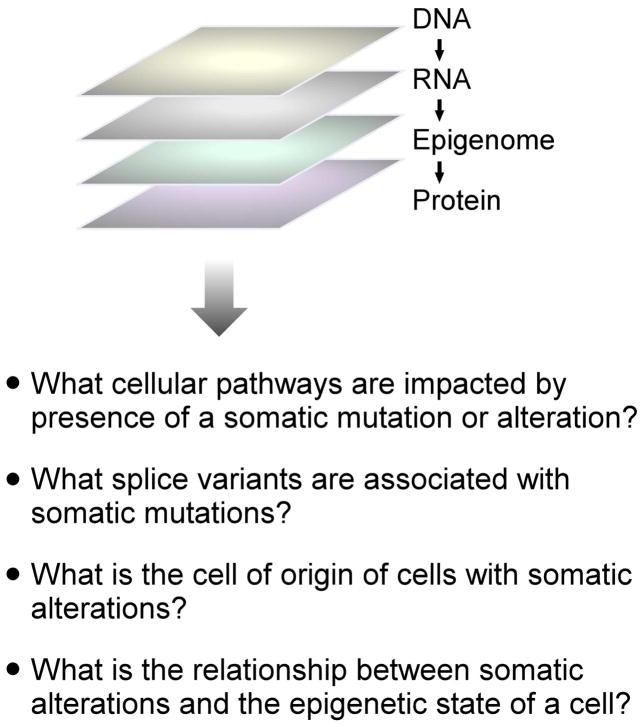
Multiomics refers to the generation and analysis of measurements of multiple molecular types in the same single cell, and has been recently reviewed by Macaulay et al. (2017). Figure 3 presents the types of questions that can be addressed using multiomic analysis.
Figure 3. Single-cell multiomic analysis of cancer.
The diagram indicates the different layers of information that could be obtained for the same set of single cells.
Three methods have been reported for obtaining both DNA and RNA information from the same single cell. Han et al. (2014) developed a microfluidics platform that physically separates cytoplasm from nucleus and then prepares sequencing libraries from these fractions. G&T-seq (Macaulay et al., 2015; Macaulay et al., 2016) also uses physical separation. In this case, beads are used to capture mRNA and separate it from the genomic DNA. In DR-Seq (Dey et al., 2015), a few rounds of MALBAC are used to amplify both cDNA and genomic DNA prior to splitting the sample to prepare an RNA sequencing library using CEL-Seq and a DNA sequencing library using MALBAC.
In order to obtain both epigenome and transcriptome information, G&T-seq has been adapted to create scM&T-seq (Angermueller et al., 2016) in which the DNA component undergoes bisulfite sequencing to characterize the methylome. In scMT-seq (Hu et al., 2016) and sc-Trio-seq (Hou et al., 2016), nucleus and cytoplasm are physically separated prior to performing sc-RNA-seq and single-cell RRBS analysis. In sc-Trio-seq, the RRBS data is computationally analyzed to determine the sCNV genotype of each cell as well.
3.3.2 RNA genotyping
Multiomic technologies that analyze DNA and RNA are poised to directly address the question of genotype/phenotype correlation in cancer, but these types of studies have not yet been reported. What has been done, though, is the determination of genotype using RNA data. Studying 77 single xenograft cells derived from a human lung adenocarcinoma, Kim et al. (2015) analyzed their scRNA-seq data to determine which single cells had the KRASG12D mutation. Their ability to detect this mutation was enhanced because KRAS was amplified in the tumor they studied. They also performed fairly deep sequencing to an average of 8.1 million reads per cell in order to increase coverage at mutation sites. Based on transcriptional signature, cells could be classifed as quiescent or proliferative. In this manner, four genotype/phenotype categories were identified: mutant KRAS/quiescent, mutant KRAS/proliferative, wild type KRAS/quiescent, and wild type KRAS/proliferative. Of these four, drug resistance was associated with the mutant/quiescent subpopulation.
Patel et al. (2014) inferred sCNVs from scRNA-seq data by averaging relative expression levels over large genomic regions. The rationale was that genes in deleted regions would have reduced expression relative to genes in diploid regions. They detected arm-level copy number variants but not smaller, focal deletions. They used this information to distinguish tumor cells from normal cells in their study of glioblastoma (see 3.2.1). Using a similar method to detect sCNVs in glioblastomas, Müller et al. (2016) associated a subclonal chromosome 13 deletion with leading edge tumor cells that had an infiltrating phenotype. The chromosome 13 deletion includes the miR-15a/16 micro-RNA cluster. Of up-regulated genes in the deletion subclone, 16% were direct validated miR-15a/16 targets and 78% were targets of transcription factors that are repressed by miR-15a/16.
Tirosh et al. (2016b) used the same sCNV inference method as Patel et al. (2014) in their study of oligodendroglioma (see 3.2.2). Surprisingly, distinct sCNV subclones within tumors displayed similar transcriptome profiles. This finding was supported by analysis of a subclonal CIC mutation. Thus, in these patients, transcriptional heterogeneity was driven by the developmental program generating astrocyte-like or oligodendrocyte-like cells, not by the genotype. Genetic heterogeneity may play a modulating role but this remains to be shown.
In a study of chronic lymphocytic leukemia (CLL), Wang et al. (2016) provide an example of using targeted, allele-specific PCR assays on single-cell RNA to detect mutations identified by bulk WES. The mutational status of the splicing factor gene SF3B1 in each cell was correlated with the results from targeted qPCR assays that quantified the expression of 96 genes and aberrant splicing of 48 transcripts. Single-cell correlation clearly demonstrated that aberrant splicing is due to SF3B1 mutations in a specific segment of the heat repeat domain. Single cells with SF3B1 mutation were confirmed to express an altered splice variant of DVL2, leading to dysregulated Notch signaling. In a separate study, Wang et al. (2017) extended the strategy of using targeted, allele-specific PCR assays on single-cell RNA to include assays for germline SNPs to enable inference of sCNVs known from bulk WES, and to enable reconstruction of phylogenies for five CLL samples.
In their study of CML, Giustacchini et al. (2017) added sensitive detection of the BCR-ABL fusion transcript to scRNA-seq by including fusion-specific primers at the reverse transcriptase and cDNA amplification steps of the Smart-seq2 protocol. Detection of BCR-ABL enabled unambiguous identification of CML cancer stem cells (BCR-ABL+) and non-malignant hematopoietic stem cells (BCR-ABL−). A distinct transcriptional signature identified a subgroup of cancer stem cells that persisted through tyrosine kinase inhibitor therapy. Analysis of non-malignant stem cells indicated that CML disrupts normal hematopoiesis.
3.3.3. Sensitivity of mutation detection
Three of the aforementioned studies used both scRNA-seq for whole transcriptome analysis and targeted assays to detect specific mutations in single-cell RNA. Their results indicate that targeted PCR assays are more sensitive than scRNA-seq for mutation detection. Wang et al. (2017) detected a specific ATM mutation in 2.1% of single cells analyzed by scRNA-seq compared to 70% for targeted PCR. In Tirosh et al. (2016b), the sensitivities for detecting a specific CIC mutation were 0.66% for sequencing and 3.9% for targeted PCR. By adding fusion-specific primers to whole-transcriptome scRNA-seq, Giustacchini et al. (2017) increased the detection rate for the BCR-ABL fusion gene in K562 cells from 25% to 100%.
3.4. Epigenomic heterogeneity
As part of their characterization of the enhancer landscape of the human hematopoietic hierarchy, Corces et al. (2016) analyzed single cells from two AML patients by ATAC-seq. The single-cell regulatory profiles were projected onto principal components derived from the normal stages of hematopoiesis in order to assess cell state identity. Epigenomic heterogeneity in AML is due both to an admixture of different cell types and the presence of individual cells with mixed regulatory programs. The exact nature of these mixed regulatory programs varies from cell to cell and from patient to patient.
In their study of K562 cells, Litzenburger et al. (2017) used single-cell ATAC-seq and scRNA-seq to identify CD24 as a cell surface marker that co-varies with accessibility of the GATA motif in chromatin and with expression levels of the GATA1 and GATA2 transcripts. Compared to CD24lo, CD24hi cells have more accessible binding sites for stem-ness transcription factors. When cells were treated with the BCR-ABL tyrosine kinase inhibitor imatinib, 2.9% CD24hi cells continued to proliferate compared to 0.6% for CD24lo cells. Identifying cell-surface surrogate markers for chromatin states represents one path for relating functional consequences to single-cell epigenomic findings.
3.5. Proteomic heterogeneity
Levine et al. (2015) used the fact that mass cytometry detects both surface and intracellular antigens to determine the correlation between surface markers and signaling-based phenotypes in their study of acute myeloid leukemia (AML). This study interrogated bone marrow aspirate samples from 16 pediatric AML patients obtained at diagnosis and from 5 healthy adults, and used a mass cytometry panel consisting of 16 surface markers and 14 antibody probes targeting phosphorylation. Sixteen different perturbations, such as treatment with various cytokines, were used to elicit signaling responses, and over 15 million single cells were evaluated. Analysis of normal cells established signaling phenotypes for the different functional states of hematopoiesis and these phenotypes correlated with the expected surface markers. By contrast, AML cells did not show consistent correlations between signaling phenotypes and surface markers. For example, AML cells with the functional state corresponding to leukemic stem cells did not necessarily have the surface markers associated with hematopoietic stem cells. Thus, the surface markers typically used in diagnostics were assessed as inadequate indicators of cellular state and function in AML. Gullaksen et al. (2017) also used mass cytometric detection of phosphorylated proteins to analyze the BCR-ABL signaling pathway in CML patients before treatment with a tyrosine kinase inhibitor and after three hours and seven days of treatment.
Chevrier et al. (2017) characterized the tumor microenvironment (TME) by using mass cytometry to analyze T cells and tumor-associated macrophages (TAMs) that had infiltrated the TME in 73 clear cell renal cell carcinoma patients. Cells were tested with panels of antibodies either for T cells or for TAMs, each panel consisting of over 30 probes. Analysis of 3.5 million cells identified 22 T cell phenotypes and 17 TAM phenotypes. One particular TAM subpopulation, designated M-5, correlated with exhausted T cells and somewhat with regulatory T cells. Poor progression-free times were associated with high frequencies of M-11 or M-13 macrophages and low frequency of the M-5 set, with the suggestion that the relative proportions of these subpopulations could serve as a predictor of progression-free survival.
In a similar study, Lavin et al. (2017) used mass cytometry to generate an immune cell atlas of early lung cancer by analyzing 29 patients with treatment-naïve Stage I non-small cell lung cancer. For each patient, cells were collected from the tumor, normal lung, and the blood, and analyzed with one of two panels of more than 30 antibodies each. For tumor and normal lung, nearby tissue was also prepared for sectioning and analyzed by MICSSS (see 2.6) in order to get spatial information. From a single patient, over 1100 tumor cells and 700 normal lung cells were analyzed by scRNA-seq. The most abundant immune cells associated with tumors were T cells and mononuclear phagocytes. Spatially, immune cells were found predominantly in the stroma and invasive margin, although some macrophages and T cells infiltrated the tumor. The scRNA-seq data identified a subpopulation of macrophages with a transcriptional phenotype distinct from normal tissue. Overall, the lung tumors were enriched for a particular class of macrophages, CD1c+ dendritic cells, regulatory T cells, and exhausted T cells compared to normal lung. Cells depleted in the tumors included CD141+ dendritic cells, CD16+ monocytes, natural killer cells, and Granzyme B+ effector cells.
4. Impact on diagnostics
4.1. Solid tumors and circulating tumor cells (CTCs)
4.1.1 Liquid biopsy
Liquid biopsies for analyzing CTCs or circulating tumor DNA (ctDNA) in blood promise to be a minimally-invasive means for cancer diagnosis, monitoring during and following treament, and prognosis (Kidess and Jeffrey, 2013). Table 1 in Calabuig-Fariñas et al. (2016) compares the utility of CTCs and ctDNA by listing the advantages and limitations of each. Although ctDNA may be more sensitive for detecting tumor-specific mutations (Shaw et al., 2017), the power of single-cell correlation means that CTCs provide more information for evaluating cancer phenotypes. The challenge for the study of CTCs is that this population is extremely rare, with abundances ranging from a few to hundreds per ml of blood (Krivacic et al., 2004; Zieglschmid et al., 2005; Racila et al., 1998).
4.1.2 Isolation of CTCs
Song et al. (2017) provide a comprehensive review on CTC isolation and include over 50 references to the many devices that have been fabricated. Other recent reviews include Zhang et al. (2016) and Hardingham et al. (2015). Isolation can be achieved using cell-surface markers, physical characteristics, or a combination of the two. Affinity reagents used to capture CTCs based on molecular markers include antibodies, peptides, and aptamers. The only clinically validated and FDA-cleared test for capturing and enumerating CTCs is the CellSearch® system that uses magnetic beads coated with antibodies to EpCAM. Label-free methods use physical characteristics such as size, density, compressibility, or polarizabilty. Most recently developed devices incorporate microfluidics to enhance capture efficiency and increase throughput. van der Toom et al. (2016) summarize the pitfalls and limitations of the most common CTC isolation and detection methods. For example, the rationale for using size selection to separate larger CTCs from smaller white blood cells is based mostly on the properties of cancer cell lines not true CTCs. The use of the marker EpCAM to identify CTCs is problematic because it labels only epithelial cells and CTCs can have variable EpCAM expression. van der Toom et al. (2016) have suggested using multiple biomarkers and especially disease-specific markers. Of particular note for high-dimension single-cell analysis is a selection-free system descrbed by Campton et al. (2015). Nucleated blood cells were spread on a microscope slide and screened to identify putative CTCs using automated immunofluorescence detection with multiple markers. A single-cell retrieval device was used to pick the putative CTCs for WGA and downstream analysis.
4.1.3. DNA analysis of CTCs
Initial efforts in the analysis of this rare population have focused on the feasibility of detecting tumor-specific alterations in CTCs. Heitzer et al. (2013) reported the detection of tumor-specific sCNVs and sSNVs in 37 CTC cells from six patients with stage IV colorectal carcinoma by the combination of comparative genomic hybridization array (copy number variation) and targeted deep sequencing for a panel of 68 genes. sSNVs identified in bulk primary tumors or metastases were found in the CTCs. For sSNVs initially detected exclusively in CTCs, ultradeep sequencing showed that most of these were, in fact, subclonal events in the matched primary tumors. Other studies, including Jiang et al. (2015) and Bingham et al. (2017), support the finding that CTCs are genetically related to primary tumors and metastases.
Lohr et al. (2014) reported a robust process for identifying true sSNVs in CTCs. They captured CTCs from 36 patients with metastatic prostate cancer (0–200 CTCs per 3.75 ml of blood). Focusing on five patients with more than 20 CTCs captured per individual, they amplified genomic DNA from more than 300 CTCs by MDA, but the libraries from only 42 CTCs survived their rigorous qualification process. After WES, sSNVs were identified using census-based calling (i.e., the sSNV is observed in two or more CTCs). For one patient, 70% of the mutations found in CTCs were present in matched primary tissue or metastatic lymph node. Of 10 early trunk and 56 metastasis-associated mutations identified in non-CTC tumor samples, 90% and 73%, respectively, were found in CTC exomes. More recently, the same group isolated circulating multiple myeloma cells and performed both single-cell DNA- and RNA-seq (Lohr et al., 2016). In this case, the DNA analysis used MDA followed by targeted sequencing of 35 loci. Both genetic and transcriptomic information confirm that CTCs are similar to bone marrow multiple myeloma cells. For genomic analysis, detailed comparison of CTCs and bone marrow biopsies found that both detected the same pattern of somatic mutations and enabled identification of actionable oncogenes. This establishes single-cell analysis of CTCs as a viable alternative to bone marrow biopsies in the monitoring and treatment of multiple myeloma.
For patients with lung adenocarcinoma and metastases in the liver, Ni et al. (2013) performed MALBAC WGA on single CTCs followed by WES for detection of sSNVs and low-depth WGS to detect sCNVs. For four patients analyzed, the sSNVs detected in CTCs overlapped with sSNVs found in primary and metastatic samples, but the pattern of sSNVs was heterogeneous from CTC to CTC. Remarkably, analysis of seven patients found that CTCs from the same patient have very similar sCNV patterns. For five of these patients, the sCNV patterns are similar between patients, with an average of 78% of the gain and loss regions shared between any two patients. In all patients, the sCNV pattern in CTCs closely resembles the metastasis rather than the primary tumor. Gao et al. (2017) extended this type of study to examine CTCs from colon, breast, gastric, and prostate cancer patients. For one of the colon cancer patients, 28 individual primary tumor cells were analyzed as well. Phylogenetic analysis indicates that the sCNVs in the primary tumor cells converge to the sCNV pattern found in the five CTCs analyzed for this patient. The pattern in CTCs was similar to the sCNV patterns in three separated metastatic lymph nodes. Across all the cancers studied, CTCs from the same patient had a similar sCNV pattern. There was also some simililarity between patients with the same type of cancer. The exception was breast cancer where different patients had distinctive patterns, perhaps reflecting the subtype diversity of breast cancer. Overall, the findings suggest that certain sCNVs are selected for during the process of metastasis.
4.1.4. RNA analysis of CTCs
The first scRNA-seq analysis of CTCs was by Ramskold et al. (2012). They analyzed six CTCs from a patient with melanoma as well as normal single cells and identified distinct gene expression patterns specific for melanoma CTCs. Ting et al. (2014) compared the transcriptome of 93 single CTCs with matched primary tumors from a mouse model of pancreatic cancer. All the CTCs clustered separately from primary tumor and tumor-derived cell lines. Compared to primary tumor, CTCs showed reduced expression of epithelial markers, heterogeneous expression of mesenchymal markers, and increased expression of stem cell markers. The finding of increased expression of extracellular matrix (ECM) genes in CTCs was confirmed by scRNA-seq analysis of human CTCs from pancreatic, breast, and prostate cancer patients. Knockdown of the ECM gene SPARC in a human pancreatic cancer cell line suppressed cell migration, invasiveness, and the ability to from metastastes when injected in mice. Thus, this study revealed novel micro-environmental signals critical to the metastasis of pancreatic cancer.
In a RNA-seq study on CTCs from breast cancer patients, Aceto et al (2014) found higher expression of the cell junction component plakoglobin in CTC clusters compared to single CTCs. Knockdown of plakoglobin in a mouse xenograft model reduced the number of CTC clusters observed and reduced lung metastases by 80%. The authors concluded that CTC clusters, groupings of primary tumor cells held together by plakoglobin-dependent adhesion, are critical mediators of breast cancer metastasis.
Miyamoto et al. (2015) examined 77 CTCs isolated from 13 patients with castration-resistant prostate cancer and could identify vast heterogeneity including androgen receptor gene (AR) mutations and splice variants. The most enriched genes comparing expression in CTCs to primary tumor were the chaperone HSP90AA1 involved in the activation and stability of AR and MALAT1, which encodes a non-coding RNA implicated in alternative splicing. CTC profiles from patients progressing under treatment with an AR inhibitor showed activation of noncanonical Wnt signaling. Moreover, the authors demonstrated that ectopic expression of Wnt5a in prostate cancer cells reduced the antiproliferative effect of the AR inhibitor. Thus, scRNA-seq of CTCs revealed signaling pathway heterogeneity that likely contributes to treatment failure.
4.2. Blood malignancies
Assays using qPCR (e.g., Slack et al., 2001) or digital PCR (e.g., Oehler et al. 2009) have often been used to monitor MRD in bulk RNA samples isolated from blood. Burger et al. (2016) exploited droplet microfluidics to create what might be called digital cellular PCR. This enables directly enumerating cancer cells in a blood sample. The throughput of droplet microfluidics enables analysis of hundreds of thousands to millions of blood cells. Single white blood cells from CLL patients were encapsulated with reagents for lysis and one-step reverse transcriptase-qPCR. The assay performed was qPCR with an allele-specific primer to detect a mutation characteristic of the patient’s cancer. For three patients who developed therapeutic resistance to the B cell receptor pathway inhibitor ibrutinib, detection of mutations in PLCG2, RPS15, or DGKA transcripts in pre-treatment samples showed that resistant subclones were present at rare frequencies prior to the initiation of ibrutinib therapy, supporting the idea that the resistant populations were positively selected by prolonged exposure to the targeted inhibitor.
Zheng et al. (2017) used droplet-based scRNA-seq to analyze bone marrow samples from two AML patients before and after hematopoietic stem cell transplant therapy. An average of 3100 single cells were analyzed per sample. After transplant, SNV analysis showed one patient (Patient 1) to have 86.2% host cells and 13.8% donor cells and a second patient (Patient 2) to have all donor cells. These results were confirmed by an independent clinical chimerism assay. The whole transcriptome data enabled cell type classification. In healthy controls, T cells dominate bone marrow mononuclear cell samples. This was also true for the donor cells in patient 1 after transplant. In the AML pretreatment samples, erythroid cells dominate. After transplant, the host cells in Patient 1 were predominantly blast cells and immature erythroid cells, consistent with the relapse diagnosis for this patient. In Patient 2, the donor cells were predominantly erythroid in nature, but there was broad distribution across the different stages of erythroid development. One unexpected finding was that monocytes were abundant in both patients before transplant, but were not detected after transplant.
5. Perspective
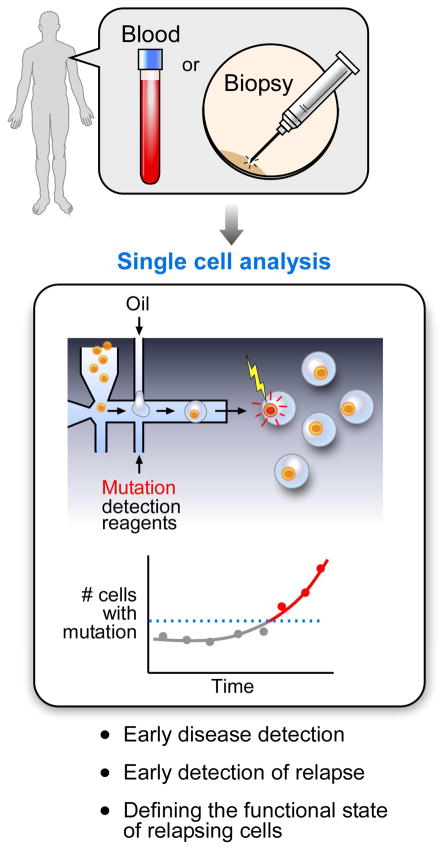
Deep sequencing of bulk samples remains the most effective approach for the unbiased and comprehensive discovery of sSNVs and sCNVs in cancer. However, high-dimension single-cell analysis, despite this limitation, is rapidly becoming a necessary tool for the study of cancer. As depicted in Figure 1, its key uses have been to confirm and refine subclone architecture, to detect and characterize less abundant cells, to enable in silico fractionation of cells, and to increase the power of correlation analysis. The ability of scRNA-seq to quantify numerous targets at single-cell resolution provides unprecedented power to distinguish different cell types and states. This is important for better classification of cancer subtypes and dissection of the complex interactions with immune cells, fibroblasts, and other cells in the tumor microenvironment. Future developments include increasing the scope of multiomic studies and greater adoption of high-dimension spatial analysis. Looking forward, we can envision the possibility of incorporating single cell analysis in clinical molecular diagnostics, in which sensitive detection of, for example, somatic mutations that confer therapeutic resistance could be detected far earlier than evidence of clinically apparent disease (Figure 4). This type of approach would be expected to facilitate efforts towards precision medicine, in which changes in the genetic content of the patient tumor would dictate how therapeutic choices are tailored.
Figure 4. Potential impact of single-cell analysis on cancer diagnostics.
An example of how single-cell analysis (in this case, using high throughput droplet approaches) could inform clinical diagnostics for early disease detection, early detection of relapse and evaluation of the functional state of the relapsed cells.
Acknowledgments
This work was supported by grants from the National Institutes of Health (NCI P01CA206978-01, R01CA182461-01, U10CA180861-01, R01CA184922-02). C.J.W. is a Scholar of the Leukemia and Lymphoma Society.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aceto N, Bardia A, Miyamoto DT, Donaldson MC, Wittner BS, Spencer JA, Yu M, Pely A, Engstrom A, Zhu H, Brannigan BW, Kapur R, Stott SL, Shioda T, Ramaswamy S, Ting DT, Lin CP, Toner M, Haber DA, Maheswaran S. Circulating tumor cell clusters are oligoclonal precursors of breast cancer metastasis. Cell. 2014;158:1110–1122. doi: 10.1016/j.cell.2014.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akrap N, Andersson D, Bom E, Gregersson P, Stahlberg A, Landberg G. Identification of Distinct Breast Cancer Stem Cell Populations Based on Single-Cell Analyses of Functionally Enriched Stem and Progenitor Pools. Stem Cell Reports. 2016;6:121–136. doi: 10.1016/j.stemcr.2015.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angermueller C, Clark SJ, Lee HJ, Macaulay IC, Teng MJ, Hu TX, Krueger F, Smallwood SA, Ponting CP, Voet T, Kelsey G, Stegle O, Reik W. Parallel single-cell sequencing links transcriptional and epigenetic heterogeneity. Nat Methods. 2016;13:229–232. doi: 10.1038/nmeth.3728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anjanappa M, Cardoso AA, Cheng L, Mohamad S, Gunawan A, Rice S, Dong Y, Li L, Sandusky GE, Srour EF, Nakshatri H. Individualized breast cancer characterization through single cell analysis of tumor and adjacent-normal cells. Cancer Res. 2017 doi: 10.1158/0008-5472.CAN-16-3308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacher R, Kendziorski C. Design and computational analysis of single-cell RNA-sequencing experiments. Genome Biol. 2016;17:63. doi: 10.1186/s13059-016-0927-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bengtsson M, Stahlberg A, Rorsman P, Kubista M. Gene expression profiling in single cells from the pancreatic islets of Langerhans reveals lognormal distribution of mRNA levels. Genome Res. 2005;15:1388–1392. doi: 10.1101/gr.3820805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bheda P, Schneider R. Epigenetics reloaded: the single-cell revolution. Trends Cell Biol. 2014;24:712–723. doi: 10.1016/j.tcb.2014.08.010. [DOI] [PubMed] [Google Scholar]
- Bingham C, Fernandez SV, Fittipaldi P, Dempsey PW, Ruth KJ, Cristofanilli M, Katherine Alpaugh R. Mutational studies on single circulating tumor cells isolated from the blood of inflammatory breast cancer patients. Breast Cancer Res Treat. 2017;163:219–230. doi: 10.1007/s10549-017-4176-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodenmiller B. Multiplexed Epitope-Based Tissue Imaging for Discovery and Healthcare Applications. Cell Syst. 2016;2:225–238. doi: 10.1016/j.cels.2016.03.008. [DOI] [PubMed] [Google Scholar]
- Bray MA, Singh S, Han H, Davis CT, Borgeson B, Hartland C, Kost-Alimova M, Gustafsdottir SM, Gibson CC, Carpenter AE. Cell Painting, a high-content image-based assay for morphological profiling using multiplexed fluorescent dyes. Nat Protoc. 2016;11:1757–1774. doi: 10.1038/nprot.2016.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burger JA, Landau DA, Taylor-Weiner A, Bozic I, Zhang H, Sarosiek K, Wang L, Stewart C, Fan J, Hoellenriegel J, Sivina M, Dubuc AM, Fraser C, Han Y, Li S, Livak KJ, Zou L, Wan Y, Konoplev S, Sougnez C, Brown JR, Abruzzo LV, Carter SL, Keating MJ, Davids MS, Wierda WG, Cibulskis K, Zenz T, Werner L, Dal Cin P, Kharchencko P, Neuberg D, Kantarjian H, Lander E, Gabriel S, O’Brien S, Letai A, Weitz DA, Nowak MA, Getz G, Wu CJ. Clonal evolution in patients with chronic lymphocytic leukaemia developing resistance to BTK inhibition. Nat Commun. 2016;7:11589. doi: 10.1038/ncomms11589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calabuig-Fariñas S, Jantus-Lewintre E, Herreros-Pomares A, Camps C. Circulating tumor cells versus circulating tumor DNA in lung cancer-which one will win? Transl Lung Cancer Res. 2016;5:466–482. doi: 10.21037/tlcr.2016.10.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campton DE, Ramirez AB, Nordberg JJ, Drovetto N, Clein AC, Varshavskaya P, Friemel BH, Quarre S, Breman A, Dorschner M, Blau S, Blau CA, Sabath DE, Stilwell JL, Kaldjian EP. High-recovery visual identification and single-cell retrieval of circulating tumor cells for genomic analysis using a dual-technology platform integrated with automated immunofluorescence staining. BMC Cancer. 2015;15:360. doi: 10.1186/s12885-015-1383-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen KH, Boettiger AN, Moffitt JR, Wang S, Zhuang X. RNA imaging. Spatially resolved, highly multiplexed RNA profiling in single cells. Science. 2015;348:aaa6090. doi: 10.1126/science.aaa6090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevrier S, Levine JH, Zanotelli VRT, Silina K, Schulz D, Bacac M, Ries CH, Ailles L, Jewett MAS, Moch H, van den Broek M, Beisel C, Stadler MB, Gedye C, Reis B, Pe’er D, Bodenmiller B. An Immune Atlas of Clear Cell Renal Cell Carcinoma. Cell. 2017;169:736–749. e718. doi: 10.1016/j.cell.2017.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark SJ, Lee HJ, Smallwood SA, Kelsey G, Reik W. Single-cell epigenomics: powerful new methods for understanding gene regulation and cell identity. Genome Biol. 2016;17:72. doi: 10.1186/s13059-016-0944-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corces MR, Buenrostro JD, Wu B, Greenside PG, Chan SM, Koenig JL, Snyder MP, Pritchard JK, Kundaje A, Greenleaf WJ, Majeti R, Chang HY. Lineage-specific and single-cell chromatin accessibility charts human hematopoiesis and leukemia evolution. Nat Genet. 2016;48:1193–1203. doi: 10.1038/ng.3646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosetto N, Bienko M, van Oudenaarden A. Spatially resolved transcriptomics and beyond. Nat Rev Genet. 2015;16:57–66. doi: 10.1038/nrg3832. [DOI] [PubMed] [Google Scholar]
- Dalerba P, Kalisky T, Sahoo D, Rajendran PS, Rothenberg ME, Leyrat AA, Sim S, Okamoto J, Johnston DM, Qian D, Zabala M, Bueno J, Neff NF, Wang J, Shelton AA, Visser B, Hisamori S, Shimono Y, van de Wetering M, Clevers H, Clarke MF, Quake SR. Single-cell dissection of transcriptional heterogeneity in human colon tumors. Nat Biotechnol. 2011;29:1120–1127. doi: 10.1038/nbt.2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demeulemeester J, Kumar P, Moller EK, Nord S, Wedge DC, Peterson A, Mathiesen RR, Fjelldal R, Zamani Esteki M, Theunis K, Fernandez Gallardo E, Grundstad AJ, Borgen E, Baumbusch LO, Borresen-Dale AL, White KP, Kristensen VN, Van Loo P, Voet T, Naume B. Tracing the origin of disseminated tumor cells in breast cancer using single-cell sequencing. Genome Biol. 2016;17:250. doi: 10.1186/s13059-016-1109-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dey SS, Kester L, Spanjaard B, Bienko M, van Oudenaarden A. Integrated genome and transcriptome sequencing of the same cell. Nat Biotechnol. 2015;33:285–289. doi: 10.1038/nbt.3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn GP, Old LJ, Schreiber RD. The immunobiology of cancer immunosurveillance and immunoediting. Immunity. 2004;21:137–148. doi: 10.1016/j.immuni.2004.07.017. [DOI] [PubMed] [Google Scholar]
- Ebinger S, Ozdemir EZ, Ziegenhain C, Tiedt S, Castro Alves C, Grunert M, Dworzak M, Lutz C, Turati VA, Enver T, Horny HP, Sotlar K, Parekh S, Spiekermann K, Hiddemann W, Schepers A, Polzer B, Kirsch S, Hoffmann M, Knapp B, Hasenauer J, Pfeifer H, Panzer-Grumayer R, Enard W, Gires O, Jeremias I. Characterization of Rare, Dormant, and Therapy-Resistant Cells in Acute Lymphoblastic Leukemia. Cancer Cell. 2016;30:849–862. doi: 10.1016/j.ccell.2016.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis JM, Zhang CZ, Maire CL, Jung J, Manzo VE, Adalsteinsson VA, Homer H, Haidar S, Blumenstiel B, Pedamallu CS, Ligon AH, Love JC, Meyerson M, Ligon KL. EGFR variant heterogeneity in glioblastoma resolved through single-nucleus sequencing. Cancer Discov. 2014 doi: 10.1158/2159-8290.CD-13-0879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao R, Davis A, McDonald TO, Sei E, Shi X, Wang Y, Tsai PC, Casasent A, Waters J, Zhang H, Meric-Bernstam F, Michor F, Navin NE. Punctuated copy number evolution and clonal stasis in triple-negative breast cancer. Nat Genet. 2016;48:1119–1130. doi: 10.1038/ng.3641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y, Ni X, Guo H, Su Z, Ba Y, Tong Z, Guo Z, Yao X, Chen X, Yin J, Yan Z, Guo L, Liu Y, Bai F, Xie XS, Zhang N. Single-cell sequencing deciphers a convergent evolution of copy number alterations from primary to circulating tumour cells. Genome Res. 2017 doi: 10.1101/gr.216788.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garvin T, Aboukhalil R, Kendall J, Baslan T, Atwal GS, Hicks J, Wigler M, Schatz MC. Interactive analysis and assessment of single-cell copy-number variations. Nat Methods. 2015;12:1058–1060. doi: 10.1038/nmeth.3578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gawad C, Koh W, Quake SR. Dissecting the clonal origins of childhood acute lymphoblastic leukemia by single-cell genomics. Proc Natl Acad Sci U S A. 2014;111:17947–17952. doi: 10.1073/pnas.1420822111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gawad C, Koh W, Quake SR. Single-cell genome sequencing: current state of the science. Nat Rev Genet. 2016;17:175–188. doi: 10.1038/nrg.2015.16. [DOI] [PubMed] [Google Scholar]
- Gierahn TM, Wadsworth MH, 2nd, Hughes TK, Bryson BD, Butler A, Satija R, Fortune S, Love JC, Shalek AK. Seq-Well: portable, low-cost RNA sequencing of single cells at high throughput. Nat Methods. 2017;14:395–398. doi: 10.1038/nmeth.4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giustacchini A, Thongjuea S, Barkas N, Woll PS, Povinelli BJ, Booth CAG, Sopp P, Norfo R, Rodriguez-Meira A, Ashley N, Jamieson L, Vyas P, Anderson K, Segerstolpe A, Qian H, Olsson-Stromberg U, Mustjoki S, Sandberg R, Jacobsen SEW, Mead AJ. Single-cell transcriptomics uncovers distinct molecular signatures of stem cells in chronic myeloid leukemia. Nat Med. 2017;23:692–702. doi: 10.1038/nm.4336. [DOI] [PubMed] [Google Scholar]
- Gullaksen SE, Skavland J, Gavasso S, Tosevski V, Warzocha K, Dumrese C, Ferrant A, Gedde-Dahl T, Hellmann A, Janssen J, Labar B, Lang A, Majeed W, Mihaylov G, Stentoft J, Stenke L, Thaler J, Thielen N, Verhoef G, Voglova J, Ossenkoppele G, Hochhaus A, Hjorth-Hansen H, Mustjoki S, Sopper S, Giles F, Porkka K, Wolf D, Gjertsen BT. Single cell immune profiling by mass cytometry of newly diagnosed chronic phase chronic myeloid leukaemia treated with nilotinib. Haematologica. 2017 doi: 10.3324/haematol.2017.167080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han L, Zi X, Garmire LX, Wu Y, Weissman SM, Pan X, Fan R. Co-detection and sequencing of genes and transcripts from the same single cells facilitated by a microfluidics platform. Sci Rep. 2014;4:6485. doi: 10.1038/srep06485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardingham JE, Grover P, Winter M, Hewett PJ, Price TJ, Thierry B. Detection and Clinical Significance of Circulating Tumor Cells in Colorectal Cancer--20 Years of Progress. Mol Med. 2015;21(Suppl 1):S25–31. doi: 10.2119/molmed.2015.00149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heath JR, Ribas A, Mischel PS. Single-cell analysis tools for drug discovery and development. Nat Rev Drug Discov. 2016;15:204–216. doi: 10.1038/nrd.2015.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heitzer E, Auer M, Gasch C, Pichler M, Ulz P, Hoffmann EM, Lax S, Waldispuehl-Geigl J, Mauermann O, Lackner C, Hofler G, Eisner F, Sill H, Samonigg H, Pantel K, Riethdorf S, Bauernhofer T, Geigl JB, Speicher MR. Complex tumor genomes inferred from single circulating tumor cells by array-CGH and next-generation sequencing. Cancer Res. 2013;73:2965–2975. doi: 10.1158/0008-5472.CAN-12-4140. [DOI] [PubMed] [Google Scholar]
- Hou Y, Guo H, Cao C, Li X, Hu B, Zhu P, Wu X, Wen L, Tang F, Huang Y, Peng J. Single-cell triple omics sequencing reveals genetic, epigenetic, and transcriptomic heterogeneity in hepatocellular carcinomas. Cell Res. 2016;26:304–319. doi: 10.1038/cr.2016.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Y, Song L, Zhu P, Zhang B, Tao Y, Xu X, Li F, Wu K, Liang J, Shao D, Wu H, Ye X, Ye C, Wu R, Jian M, Chen Y, Xie W, Zhang R, Chen L, Liu X, Yao X, Zheng H, Yu C, Li Q, Gong Z, Mao M, Yang X, Yang L, Li J, Wang W, Lu Z, Gu N, Laurie G, Bolund L, Kristiansen K, Wang J, Yang H, Li Y, Zhang X, Wang J. Single-cell exome sequencing and monoclonal evolution of a JAK2-negative myeloproliferative neoplasm. Cell. 2012;148:873–885. doi: 10.1016/j.cell.2012.02.028. [DOI] [PubMed] [Google Scholar]
- Hu P, Zhang W, Xin H, Deng G. Single Cell Isolation and Analysis. Front Cell Dev Biol. 2016;25:116. doi: 10.3389/fcell.2016.00116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Huang K, An Q, Du G, Hu G, Xue J, Zhu X, Wang CY, Xue Z, Fan G. Simultaneous profiling of transcriptome and DNA methylome from a single cell. Genome Biol. 2016;17:88. doi: 10.1186/s13059-016-0950-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hug H, Schuler R. Measurement of the number of molecules of a single mRNA species in a complex mRNA preparation. J Theor Biol. 2003;221:615–624. doi: 10.1006/jtbi.2003.3211. [DOI] [PubMed] [Google Scholar]
- Jiang R, Lu YT, Ho H, Li B, Chen JF, Lin M, Li F, Wu K, Wu H, Lichterman J, Wan H, Lu CL, OuYang W, Ni M, Wang L, Li G, Lee T, Zhang X, Yang J, Rettig M, Chung LW, Yang H, Li KC, Hou Y, Tseng HR, Hou S, Xu X, Wang J, Posadas EM. A comparison of isolated circulating tumor cells and tissue biopsies using whole-genome sequencing in prostate cancer. Oncotarget. 2015;6:44781–44793. doi: 10.18632/oncotarget.6330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidess E, Jeffrey SS. Circulating tumor cells versus tumor-derived cell-free DNA: rivals or partners in cancer care in the era of single-cell analysis? Genome Med. 2013;5:70. doi: 10.1186/gm474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Zheng S, Amini SS, Virk SM, Mikkelsen T, Brat DJ, Grimsby J, Sougnez C, Muller F, Hu J, Sloan AE, Cohen ML, Van Meir EG, Scarpace L, Laird PW, Weinstein JN, Lander ES, Gabriel S, Getz G, Meyerson M, Chin L, Barnholtz-Sloan JS, Verhaak RG. Whole-genome and multisector exome sequencing of primary and post-treatment glioblastoma reveals patterns of tumor evolution. Genome Res. 2015;25:316–327. doi: 10.1101/gr.180612.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kivioja T, Vaharautio A, Karlsson K, Bonke M, Enge M, Linnarsson S, Taipale J. Counting absolute numbers of molecules using unique molecular identifiers. Nat Methods. 2011;9:72–74. doi: 10.1038/nmeth.1778. [DOI] [PubMed] [Google Scholar]
- Klein AM, Mazutis L, Akartuna I, Tallapragada N, Veres A, Li V, Peshkin L, Weitz DA, Kirschner MW. Droplet barcoding for single-cell transcriptomics applied to embryonic stem cells. Cell. 2015;161:1187–1201. doi: 10.1016/j.cell.2015.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolodziejczyk AA, Kim JK, Svensson V, Marioni JC, Teichmann SA. The technology and biology of single-cell RNA sequencing. Mol Cell. 2015;58:610–620. doi: 10.1016/j.molcel.2015.04.005. [DOI] [PubMed] [Google Scholar]
- Krivacic RT, Ladanyi A, Curry DN, Hsieh HB, Kuhn P, Bergsrud DE, Kepros JF, Barbera T, Ho MY, Chen LB, Lerner RA, Bruce RH. A rare-cell detector for cancer. Proc Natl Acad Sci U S A. 2004;101:10501–10504. doi: 10.1073/pnas.0404036101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landau DA, Carter SL, Stojanov P, McKenna A, Stevenson K, Lawrence MS, Sougnez C, Stewart C, Sivachenko A, Wang L, Wan Y, Zhang W, Shukla SA, Vartanov A, Fernandes SM, Saksena G, Cibulskis K, Tesar B, Gabriel S, Hacohen N, Meyerson M, Lander ES, Neuberg D, Brown JR, Getz G, Wu CJ. Evolution and impact of subclonal mutations in chronic lymphocytic leukemia. Cell. 2013;152:714–726. doi: 10.1016/j.cell.2013.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavin Y, Kobayashi S, Leader A, Amir ED, Elefant N, Bigenwald C, Remark R, Sweeney R, Becker CD, Levine JH, Meinhof K, Chow A, Kim-Shulze S, Wolf A, Medaglia C, Li H, Rytlewski JA, Emerson RO, Solovyov A, Greenbaum BD, Sanders C, Vignali M, Beasley MB, Flores R, Gnjatic S, Pe’er D, Rahman A, Amit I, Merad M. Innate Immune Landscape in Early Lung Adenocarcinoma by Paired Single-Cell Analyses. Cell. 2017;169:750–765. e717. doi: 10.1016/j.cell.2017.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson DA, Bhakta NR, Kessenbrock K, Prummel KD, Yu Y, Takai K, Zhou A, Eyob H, Balakrishnan S, Wang CY, Yaswen P, Goga A, Werb Z. Single-cell analysis reveals a stem-cell program in human metastatic breast cancer cells. Nature. 2015;526:131–135. doi: 10.1038/nature15260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Daugharthy ER, Scheiman J, Kalhor R, Ferrante TC, Terry R, Turczyk BM, Yang JL, Lee HS, Aach J, Zhang K, Church GM. Fluorescent in situ sequencing (FISSEQ) of RNA for gene expression profiling in intact cells and tissues. Nat Protoc. 2015;10:442–458. doi: 10.1038/nprot.2014.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung ML, Davis A, Gao R, Casasent A, Wang Y, Sei E, Vilar E, Maru D, Kopetz S, Navin NE. Single-cell DNA sequencing reveals a late-dissemination model in metastatic colorectal cancer. Genome Res. 2017 doi: 10.1101/gr.209973.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine JH, Simonds EF, Bendall SC, Davis KL, Amir el AD, Tadmor MD, Litvin O, Fienberg HG, Jager A, Zunder ER, Finck R, Gedman AL, Radtke I, Downing JR, Pe’er D, Nolan GP. Data-Driven Phenotypic Dissection of AML Reveals Progenitor-like Cells that Correlate with Prognosis. Cell. 2015;162:184–197. doi: 10.1016/j.cell.2015.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Courtois ET, Sengupta D, Tan Y, Chen KH, Goh JJL, Kong SL, Chua C, Hon LK, Tan WS, Wong M, Choi PJ, Wee LJK, Hillmer AM, Tan IB, Robson P, Prabhakar S. Reference component analysis of single-cell transcriptomes elucidates cellular heterogeneity in human colorectal tumors. Nat Genet. 2017 doi: 10.1038/ng.3818. [DOI] [PubMed] [Google Scholar]
- Li Y, Xu X, Song L, Hou Y, Li Z, Tsang S, Li F, Im KM, Wu K, Wu H, Ye X, Li G, Wang L, Zhang B, Liang J, Xie W, Wu R, Jiang H, Liu X, Yu C, Zheng H, Jian M, Nie L, Wan L, Shi M, Sun X, Tang A, Guo G, Gui Y, Cai Z, Li J, Wang W, Lu Z, Zhang X, Bolund L, Kristiansen K, Wang J, Yang H, Dean M, Wang J. Single-cell sequencing analysis characterizes common and cell-lineage-specific mutations in a muscle-invasive bladder cancer. Gigascience. 2012;1:12. doi: 10.1186/2047-217X-1-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litzenburger UM, Buenrostro JD, Wu B, Shen Y, Sheffield NC, Kathiria A, Greenleaf WJ, Chang HY. Single-cell epigenomic variability reveals functional cancer heterogeneity. Genome Biol. 2017;18:15. doi: 10.1186/s13059-016-1133-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Trapnell C. Single-cell transcriptome sequencing: recent advances and remaining challenges. F1000Res. 2016;5 doi: 10.12688/f1000research.7223.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ. In: Eukaryotic Single-Cell mRNA Sequencing. in Field Guidelines for Genetic Experimental Designs in High-Throughput Sequencing. Aransay AM, Lavín Trueba JL, editors. Switzerland: Springer International Publishing; 2016. pp. 343–365. [DOI] [Google Scholar]
- Livak KJ, Wills QF, Tipping AJ, Datta K, Mittal R, Goldson AJ, Sexton DW, Holmes CC. Methods for qPCR gene expression profiling applied to 1440 lymphoblastoid single cells. Methods. 2013;59:71–79. doi: 10.1016/j.ymeth.2012.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohr JG, Adalsteinsson VA, Cibulskis K, Choudhury AD, Rosenberg M, Cruz-Gordillo P, Francis JM, Zhang CZ, Shalek AK, Satija R, Trombetta JJ, Lu D, Tallapragada N, Tahirova N, Kim S, Blumenstiel B, Sougnez C, Lowe A, Wong B, Auclair D, Van Allen EM, Nakabayashi M, Lis RT, Lee GS, Li T, Chabot MS, Ly A, Taplin ME, Clancy TE, Loda M, Regev A, Meyerson M, Hahn WC, Kantoff PW, Golub TR, Getz G, Boehm JS, Love JC. Whole-exome sequencing of circulating tumor cells provides a window into metastatic prostate cancer. Nat Biotechnol. 2014;32:479–484. doi: 10.1038/nbt.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohr JG, Kim S, Gould J, Knoechel B, Drier Y, Cotton MJ, Gray D, Birrer N, Wong B, Ha G, Zhang CZ, Guo G, Meyerson M, Yee AJ, Boehm JS, Raje N, Golub TR. Genetic interrogation of circulating multiple myeloma cells at single-cell resolution. Sci Transl Med. 2016;8:363ra147. doi: 10.1126/scitranslmed.aac7037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Xue Q, Eisele MR, Sulistijo ES, Brower K, Han L, Amir el AD, Pe’er D, Miller-Jensen K, Fan R. Highly multiplexed profiling of single-cell effector functions reveals deep functional heterogeneity in response to pathogenic ligands. Proc Natl Acad Sci U S A. 2015;112:E607–615. doi: 10.1073/pnas.1416756112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubeck E, Coskun AF, Zhiyentayev T, Ahmad M, Cai L. Single-cell in situ RNA profiling by sequential hybridization. Nat Methods. 2014;11:360–361. doi: 10.1038/nmeth.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macaulay IC, Haerty W, Kumar P, Li YI, Hu TX, Teng MJ, Goolam M, Saurat N, Coupland P, Shirley LM, Smith M, Van der Aa N, Banerjee R, Ellis PD, Quail MA, Swerdlow HP, Zernicka-Goetz M, Livesey FJ, Ponting CP, Voet T. G&T-seq: parallel sequencing of single-cell genomes and transcriptomes. Nat Methods. 2015;12:519–522. doi: 10.1038/nmeth.3370. [DOI] [PubMed] [Google Scholar]
- Macaulay IC, Ponting CP, Voet T. Single-Cell Multiomics: Multiple Measurements from Single Cells. Trends Genet. 2017;33:155–168. doi: 10.1016/j.tig.2016.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macaulay IC, Svensson V, Labalette C, Ferreira L, Hamey F, Voet T, Teichmann SA, Cvejic A. Single-Cell RNA-Sequencing Reveals a Continuous Spectrum of Differentiation in Hematopoietic Cells. Cell Rep. 2016;14:966–977. doi: 10.1016/j.celrep.2015.12.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macaulay IC, Voet T. Single cell genomics: advances and future perspectives. PLoS Genet. 2014;10:e1004126. doi: 10.1371/journal.pgen.1004126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macosko EZ, Basu A, Satija R, Nemesh J, Shekhar K, Goldman M, Tirosh I, Bialas AR, Kamitaki N, Martersteck EM, Trombetta JJ, Weitz DA, Sanes JR, Shalek AK, Regev A, McCarroll SA. Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets. Cell. 2015;161:1202–1214. doi: 10.1016/j.cell.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann KM, Newberg JY, Black MA, Jones DJ, Amaya-Manzanares F, Guzman-Rojas L, Kodama T, Ward JM, Rust AG, van der Weyden L, Yew CC, Waters JL, Leung ML, Rogers K, Rogers SM, McNoe LA, Selvanesan L, Navin N, Jenkins NA, Copeland NG, Mann MB. Analyzing tumor heterogeneity and driver genes in single myeloid leukemia cells with SBCapSeq. Nat Biotechnol. 2016;34:962–972. doi: 10.1038/nbt.3637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mato Prado M, Frampton AE, Stebbing J, Krell J. Single-cell sequencing in cancer research. Expert Rev Mol Diagn. 2016;16:1–5. doi: 10.1586/14737159.2016.1115345. [DOI] [PubMed] [Google Scholar]
- Miyamoto DT, Zheng Y, Wittner BS, Lee RJ, Zhu H, Broderick KT, Desai R, Fox DB, Brannigan BW, Trautwein J, Arora KS, Desai N, Dahl DM, Sequist LV, Smith MR, Kapur R, Wu CL, Shioda T, Ramaswamy S, Ting DT, Toner M, Maheswaran S, Haber DA. RNA-Seq of single prostate CTCs implicates noncanonical Wnt signaling in antiandrogen resistance. Science. 2015;349:1351–1356. doi: 10.1126/science.aab0917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mroz EA, Rocco JW. MATH, a novel measure of intratumor genetic heterogeneity, is high in poor-outcome classes of head and neck squamous cell carcinoma. Oral Oncol. 2013;49:211–215. doi: 10.1016/j.oraloncology.2012.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller S, Diaz A. Single-Cell mRNA Sequencing in Cancer Research: Integrating the Genomic Fingerprint. Front Genet. 2017;8:73. doi: 10.3389/fgene.2017.00073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller S, Liu SJ, Di Lullo E, Malatesta M, Pollen AA, Nowakowski TJ, Kohanbash G, Aghi M, Kriegstein AR, Lim DA, Diaz A. Single-cell sequencing maps gene expression to mutational phylogenies in PDGF- and EGF-driven gliomas. Mol Syst Biol. 2016;12:889. doi: 10.15252/msb.20166969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray E, Cho JH, Goodwin D, Ku T, Swaney J, Kim SY, Choi H, Park YG, Park JY, Hubbert A, McCue M, Vassallo S, Bakh N, Frosch MP, Wedeen VJ, Seung HS, Chung K. Simple, Scalable Proteomic Imaging for High-Dimensional Profiling of Intact Systems. Cell. 2015;163:1500–1514. doi: 10.1016/j.cell.2015.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navin N, Kendall J, Troge J, Andrews P, Rodgers L, McIndoo J, Cook K, Stepansky A, Levy D, Esposito D, Muthuswamy L, Krasnitz A, McCombie WR, Hicks J, Wigler M. Tumour evolution inferred by single-cell sequencing. Nature. 2011;472:90–94. doi: 10.1038/nature09807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navin NE. Delineating cancer evolution with single-cell sequencing. Sci Transl Med. 2015a;7:296fs229. doi: 10.1126/scitranslmed.aac8319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navin NE. The first five years of single-cell cancer genomics and beyond. Genome Res. 2015b;25:1499–1507. doi: 10.1101/gr.191098.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen A, Yoshida M, Goodarzi H, Tavazoie SF. Highly variable cancer subpopulations that exhibit enhanced transcriptome variability and metastatic fitness. Nat Commun. 2016;7:11246. doi: 10.1038/ncomms11246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni X, Zhuo M, Su Z, Duan J, Gao Y, Wang Z, Zong C, Bai H, Chapman AR, Zhao J, Xu L, An T, Ma Q, Wang Y, Wu M, Sun Y, Wang S, Li Z, Yang X, Yong J, Su XD, Lu Y, Bai F, Xie XS, Wang J. Reproducible copy number variation patterns among single circulating tumor cells of lung cancer patients. Proc Natl Acad Sci U S A. 2013;110:21083–21088. doi: 10.1073/pnas.1320659110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichterwitz S, Chen G, Aguila Benitez J, Yilmaz M, Storvall H, Cao M, Sandberg R, Deng Q, Hedlund E. Laser capture microscopy coupled with Smart-seq2 for precise spatial transcriptomic profiling. Nat Commun. 2016;7:12139. doi: 10.1038/ncomms12139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oehler VG, Qin J, Ramakrishnan R, Facer G, Ananthnarayan S, Cummings C, Deininger M, Shah N, McCormick F, Willis S, Daridon A, Unger M, Radich JP. Absolute quantitative detection of ABL tyrosine kinase domain point mutations in chronic myeloid leukemia using a novel nanofluidic platform and mutation-specific PCR. Leukemia. 2009;23:396–399. doi: 10.1038/leu.2008.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paguirigan AL, Smith J, Meshinchi S, Carroll M, Maley C, Radich JP. Single-cell genotyping demonstrates complex clonal diversity in acute myeloid leukemia. Sci Transl Med. 2015;7:281re282. doi: 10.1126/scitranslmed.aaa0763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papaemmanuil E, Gerstung M, Malcovati L, Tauro S, Gundem G, Van Loo P, Yoon CJ, Ellis P, Wedge DC, Pellagatti A, Shlien A, Groves MJ, Forbes SA, Raine K, Hinton J, Mudie LJ, McLaren S, Hardy C, Latimer C, Della Porta MG, O’Meara S, Ambaglio I, Galli A, Butler AP, Walldin G, Teague JW, Quek L, Sternberg A, Gambacorti-Passerini C, Cross NC, Green AR, Boultwood J, Vyas P, Hellstrom-Lindberg E, Bowen D, Cazzola M, Stratton MR, Campbell PJ Chronic Myeloid Disorders Working Group of the International Cancer Genome C. Clinical and biological implications of driver mutations in myelodysplastic syndromes. Blood. 2013;122:3616–3627. doi: 10.1182/blood-2013-08-518886. quiz 3699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papaemmanuil E, Rapado I, Li Y, Potter NE, Wedge DC, Tubio J, Alexandrov LB, Van Loo P, Cooke SL, Marshall J, Martincorena I, Hinton J, Gundem G, van Delft FW, Nik-Zainal S, Jones DR, Ramakrishna M, Titley I, Stebbings L, Leroy C, Menzies A, Gamble J, Robinson B, Mudie L, Raine K, O’Meara S, Teague JW, Butler AP, Cazzaniga G, Biondi A, Zuna J, Kempski H, Muschen M, Ford AM, Stratton MR, Greaves M, Campbell PJ. RAG-mediated recombination is the predominant driver of oncogenic rearrangement in ETV6-RUNX1 acute lymphoblastic leukemia. Nat Genet. 2014;46:116–125. doi: 10.1038/ng.2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker JS, Mullins M, Cheang MC, Leung S, Voduc D, Vickery T, Davies S, Fauron C, He X, Hu Z, Quackenbush JF, Stijleman IJ, Palazzo J, Marron JS, Nobel AB, Mardis E, Nielsen TO, Ellis MJ, Perou CM, Bernard PS. Supervised risk predictor of breast cancer based on intrinsic subtypes. J Clin Oncol. 2009;27:1160–1167. doi: 10.1200/JCO.2008.18.1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel AP, Tirosh I, Trombetta JJ, Shalek AK, Gillespie SM, Wakimoto H, Cahill DP, Nahed BV, Curry WT, Martuza RL, Louis DN, Rozenblatt-Rosen O, Suva ML, Regev A, Bernstein BE. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science. 2014;344:1396–1401. doi: 10.1126/science.1254257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirion OB, Zhu X, Ching T, Garmire L. Single-Cell Transcriptomics Bioinformatics and Computational Challenges. Front Genet. 2016;7:163. doi: 10.3389/fgene.2016.00163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter NE, Ermini L, Papaemmanuil E, Cazzaniga G, Vijayaraghavan G, Titley I, Ford A, Campbell P, Kearney L, Greaves M. Single cell mutational profiling and clonal phylogeny in cancer. Genome Res. 2013 doi: 10.1101/gr.159913.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakadan SM, Shalek AK, Weitz DA. Scaling by shrinking: empowering single-cell ‘omics’ with microfluidic devices. Nat Rev Genet. 2017 doi: 10.1038/nrg.2017.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quek L, Otto GW, Garnett C, Lhermitte L, Karamitros D, Stoilova B, Lau IJ, Doondeea J, Usukhbayar B, Kennedy A, Metzner M, Goardon N, Ivey A, Allen C, Gale R, Davies B, Sternberg A, Killick S, Hunter H, Cahalin P, Price A, Carr A, Griffiths M, Virgo P, Mackinnon S, Grimwade D, Freeman S, Russell N, Craddock C, Mead A, Peniket A, Porcher C, Vyas P. Genetically distinct leukemic stem cells in human CD34- acute myeloid leukemia are arrested at a hemopoietic precursor-like stage. J Exp Med. 2016;213:1513–1535. doi: 10.1084/jem.20151775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Racila E, Euhus D, Weiss AJ, Rao C, McConnell J, Terstappen LW, Uhr JW. Detection and characterization of carcinoma cells in the blood. Proc Natl Acad Sci U S A. 1998;95:4589–4594. doi: 10.1073/pnas.95.8.4589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramskold D, Luo S, Wang YC, Li R, Deng Q, Faridani OR, Daniels GA, Khrebtukova I, Loring JF, Laurent LC, Schroth GP, Sandberg R. Full-length mRNA-Seq from single-cell levels of RNA and individual circulating tumor cells. Nat Biotechnol. 2012;30:777–782. doi: 10.1038/nbt.2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remark R, Merghoub T, Grabe N, Litjens G, Damotte D, Wolchuk JD, Merad M, Gnjatic S. In-depth tissue profiling using multiplexed immunohistochemical consecutive staining on single slide. Sci Immuol. 2016;1:aaf6925. doi: 10.1126/sciimmunol.aaf6925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross EM, Markowetz F. OncoNEM: inferring tumor evolution from single-cell sequencing data. Genome Biol. 2016;17:69. doi: 10.1186/s13059-016-0929-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saadatpour A, Lai S, Guo G, Yuan GC. Single-Cell Analysis in Cancer Genomics. Trends Genet. 2015;31:576–586. doi: 10.1016/j.tig.2015.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt F, Efferth T. Tumor Heterogeneity, Single-Cell Sequencing, and Drug Resistance. Pharmaceuticals (Basel) 2016;9 doi: 10.3390/ph9020033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw JA, Guttery DS, Hills A, Fernandez-Garcia D, Page K, Rosales BM, Goddard KS, Hastings RK, Luo J, Ogle O, Woodley L, Ali S, Stebbing J, Coombes RC. Mutation Analysis of Cell-Free DNA and Single Circulating Tumor Cells in Metastatic Breast Cancer Patients with High Circulating Tumor Cell Counts. Clin Cancer Res. 2017;23:88–96. doi: 10.1158/1078-0432.CCR-16-0825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slack JL, Bi W, Livak KJ, Beaubier N, Yu M, Clark M, Kim SH, Gallagher RE, Willman CL. Pre-clinical validation of a novel, highly sensitive assay to detect PML-RARalpha mRNA using real-time reverse-transcription polymerase chain reaction. J Mol Diagn. 2001;3:141–149. doi: 10.1016/s1525-1578(10)60665-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y, Tian T, Shi Y, Liu W, Zou Y, Khajvand T, Wang S, Zhu Z, Yang C. Enrichment and single-cell analysis of circulating tumor cells. Chem Sci. 2017;8:1736–1751. doi: 10.1039/c6sc04671a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitzer MH, Nolan GP. Mass Cytometry: Single Cells, Many Features. Cell. 2016;165:780–791. doi: 10.1016/j.cell.2016.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stegle O, Teichmann SA, Marioni JC. Computational and analytical challenges in single-cell transcriptomics. Nat Rev Genet. 2015;16:133–145. doi: 10.1038/nrg3833. [DOI] [PubMed] [Google Scholar]
- Stubbington MJ, Lonnberg T, Proserpio V, Clare S, Speak AO, Dougan G, Teichmann SA. T cell fate and clonality inference from single-cell transcriptomes. Nat Methods. 2016;13:329–332. doi: 10.1038/nmeth.3800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Y, Shi Q, Wei W. Single cell proteomics in biomedicine: High-dimensional data acquisition, visualization, and analysis. Proteomics. 2017:17. doi: 10.1002/pmic.201600267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun HJ, Chen J, Ni B, Yang X, Wu YZ. Recent advances and current issues in single-cell sequencing of tumors. Cancer Lett. 2015;365:1–10. doi: 10.1016/j.canlet.2015.04.022. [DOI] [PubMed] [Google Scholar]
- Szulwach KE, Livak KJ. Eukaryotic Single-Cell DNA Sequencing. In: Aransay AM, Lavín Trueba JL, editors. Field Guidelines for Genetic Experimental Designs in High-Throughput Sequencing. Switzerland: Springer International Publishing; 2016. pp. 367–384. [DOI] [Google Scholar]
- Tellez-Gabriel M, Ory B, Lamoureux F, Heymann MF, Heymann D. Tumour Heterogeneity: The Key Advantages of Single-Cell Analysis. Int J Mol Sci. 2016:17. doi: 10.3390/ijms17122142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ting DT, Wittner BS, Ligorio M, Vincent Jordan N, Shah AM, Miyamoto DT, Aceto N, Bersani F, Brannigan BW, Xega K, Ciciliano JC, Zhu H, MacKenzie OC, Trautwein J, Arora KS, Shahid M, Ellis HL, Qu N, Bardeesy N, Rivera MN, Deshpande V, Ferrone CR, Kapur R, Ramaswamy S, Shioda T, Toner M, Maheswaran S, Haber DA. Single-cell RNA sequencing identifies extracellular matrix gene expression by pancreatic circulating tumor cells. Cell Rep. 2014;8:1905–1918. doi: 10.1016/j.celrep.2014.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirosh I, Izar B, Prakadan SM, Wadsworth MH, II, Treacy D, Trombetta JJ, Rotem A, Rodman C, Lian C, Murphy G, Fallahi-Sichani M, Dutton-Regester K, Lin JR, Cohen O, Shah P, Lu D, Genshaft AS, Hughes TK, Ziegler CG, Kazer SW, Gaillard A, Kolb KE, Villani AC, Johannessen CM, Andreev AY, Van Allen EM, Bertagnolli M, Sorger PK, Sullivan RJ, Flaherty KT, Frederick DT, Jane-Valbuena J, Yoon CH, Rozenblatt-Rosen O, Shalek AK, Regev A, Garraway LA. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science. 2016;352:189–196. doi: 10.1126/science.aad0501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirosh I, Venteicher AS, Hebert C, Escalante LE, Patel AP, Yizhak K, Fisher JM, Rodman C, Mount C, Filbin MG, Neftel C, Desai N, Nyman J, Izar B, Luo CC, Francis JM, Patel AA, Onozato ML, Riggi N, Livak KJ, Gennert D, Satija R, Nahed BV, Curry WT, Martuza RL, Mylvaganam R, Iafrate AJ, Frosch MP, Golub TR, Rivera MN, Getz G, Rozenblatt-Rosen O, Cahill DP, Monje M, Bernstein BE, Louis DN, Regev A, Suva ML. Single-cell RNA-seq supports a developmental hierarchy in human oligodendroglioma. Nature. 2016;539:309–313. doi: 10.1038/nature20123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Cacchiarelli D, Grimsby J, Pokharel P, Li S, Morse M, Lennon NJ, Livak KJ, Mikkelsen TS, Rinn JL. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat Biotechnol. 2014;32:381–386. doi: 10.1038/nbt.2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Toom EE, Verdone JE, Gorin MA, Pienta KJ. Technical challenges in the isolation and analysis of circulating tumor cells. Oncotarget. 2016;7:62754–62766. doi: 10.18632/oncotarget.11191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Loo P, Voet T. Single cell analysis of cancer genomes. Curr Opin Genet Dev. 2014;24:82–91. doi: 10.1016/j.gde.2013.12.004. [DOI] [PubMed] [Google Scholar]
- Wagner A, Regev A, Yosef N. Revealing the vectors of cellular identity with single-cell genomics. Nat Biotechnol. 2016;34:1145–1160. doi: 10.1038/nbt.3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Brooks AN, Fan J, Wan Y, Gambe R, Li S, Hergert S, Yin S, Freeman SS, Levin JZ, Fan L, Seiler M, Buonamici S, Smith PG, Chau KF, Cibulskis CL, Zhang W, Rassenti LZ, Ghia EM, Kipps TJ, Fernandes S, Bloch DB, Kotliar D, Landau DA, Shukla SA, Aster JC, Reed R, DeLuca DS, Brown JR, Neuberg D, Getz G, Livak KJ, Meyerson MM, Kharchenko PV, Wu CJ. Transcriptomic Characterization of SF3B1 Mutation Reveals Its Pleiotropic Effects in Chronic Lymphocytic Leukemia. Cancer Cell. 2016;30:750–763. doi: 10.1016/j.ccell.2016.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Fan J, Francis JM, Georghiou G, Hergert S, Li S, Gambe R, Zhou CW, Yang C, Xiao S, Cin PD, Bowden M, Kotliar D, Shukla SA, Brown JR, Neuberg D, Alessi DR, Zhang CZ, Kharchenko PV, Livak KJ, Wu CJ. Integrated single-cell genetic and transcriptional analysis suggests novel drivers of chronic lymphocytic leukemia. Genome Res. 2017;27:1300–1311. doi: 10.1101/gr.217331.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Navin NE. Advances and applications of single-cell sequencing technologies. Mol Cell. 2015;58:598–609. doi: 10.1016/j.molcel.2015.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Waters J, Leung ML, Unruh A, Roh W, Shi X, Chen K, Scheet P, Vattathil S, Liang H, Multani A, Zhang H, Zhao R, Michor F, Meric-Bernstam F, Navin NE. Clonal evolution in breast cancer revealed by single nucleus genome sequencing. Nature. 2014;512:155–160. doi: 10.1038/nature13600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wills QF, Higgs DR, Mead AJ. Studying epigenomics in single cells: what is feasible and what can we learn? Epigenomics. 2015;7:1231–1234. doi: 10.2217/epi.15.93. [DOI] [PubMed] [Google Scholar]
- Wills QF, Livak KJ, Tipping AJ, Enver T, Goldson AJ, Sexton DW, Holmes C. Single-cell gene expression analysis reveals genetic associations masked in whole-tissue experiments. Nat Biotechnol. 2013;31:748–752. doi: 10.1038/nbt.2642. [DOI] [PubMed] [Google Scholar]
- Wills QF, Mead AJ. Application of single-cell genomics in cancer: promise and challenges. Hum Mol Genet. 2015;24:R74–84. doi: 10.1093/hmg/ddv235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winterhoff BJ, Maile M, Mitra AK, Sebe A, Bazzaro M, Geller MA, Abrahante JE, Klein M, Hellweg R, Mullany SA, Beckman K, Daniel J, Starr TK. Single cell sequencing reveals heterogeneity within ovarian cancer epithelium and cancer associated stromal cells. Gynecol Oncol. 2017;144:598–606. doi: 10.1016/j.ygyno.2017.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Hou Y, Yin X, Bao L, Tang A, Song L, Li F, Tsang S, Wu K, Wu H, He W, Zeng L, Xing M, Wu R, Jiang H, Liu X, Cao D, Guo G, Hu X, Gui Y, Li Z, Xie W, Sun X, Shi M, Cai Z, Wang B, Zhong M, Li J, Lu Z, Gu N, Zhang X, Goodman L, Bolund L, Wang J, Yang H, Kristiansen K, Dean M, Li Y, Wang J. Single-cell exome sequencing reveals single-nucleotide mutation characteristics of a kidney tumor. Cell. 2012;148:886–895. doi: 10.1016/j.cell.2012.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye B, Gao Q, Zeng Z, Stary CM, Jian Z, Xiong X, Gu L. Single-Cell Sequencing Technology in Oncology: Applications for Clinical Therapies and Research. Anal Cell Pathol (Amst) 2016;2016:9369240. doi: 10.1155/2016/9369240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C, Yu J, Yao X, Wu WK, Lu Y, Tang S, Li X, Bao L, Li X, Hou Y, Wu R, Jian M, Chen R, Zhang F, Xu L, Fan F, He J, Liang Q, Wang H, Hu X, He M, Zhang X, Zheng H, Li Q, Wu H, Chen Y, Yang X, Zhu S, Xu X, Yang H, Wang J, Zhang X, Sung JJ, Li Y, Wang J. Discovery of biclonal origin and a novel oncogene SLC12A5 in colon cancer by single-cell sequencing. Cell Res. 2014;24:701–712. doi: 10.1038/cr.2014.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zafar H, Wang Y, Nakhleh L, Navin N, Chen K. Monovar: single-nucleotide variant detection in single cells. Nat Methods. 2016;13:505–507. doi: 10.1038/nmeth.3835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang CZ, Spektor A, Cornils H, Francis JM, Jackson EK, Liu S, Meyerson M, Pellman D. Chromothripsis from DNA damage in micronuclei. Nature. 2015;522:179–184. doi: 10.1038/nature14493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Chen K, Fan ZH. Circulating Tumor Cell Isolation and Analysis. Adv Clin Chem. 2016;75:1–31. doi: 10.1016/bs.acc.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Marjani SL, Hu Z, Weissman SM, Pan X, Wu S. Single-Cell Sequencing for Precise Cancer Research: Progress and Prospects. Cancer Res. 2016;76:1305–1312. doi: 10.1158/0008-5472.CAN-15-1907. [DOI] [PubMed] [Google Scholar]
- Zheng C, Zheng L, Yoo JK, Guo H, Zhang Y, Guo X, Kang B, Hu R, Huang JY, Zhang Q, Liu Z, Dong M, Hu X, Ouyang W, Peng J, Zhang Z. Landscape of Infiltrating T Cells in Liver Cancer Revealed by Single-Cell Sequencing. Cell. 2017;169:1342–1356. e1316. doi: 10.1016/j.cell.2017.05.035. [DOI] [PubMed] [Google Scholar]
- Zheng GX, Terry JM, Belgrader P, Ryvkin P, Bent ZW, Wilson R, Ziraldo SB, Wheeler TD, McDermott GP, Zhu J, Gregory MT, Shuga J, Montesclaros L, Underwood JG, Masquelier DA, Nishimura SY, Schnall-Levin M, Wyatt PW, Hindson CM, Bharadwaj R, Wong A, Ness KD, Beppu LW, Deeg HJ, McFarland C, Loeb KR, Valente WJ, Ericson NG, Stevens EA, Radich JP, Mikkelsen TS, Hindson BJ, Bielas JH. Massively parallel digital transcriptional profiling of single cells. Nat Commun. 2017;8:14049. doi: 10.1038/ncomms14049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu S, Qing T, Zheng Y, Jin L, Shi L. Advances in single-cell RNA sequencing and its applications in cancer research. Oncotarget. 2017 doi: 10.18632/oncotarget.17893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zieglschmid V, Hollmann C, Bocher O. Detection of disseminated tumor cells in peripheral blood. Crit Rev Clin Lab Sci. 2005;42:155–196. doi: 10.1080/10408360590913696. [DOI] [PubMed] [Google Scholar]