Abstract
Background
Non-functioning pancreatic neuroendocrine tumors (NFPanNETs) may be sporadic or inherited due to germline mutations associated with von Hippel-Lindau disease (VHL) or multiple endocrine neoplasia type 1 (MEN1). The clinical behavior of NFPanNETs is difficult to predict, even in same stage and grade tumors. Herein we analyzed genotype-specific patterns of transcriptional messenger RNA (mRNA) levels of NFPanNETs in order to understand the molecular features that determine PanNET phenotype.
Methods
32 samples were included for genome-wide mRNA gene expression analysis [9 VHL-, 10 MEN1-, 9 sporadic NFPanNETs and 4 purified normal islet cells (NIC) samples]. Validation of genes was performed by Real-Time PCR and immunohistochemistry. Gene expression profiles were analyzed by tumor genotype and pathway analysis was curated.
Results
Consensus clustering of mRNA expression showed separate clustering of NIC, VHL- and MEN1-associated NFPanNETs, while some sporadic tumors clustered with MEN1. Four of 5 “MEN1-like” sporadic PanNET subtypes had loss of heterozygosity at the MEN1 gene locus. Pathway analysis showed subtype-specific pathway activation comprising angiogenesis and immune response in VHL; neuronal development in MEN1, protein ubiquitination in the new MEN1/Sporadic subtype; and cytokinesis, cilium/microtubule development in sporadic NFPanNETs. Among many genes, PDGFRB, Lef-1, CDK4, and CDK6 were found to be upregulated in VHL and MEN1 NFPanNETs, respectively, providing potential subtype-specific treatment targets.
Conclusions
Distinct mRNA expression patterns were identified in sporadic-, VHL- and MEN1- associated NFPanNETs. Our results uncover new pathways involved in NFPanNETs that are subtype-specific and provide potential new diagnostic or therapeutic targets based on tumor subtype.
Keywords: Pancreas, Neuroendocrine, VHL, Sporadic, MEN1
INTRODUCTION
Pancreatic neuroendocrine tumors (PanNETs) are characterized by a distinctive histology and evidence of neuroendocrine differentiation. In the United States, it is estimated that 3–5 cases per 100,000 inhabitants occur annually, and the incidence of these neoplasms continues to increase (1). PanNETs can be divided into functioning (produce clinical findings related to excess hormone production) and non-functioning tumors (NFPanNET), the latter being much more prevalent. While functioning PanNETs require treatment because of the morbidity and mortality caused by excess hormone production, small NFPanNETs do not always require treatment.
PanNETs can occur sporadically or in inherited cancer syndromes such as von Hippel-Lindau syndrome (VHL), Multiple Endocrine Neoplasia Type 1 (MEN1), Neurofibromatosis type 1 (NF1), Tuberous sclerosis complex (TSC) and Cowden syndrome (CS) (2-6). Nine to 17% and 30-80% of patients with VHL and MEN1, respectively, develop PanNETs during their lifetime (4, 7). By contrast, the penetrance of PanNET in NF1, TSC and CS is lower and these latter syndromes are relatively rare.
Hereditary PanNETs often behave less aggressively than their sporadic counterparts, but the reasons underlying these differences in clinical behavior are not well defined (4, 8). The clinical management of hereditary PanNETs is complicated by the presence of multiple lesions and the risks associated with multiple surgeries. Current recommendations for resection of hereditary PanNETs are mostly based on the risk of metastatic disease, using tumor growth and size as a surrogate marker for aggressiveness (4, 7). However, these criteria are imperfect and do not accurately predict the metastatic potential and/or aggressive behavior of all patients with PanNETs and VHL or MEN1 (7, 9-12).
The genes responsible for the major inherited predisposition syndromes (VHL, MEN1) are known, and a large number of PanNETs have been sequenced (13). Sequencing of sporadic PanNETs have identified a multitude of somatic mutations, including MEN1 in up to 44% of cases, DAXX, ATRX, PTEN, TSC2 and PIK3CA as well as mutations or promoter hypermethylation in VHL in up to 25% of cases (13-16). Despite these advances, the transcriptional changes and downstream pathway activations promoting early tumorigenesis are largely unknown, but could be useful to identify targets for diagnosis and treatment of both hereditary and sporadic PanNETs (4, 7, 17).
In this study, we sought to investigate transcriptional levels of messenger RNA (mRNA) by DNA microarray from hereditary (MEN1, VHL) and sporadic early stage NFPanNETs in order to discover dysregulated pathways associated with tumorigenesis and elucidate possible transcriptional targets that may be deregulated across all or just in one particular genotype of NFPanNETs.
MATERIAL AND METHODS
Sample Selection
Thirty NFPanNETs and six normal islet cell samples were initially selected for this study. All tumors were chosen to be early stage NFPanNETs in order to account for stage mismatch when comparing genotypes as well as reflect common actual presentations of NFPanNETs in MEN1 and VHL patients. 32 samples were ultimately included in this analysis; one sporadic sample was excluded due to advanced stage including metastatic disease, and one VHL and 2 normal islet cell samples were excluded due to poor RNA quality. The final analyzed group comprised 10 NFPanNET from MEN1 patients, 9 NFPanNET from VHL patients, 9 sporadic NFPanNETs and 4 purified normal islet cell samples (NIC) (Figure 1). All the MEN1 and VHL cases had a germline mutation in the respective syndromal gene. The functional status of the tumor was determined by clinical features and symptoms, and biochemical testing (fasting gastrin, insulin, vasoactive intestinal peptide, somatostatin and glucagon). Immunohistochemistry (IHC) staining of paraffin-embedded tumor slides for gastrin and insulin was also performed in MEN1 NFPanNETs. Tissues were obtained from pancreatectomies (pancreaticoduodenectomy or distal pancreatectomy specimens) or NFPanNET enucleations performed at our or the participating institution between 2010 and 2014. These samples were fresh-frozen after procurement and stored at −80°C. Serial tissue sample sections were used for RNA extraction, stained with hematoxylin and eosin, and reviewed by a pathologist to confirm the diagnosis and ensure a neoplastic nuclei content of more than 80%.
Figure 1.
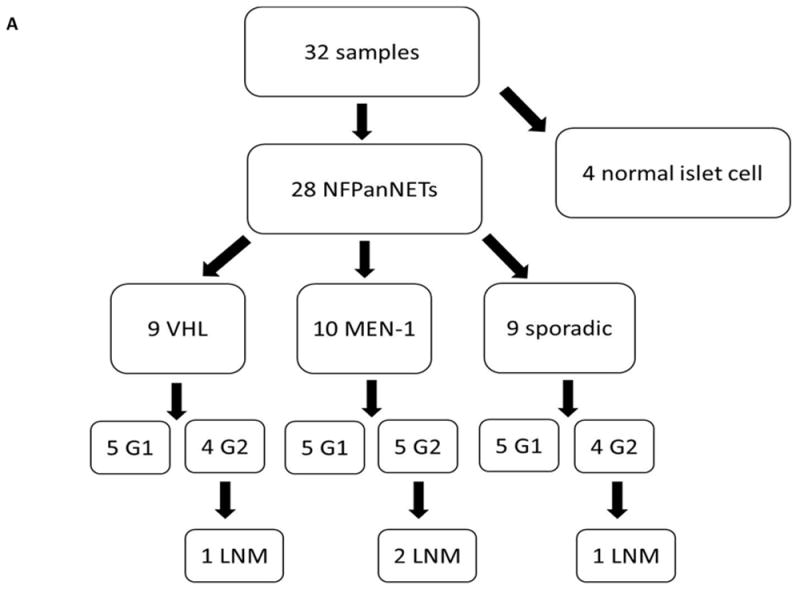
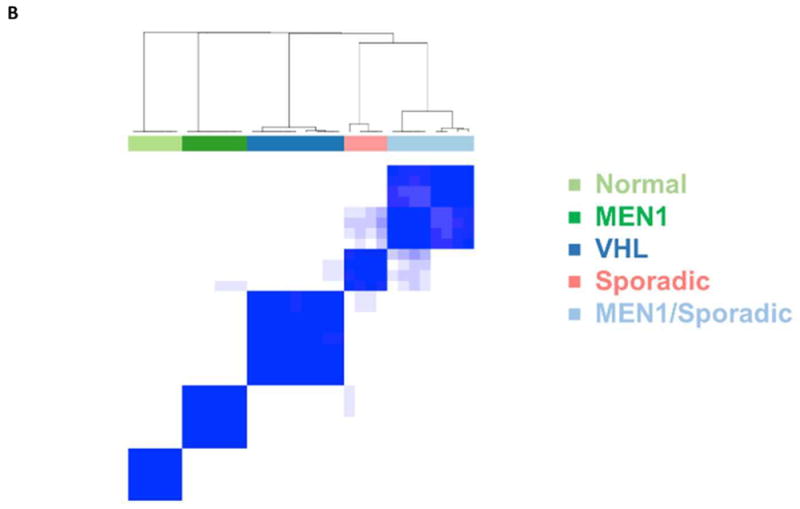
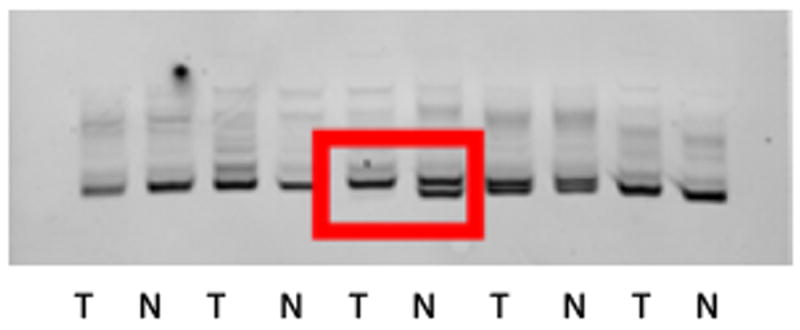
A. Breakdown of PanNET and normal islet cell samples analyzed by cDNA microarray (G=Grade, LNM=Lymph Node Metastases). B. Unsupervised hierarchical clustering of microarray data defines five distinct groups. C. Representative image of loss of heterozygosity for the MEN1 gene in a sporadic NFPanNET using microsatellite marker DS11S956 (red box).
T: Tumor, N: Normal
Primary human pancreatic islets of Langerhans were purchased from the Division of Transplantation Surgery at the University of Alabama at Birmingham. Pancreata were recovered, with informed consent, from cadaveric donors after in situ vascular perfusion with University of Wisconsin solution at 4°C, as part of a multiorgan procurement. Pancreata were immediately transported to the islet isolation laboratory for processing. Islets were isolated by a semiautomated method and purified using the Cobe 2991 cell processor (Gambro BCT, Lakewood, CO). The number of islets within each size class was converted to the standard number of islets of 150-μm diameter, equal in volume to the sample. Purity was assessed by comparing the relative quantity of DTZ-stained endocrine tissue with unstained exocrine tissue. Only islet isolations with greater than 90% viability and greater than 60% purity were used.
The study was approved by the Office for Human Research Protections at the Department of Health and Human Services. Patient information was collected prospectively under an Institutional Review Board–approved protocol at the NIH (Bethesda, MD) after obtaining written informed consent.
RNA extraction
Total RNA was extracted from ~20-30 mg of tissue using the Total RNA Purification Kit (17200, Norgen Biotek Corp., Thorold, ON, Canada) and from purified normal islet cells using the AllPrep DNA/RNA mini Kit (Qiagen, Cat # 80204). Total RNA yield was assessed via NanoDrop (NanoDrop Technologies, Wilmington, DE) and Bioanalyzer 2100 and RNA 6000 Nano/Pico LabChip (Agilent Technologies, Palo Alto, CA). cDNA was obtained via reverse transcription via the M-MLV RT protocol (Thermo Fisher Scientific Inc).
Messenger RNA microarray
Samples
RNA quantity and quality were measured by NanoDrop ND-1000. RNA integrity was assessed by standard denaturing agarose gel electrophoresis.
DNA microarray
The Whole Human Genome Oligo Microarray was a broad view that represents all known genes and transcripts in the human genome. Sequences were compiled from a broad source survey, and then verified and optimized by alignment to the assembled human genome.
RNA labeling and array hybridization
Sample labeling and array hybridization were performed according to the Agilent One-Color Microarray-Based Gene Expression Analysis protocol (Agilent Technology). Briefly, total RNA from each sample was linearly amplified and labeled with Cy3-UTP. The Labeled cRNAs were purified by RNeasy Mini Kit (Qiagen). The concentration and specific activity of the labeled cRNAs (pmol Cy3/μg cRNA) were measured by NanoDrop ND-1000. 1 μg of each labeled cRNA was fragmented by adding 11 μl 10 × Blocking Agent and 2.2 μl of 25×Fragmentation Buffer, then heated at 60 °C for 30 min, and finally 55 μl 2 × GE Hybridization buffer was added to dilute the labeled cRNA. 100μl of hybridization solution was dispensed into the gasket slide and assembled to the gene expression microarray slide. The slides were incubated for 17 hours at 65°C in an Agilent Hybridization Oven. The hybridized arrays were washed, fixed and scanned with using the Agilent DNA Microarray Scanner (part number G2505C).
Data analysis
Agilent Feature Extraction software (version 11.0.1.1) was used to analyze the acquired array images. Quantile normalization and subsequent data processing were performed using the GeneSpring GX v12.1 software (Agilent Technologies). Raw data were normalized using quantile normalization, followed by cyclic loess using the Bioconductor limma package (v. 3.30.11) (18). Consensus Clustering was performed on all gene expression data using Bioconductor package ConsensusClusterPlus (v. 1.38) (19). We evaluated from k=2-6 clusters and selected k=5 which showed the maximum area under the curve in the consensus cumulative distribution function plot. Subtype-unique expressed genes were identified by selecting genes wherein the 90th percentile confidence interval (CI) within each subtype exceeded the 90th percentile CI of expression for all other subtypes including normal. Subtype-unique genelists were subjected to gene category enrichment analysis using a Fisher’s Exact Test on a variety of publicly available databases including Gene Ontology (GO) (https://www.geneontology.org), Kyoto Encyclopedia of Genes and Genomes (KEGG) (https://www.genome.jp/kegg/pathway.html), Protein ANalysis THrough Evolutionary Relationships (PANTHER) (https://www.pantherdb.org), PC REACTOME (https://www.reactome.org), Inter Partes Review (IPR), PC NCI NATURE (http://pid.nci.nih.gov) and the National Center for Biotechnology Information (NCBI) (https://www.ncbi.nlm.nih.gov) database.
Group comparisons of categorical variables were performed using Pearson’s Chi-Square testing or Fisher exact probability test, and the comparison of continuous variables was performed with a two-tailed t-test or Wilcoxon Rank test. All P-values were considered statistically significant if P was less than 0.05. GraphPad Prism (GraphPad Software Inc. La Jolla, CA) was used for statistical analysis.
Ion Torrent Next Generation Sequencing Analysis
Primers for genes involved in NFPanNETs (MEN1, ATRX, DAXX, VHL) were designed for a custom Ampliseq Panel using Life Technologies software, Ion Ampliseq Designer. Ampliseq libraries were prepared from DNA samples extracted from NFPanNET samples using the RNA/DNA/Protein Purification Plus Kit (47700, Norgen Biotek Corp., Thorold, ON, Canada). DNA sample concentration were determined using Nanodrop (Thermo Fisher’s ND8000, Waltham, MA). Final sample qualification and quantitation was established with Picogreen analysis (standard curve assay) on a Molecular Devices Gemini XPS Plate Reader and Softmax Pro Software (Sunnyvale, CA). All samples and controls were run in triplicate.
Ampliseq libraries were prepared for each DNA sample. To create libraries, each DNA sample was first amplified according to the manufacturer’s protocol. Each library was then assigned a unique barcode-adapter sequence and ligated onto the libraries. The final libraries were qualified and quantitated using Life Technologies’ Ion Library Quantitation Kit (Item #4468802). Libraries were then cycled and analyzed on an ABI PRISM® 7900HT Sequence Detection System. Using the quantitation values from the real time PCR analysis, all libraries were then normalized with 1X Low TE Buffer to either 50pMol or 100pMol to prepare for emulsion PCR. Libraries were then combined into pools of 9 for a total of 4 pools. Combined with an Ion 318 chip, roughly 300X coverage per amplicon was achieved. The clonal emulsion amplification of Ampliseq libraries was sequenced on the Ion Torrent PGM platform, using Life Technologies OT2 system (Waltham, MA). The OT2 system is automated, and when used in conjunction with the Enrichment Station (ES), generates template positive Ion PGM Template One Touch 2 200 Ion Sphere Particles for semiconductor sequencing. All analysis and data QC was completed with Life Technologies Ion Torrent Suite v5.0.4.
Real Time PCR
Gene expression levels were measured using specific primers and probes. Briefly, 500 to 1,000 ng of total RNA was reverse transcribed using a High Capacity Reverse Transcription cDNA kit (Cat #4374967, Applied Biosystems, Thermo Fisher Scientific), and the resulting cDNA was diluted and amplified according to the manufacturer’s instructions. GAPDH was used as an endogenous control. Gene expression levels were calculated using SDS 2.3 software (Applied Biosystems, Thermo Fisher Scientific).
Immunohistochemistry
Sections were deparaffinized and rehydrated, and antigen retrieval was performed with citrate buffer in a water bath at 120C. The sections were incubated with the anti-PDGFR-Beta antibody (1:100; Abcam), overnight at 4C, followed by incubation with a biotinylated secondary antibody for 1 hour at room temperature. The slides were developed with diaminobenzidine [DAB; EnVisionþ Kit system HRP (DAB), Dako] and counterstained with hematoxylin. The slides were scanned at a 20x magnification using a ScanScope XT digital slide scanner (Aperio Technologies, Leica) to create whole slide image data files at a resolution of 0.5 mm/pixel, and they were viewed using ImageScope software (Aperio Technologies). All IHC slides were reviewed and scored by a pathologist unaware of the sample groups. Tumor staining of more than 50% was considered positive. Positive staining were graded as weakly positive (+), moderately positive (++) or strongly positive (+++).
DNA extraction and Loss of Heterozygosity (LOH) Analysis
DNA was extracted from tissue and leukocytes using the Qiagen Blood and Cell Culture DNA Kit (Qiagen) following the manufacturer’s protocol. Primers for the polymorphic microsatellite markers D11S956, D11S4939, D11S4946, PYGM and D11S987 were identical to those published previously (20). PCR was performed in a total volume of 100 ml containing 30 pmol of each primer, 2.5 U TAQ polymerase (Perkin Elmer, Foster City, USA) and 100–400 ng DNA. The cycler program was as follows: initial denaturation at 95.8C for 5 min, 30 cycles of 55.8 for 1.5 min, 72 8C for 1.5 min, 94 8C for 1.5 min, final extension at 72 8C for 5 min. Primer sequence for
D11S956, D11S4939, D11S4946, PYGM and D11S987 PCR conditions were used as published previously with minor modifications (21). The PCR product was purified with chloroform. Purified PCR product was dried in a speed-vac, redissolved in 10 μl formamide and 10 μl H2O and loaded onto a 6% TBE polyacrylamide gel. Electrophoresis was performed at 150 V for 90 min. After staining with ethidium bromide for 10 min the amplification products were visualized on a UV transilluminator. LOH was determined by comparing tumor DNA to germline DNA pattern.
RESULTS
Histopathological Characteristics
Twenty-eight patient samples with NFPanNETs were selected according to the criteria mentioned in the methods section. 50% of samples were WHO grade 1 and 50% WHO grade 2 tumors in each subgroup (VHL, MEN1 and sporadic). Two patients in the VHL and one patient in the MEN1 and sporadic group had lymph node metastases at the time of diagnosis. There were no statistically significant differences when comparing clinical and histopathological characteristics between subgroups, except for tumor size which was significantly larger in sporadic tumors as compared to VHL (p=0.01) and MEN1 NFPanNET samples (p=0.002) (Figure 1A, Supplemental Table 1).
Consensus cluster analysis
Consensus cluster analysis of gene expression levels showed normal islet cells and VHL tumor samples clustered in separate groups while sporadic and MEN1 samples clustered alone or in a fifth MEN1/sporadic group (Figure 1B). We did not find any clustering by patient demographic and histopathological data or MEN1 mutation locus between the MEN1, sporadic and MEN1/sporadic groups. Ion Torrent NGS analysis showed two non-synonymous and one stopgain exonic mutations in MEN1 in three separate sporadic NFPanNET samples that clustered with the MEN1 and loss of heterozygosity (LOH) analysis showed LOH at the MEN1 locus in 4 of 5 samples (Figure 1C), possibly explaining why these samples clustered into the 5th group on unsupervised cluster analysis and confirming the common occurrence of MEN1 mutations in sporadic PanNETs as previously reported (13). Since the transcription/chromatin remodeling genes DAXX (death-domain-associated protein) and ATRX (α thalassemia/mental retardation syndrome X-linked) have previously been found to be commonly mutated in NFPanNETs (13), we evaluated whether somatic mutation in these genes influenced transcriptional clustering. Four ATRX and no DAXX mutations were found in MEN1 and sporadic NFPanNET samples. Two non-synonymous exonic ATRX mutations were found in the sporadic only group, one non-synonymous exonic mutation in the MEN1/sporadic group and one non-synonymous exonic mutation in the MEN1 group, suggesting that ATRX and DAXX mutations were not associated with the transcriptional clustering of tumor samples.
Differentially expressed genes in NFPanNETs according to genotype
A limited list of the most highly upregulated genes in NFPanNETs according to subtype are summarized in Supplemental Figure 1. The most commonly upregulated genes in all three subtypes when compared to NIC was Kruppel-Like Factor 12 (KLF12). KLF12 is a developmentally-regulated transcription factor, which has been shown to promote cell cycle transition and regulate anoikis through S phase and therefore cell proliferation. It was the gene with the highest predicted connectivity in a recent study evaluating integrated mRNA-miRNA analysis in PanNETs (22, 23). We randomly selected two overexpressed genes, KLF-12 and Cyclin D1 Binding Protein 1 (CCNDBP1) and one underexpressed gene Matrix Metallopeptidase 1 (MMP1) for validation by RT-qPCR (Supplemental Figure 2).
We also found genotype-specific differences in gene expression when comparing one subgroup to all others (Supplemental Figure 1). In the VHL group, we found overexpression of the neurogenic locus notch homolog protein 3 mRNA (Notch 3), which has previously been shown to be upregulated in the vascular endothelium of VHL -associated central nervous system hemangioblastomas, as well as overexpression of Insulin-like growth factor 2 (IGF-2) mRNA which was shown to be upregulated in the PanNET cell lines BON-1 and QPG-1 (24, 25). In the MEN1 group, overexpression of Ryanodine Receptor 3 (RYR3) mRNA was common, and has been described to play an important role for insulin secretion in pancreatic beta-cells (26). Fibroblast growth factor 13 (FGF-13), previously associated with aggressive tumor behavior and shorter disease-free survival, interestingly was found to be significantly upregulated in the sporadic and MEN1 subgroups only (27).
Gene Ontology (GO) Pathway Analysis
GO pathway analysis yielded subtype-specific pathway alterations. Within the twenty most upregulated pathways in VHL NFPanNETs, those relating to angiogenesis and immune response were found to be highly upregulated (Figure 2). Pathways involved in cilium and microtubule formation, both part of the cell’s spindle cell apparatus and crucial to cell division, were found to be upregulated in sporadic NFPanNETs, which could explain the reported aggressive behavior and rapid growth of sporadic tumors compared to their hereditary counterparts (Figure 3) (28). Gene enrichment in pathways comprising ion channel activity and neuron development, both previously described as upregulated in PanNETs, were most prevalent in MEN1 NFPanNETs only (Figure 4) (29). Protein ubiquitination pathways, previously found to be involved in blockade of the proteasome degradation machinery and thus enhancing glucose-stimulated insulin secretion and promoting certain gene expression patterns, was most upregulated in MEN1/Sporadic NFPanNET subtype (Figure 5) (30) .
Figure 2.
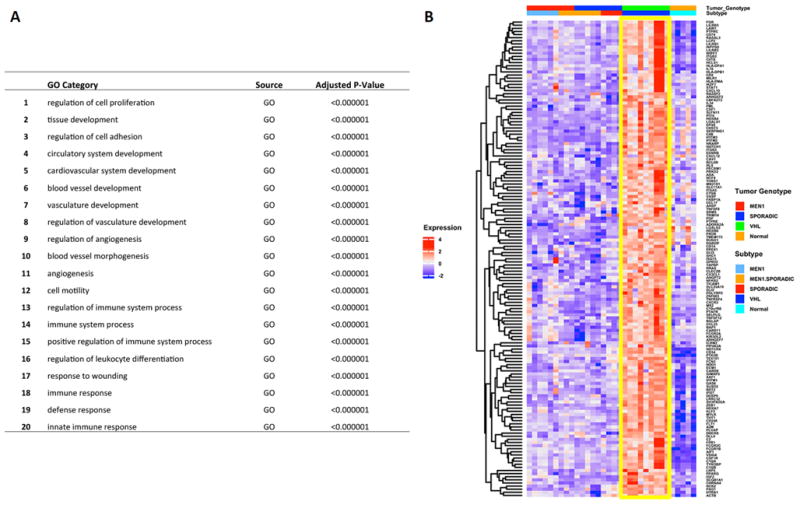
Messenger RNA enrichment analysis (GO, Gene Ontology): Top 20 upregulated pathways in VHL vs. other NFPanNETs and NIC (Figure A) shows predominance of genes involved in angiogenesis and immune response (Figure B, yellow box).
Figure 3.
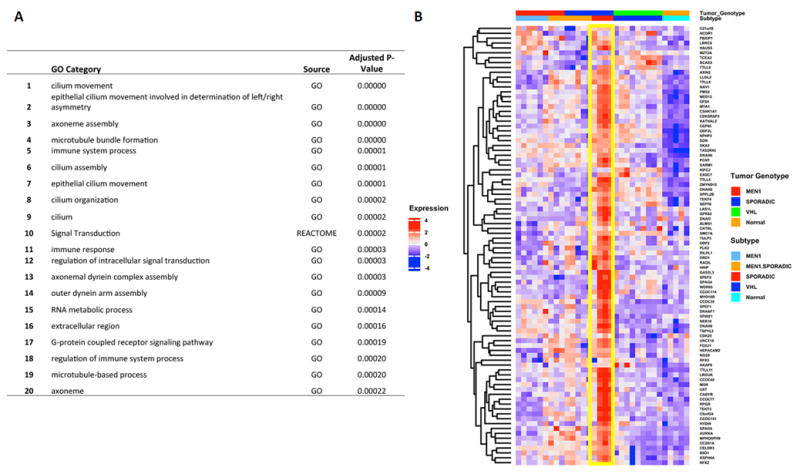
Messenger RNA enrichment analysis (REACTOME and GO, Gene Ontology): Top 20 upregulated pathways in Sporadic vs. other NFPanNETs and NIC (Figure A) shows predominance of genes involved in cilium and microtubule processes (Figure B, yellow box).
Figure 4.
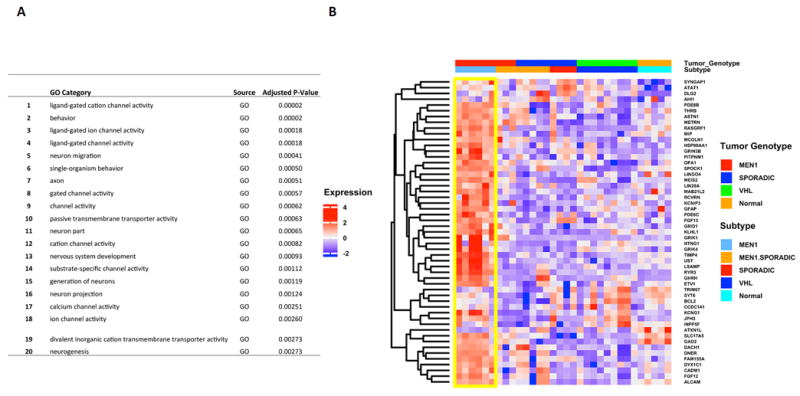
Messenger RNA enrichment analysis (GO, Gene Ontology): Top 20 upregulated pathways in MEN1 vs. other NFPanNETs and NIC (Figure A) shows predominance of genes involved in ion channel activity and neural development (Figure B, yellow box).
GO: Gene Ontology
Figure 5.
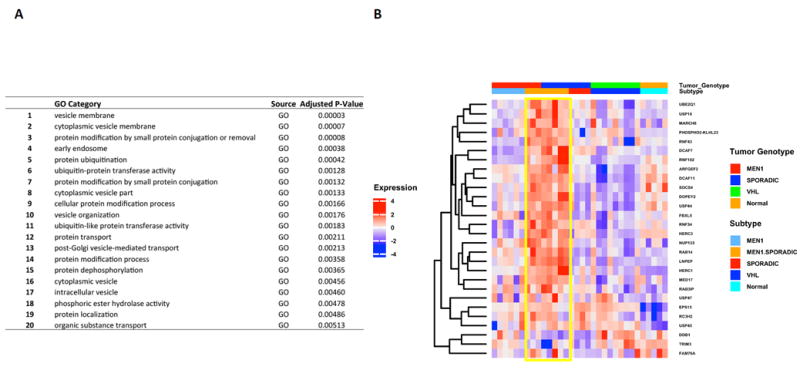
Messenger RNA enrichment analysis (GO, Gene Ontology): Top 20 upregulated pathways in MEN1/Sporadic vs. other NFPanNETs and NIC (Figure A) shows predominance of genes involved in protein ubiquitination (Figure B, yellow box).
Validation of selected genotype-specific mRNA targets
In order to further validate our microarray analysis, we compared our expression data to a previous study that compared mRNA expression of VHL PanNETs to sporadic PanNETs (31). Nineteen genes previously reported to be upregulated and 3 genes previously reported to be downregulated had similar expression levels in our dataset when comparing VHL to sporadic NFPanNETs (Supplemental Figure 3).
Next, we selected a small subset of genotype-specific genes for validation based on their potential translational relevance due to their ability to be targeted for treatment. Platelet-Derived-Growth-Factor-Receptor Beta (PDGFRβ) was found to be upregulated (>2 fold) in VHL PanNETs and this was confirmed by TaqMan RT-PCR (Figure 6 B and C). By immunohistochemistry, we found that PDGFRβ was significantly overexpressed in VHL- vs. MEN1 and sporadic NFPanNETs (p=0.03), but that mostly the tumor stroma and not the tumor cells stained positive in VHL samples (Figure 6 A). To test the hypothesis that VHL NFPanNETs have greater stromal content than other PanNETs, we used the ESTIMATE (Estimation of STromal and Immune cells in Malignant Tumours using Expression data) gene signature model as previously described (32). We found significant overexpression of the 141 stromal gene signature panel in VHL tumors as compared to the MEN1 and sporadic NFPanNET genotype (P<0.05) (Figure 6 D).
Figure 6.
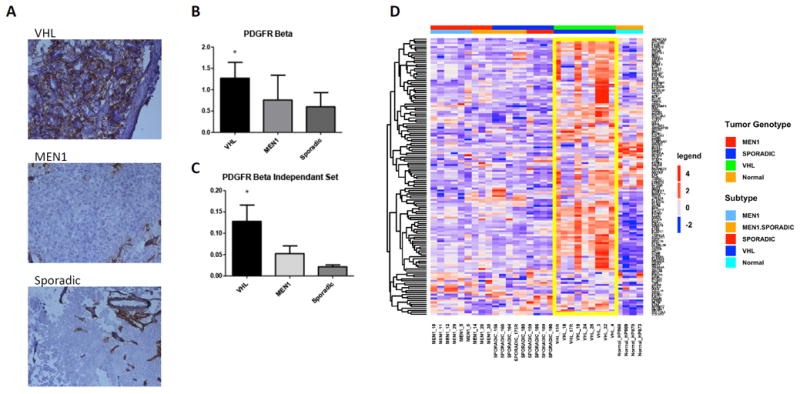
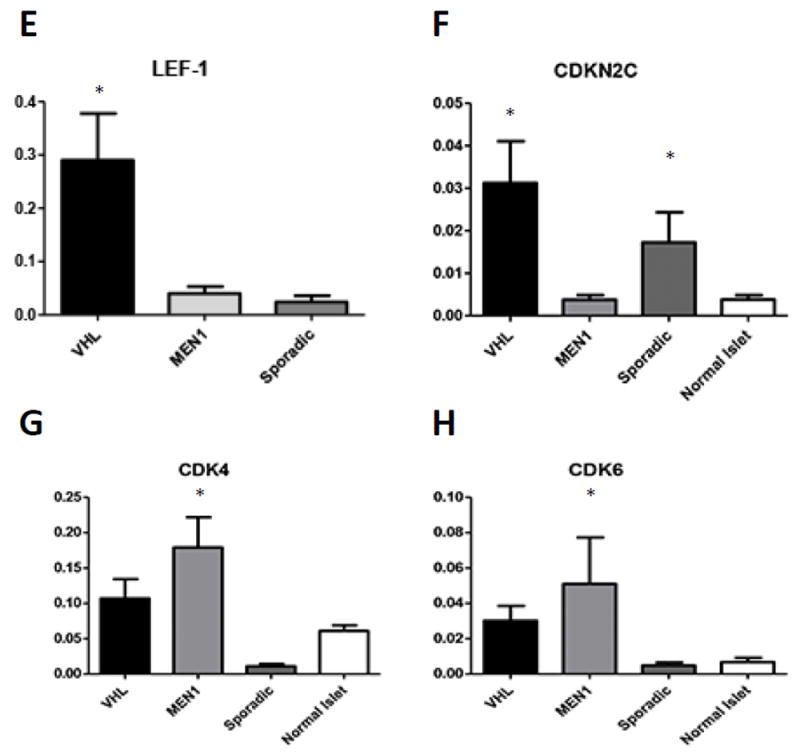
A. Immunohistochemical analysis of PDGFR-Beta in VHL, MEN1 and Sporadic PanNETs shows significantly increased expression in VHL PanNET cells (p<0.05). B. RT-qPCR of PDGFR-Beta confirms microarray analysis and shows significantly increased mRNA levels in the initial cohort. C. Independent validation of PDGFR-Beta expression via RT-qPCR in a separate set of 19 NFPanNETs and four NIC (p<0.05). C. The ESTIMATE (Estimation of Stromal and Immune cells in Malignant Tumours using Expression data) gene signature model confirms increased stromal signature in VHL PanNETs (yellow box). E. RT-qPCR expression analysis showing overexpression of the transcription factor Lef-1 in VHL NFPanNETs when compared to sporadic and MEN1 NFPanNETs. F. RT-qPCR expression analysis showing overexpression the CDK4/CDK6 inhibitor CDKN2C in MEN1 NFPanNETs when compared to sporadic (p=0.06) and VHL NFPanNETs (p=0.02). G. RT-qPCR expression analysis showing overexpression of CDK4 in MEN1 NFPanNETs when compared to sporadic NFPanNETs (p=0.0009) and normal islet cells (p=0.05). H. RT-qPCR expression analysis showing overexpression of CDK6 in MEN1 NFPanNETs when compared to sporadic NFPanNETs (p=0.05).
Additional genes that were found to be upregulated on gene expression analysis and validated by RT-PCR included overexpression of the transcription factor lymphoid enhancer-binding factor-1 (Lef-1) in VHL NFPanNETs and overexpression of Cyclin-dependent kinase 4 and 6 (CDK4, CDK6) in MEN1 NFPanNETs (Figure 6 E, G, H). The tumor suppressor CDKN1B was also found to be downregulated in MEN1 NFPanNETs when compared to other genotypes (Figure 6 F). When looking at CD4 and CD6 gene expression according to groups from hierarchical clustering, we found that CD4 levels were higher in the MEN1 and MEN1/Sporadic genotype when compared to the sporadic, non-MEN1 genotype, although not statistically significant (Supplemental Figure 4).
We next evaluated the differentially expressed genes in an independent cohort of previously analyzed PanNETs using publicly available microarray data via the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO) repository (https://www.ncbi.nlm.nih.gov/geo). In a previously published study comparing cDNA microarray data from 56 NFPanNETs to normal pancreatic islet cells (33), we randomly selected 15 NFPanNET samples from this microarray dataset for cross-validation and found that CDK4 expression was highest among all samples (average 6.9-fold compared to normal pancreatic islet cells) and Notch3 expression was lowest (average 3.8 fold when compared to normal pancreatic islet cells). Lef1, CDK6, PDGFRB and IGF2 were also overexpressed when compared to normal pancreatic islet cells (fold change range from 3.8 to 4.9-fold).
DISCUSSION
The molecular mechanisms that give rise to NFPanNETs are complex and not completely understood. Inactivating germline or somatic mutations in tumor suppressor genes such as VHL or MEN1 contribute significantly to the tumorigenesis of NFPanNETs. However, additional events involved in proliferative and antiapoptotic signals, and in certain cases evasion of the immune system and modification in the tumor microenvironments are important in NFPanNETs initation and or progression. Proliferative and antiapoptotic signals are probably genotype-specific as shown in this study and arise from aberrant regulation of signal transduction/growth (PI3K/AKT pathway), hypoxia/angiogenesis (VHL mutation), cell-cycle progression (downregulation of cyclin-dependent kinase inhibitors) and/or genome stabilization pathways.
Previous studies looking at transcriptional alterations in PanNETs have mostly focused on comparing “benign” to “malignant” PanNETs, defining these categories by absence or presence of metastatic disease (27, 29, 34, 35). Most of these studies also comprised relatively small samples sizes and/or a heterogenous PanNET cohorts including functioning and non-functioning tumors or used normal pancreatic tissues as control (29, 34, 35). Our analysis aims at identifying transcriptional alterations based on tumor genotype. Indeed, we discovered that there is a subset of sporadic tumors that cluster with MEN1 tumors, which reveals an additional way to classify tumors according to gene expression phenotypes. Additionally, our sample selection was purposely chosen to be as homogenous as possible in order to uncover differences in pathways according to tumor genotype rather than differences in stage-dependent tumor biology. We did not find any statistically significant differential gene expression across tumor grades and samples with lymph node metastases. This is likely due to the fact that most tumors had a low Ki-67 index within the G2 group and low rate of lymph node metastases in each of the subgroups to perform an adequately powered analysis. However, we believe that by choosing a homogenous group of stage I/II NFPanNETs for each genotype, we uncovered transcriptional alterations that may occur at earlier stages in NFPanNET tumorigenesis.
Mutations in the tumor suppressor VHL gene cause E3 ubiquitin ligase inactivity and subsequent upregulation of the hypoxia inducible factor 1alpha (HIF1α) protein by inhibiting its degradation (17). HIF1α is a transcription factor that induces the expression of a number of angiogenesis related factor, as was shown in this study, but also genes that promote macrophage and neutrophil energy generation, inflammatory and bactericidal activities, and cell survival (17, 36). The exact mechanisms by which VHL mutated cells influence the immune systems response are complex and poorly understood. However, some data suggest that the pro-angiogenesis state of VHL mutated cells is central for immune response evasion of tumor cells. It has been previously shown that the high pro-angiogenic status of clear cell renal cell cancers (RCC), which commonly have VHL mutations, is associated with an accumulation of regulatory T-cells and myeloid-derived suppressor cells in the local tumor microenvironment, in particular in the tumor invasion zone (37). This might allow cells to evade immune surveillance thereby promoting disease progression. Another recent study in RCC also suggests that VHL-dependent alterations of the RCC secretome modulate the T-cell activation by negatively interfering with T-cell proliferation and cytokine secretion (29). VEGF overexpression has also been directly linked to immunosuppression by inhibiting the maturation of dendritic cells via a nuclear factor κB-dependent pathway (mediated by VEGFR-1 signaling), and to promote the induction of regulatory CD4+ CD25+ T cells within the tumor microenvironment (38). Lastly, increased adenosine concentration found in VHL mutated cells is involved in impaired T cell-mediated tumor rejection and supports tumor angiogenesis (39, 40).
PDGFRβ is a tyrosine kinase inhibitor involved in angiogenesis, autocrine stimulation of tumor cells and regulation of interstitial fluid pressure (41-43). There are only few reports analyzing the expression of PDGFRB in PanNETs, and all found that functioning and non-functioning PanNET overexpress PDGFRβ mRNA and protein (35, 41, 44). In this study, we demonstrate that the VHL NFPanNET genotype expresses the highest PDGFRβ mRNA and protein levels when compared to the MEN1 and sporadic genotypes and that the PDGFRB protein staining is mostly positive in the VHL stroma. PDGFRβ in tumor stroma is important in regulating interstitial fluid pressure and inhibition of this protein may allow for greater uptake of other chemotherapy drugs (43). Thus, we propose that NFPanNETs with alterations in the VHL promoter or mutation in the VHL gene may be particularly susceptible to PDGFRB antagonist treatment.
Since tyrosine kinase inhibitors currently used for treatment of advanced PanNETs can be associated with development of resistance over time, a combination of anti-angiogenic and immune-based treatment could be beneficial in those tumors expressing high levels of angiogenic and immune infiltrating cells. Based on the current study results, we believe that VHL-associated, locally advanced and metastatic NFPanNETs may therefore be especially good candidate for such combination therapies.
Singhi and colleagues analyzed the expression of LEF1 in pancreatic neoplasms and found no expression in 44 well-differentiated neuroendocrine tumors (45). This is in contradiction with our findings. It is unclear whether the PanNET samples studied were all sporadic and did not have alterations in the VHL gene or expression level. If the transcription factor LEF1 is further confirmed to be upregulated in VHL NFPanNETs only, it could potentially be targeted by currently available small-molecule inhibitor used in chronic lymphocytic leukemia (46).
Cyclin-dependent kinase inhibitor 1B (CDKN1B, p27Kip1) is an enzyme inhibitor that we found to be underexpressed in the MEN1 NFPanNET genotype. CDKN1B is considered a tumor suppressor because it functions as a cell cycle regulator by in part preventing the activation of cyclin D-CDK4 complex. Its inactivation can be accomplished post-transcription by the activation of several pathways including the Ras-mitogen activated protein kinase pathway (MAPK), which we found to be upregulated in MEN1 NFPanNETs (47).
In our study we also demonstrated that Cyclin-dependent kinase 4 (CDK4) and Cyclin-dependent kinase 6 (CDK6) gene expression is higher in MEN1 NFPanNETs genotype. Tang and associates analyzed expression of CDK4 and CDK6 in functioning and non-functioning PanNETs and found high CDK4 expression levels and increased copy number of CDK4 and CDK6. They also found that the human PanNET cell line QGP1 was inhibited in a xenograft mouse model by a CDK4/6 inhibitor, PD0332991 (48). In another study, knockdown of CDK6 enhanced glioma sensitivity to the chemotherapeutic temozolomide, which has been shown to be effective in PanNETs (49, 50). Inhibitors of CDK6 are currently in clinical trials for other malignancies and therefore understanding which subset of NFPanNETs could benefit from CDK4/CDK6 inhibition and potentially enhancing temozolomide efficacy with CDK6 inhibitors would be of interest and should be investigated further.
Our study has several limitations. We used human islets cells for some of the comparisons and they were not cell sorted to distinguish the specific cell type population. This may have influenced the results comparing NFPanNETs to normal islet cells as NFPanNETs are generally thought to originate from one cell type. Newer methods to isolate single cell population have recently been described and could be applied in the future (51). Furthermore, even though we confirmed overexpression of LEF1, CDK4, CDK6, NOTCH3, RYR3 and IGF2 in an independent cohort of NFPanNETs, we cannot reliably determine that these genes can segregate NFPanNETs by genotype, as this particular validation set does not have data on MEN1 or VHL mutation status. Additional future studies will be needed for this purpose. Lastly, even though we validated highly differentially expressed genes that may have biologic significance and could be targeted for therapy at the protein level (such as PDGFRB), we cannot be certain that this applies to all of the differentially expressed genes identified in this study.
In summary, we have demonstrated genotype-specific differences in mRNA expression patterns in sporadic-, VHL- and MEN1-NFPanNETs, and suggest that genotype and subtype-specific activation of pathways may contribute to NFPanNETs tumorigenesis. These findings implicate specific genes and pathways that could be exploited to identify new diagnostic or therapeutic pathways and allow for subtype-specific treatments of NFPanNETs in the future.
Supplementary Material
Supplemental Figure 1: Gene expression profile (10 most upregulated genes) according to the VHL-, MEN1-, Sporadic-, MEN1/Sporadic NFPanNETs and Normal Islet cells groups shows distinctive genotype-specific transcriptional alterations (yellow boxes).
Supplemental Figure 2. A. KLF12 mRNA overexpression in VHL vs. NIC (p=0.021), MEN1 vs. NIC (p=0.027) and Sporadic vs. NIC (p=0.007). B. CCNDBP1 mRNA overexpression in VHL vs. NIC (p=0.047), MEN1 vs. NIC (p=0.13) and Sporadic vs. NIC (p=0.05). C. MMP1 mRNA underexpression in VHL vs. NIC (p=0.04), MEN1 vs. NIC (p=0.04) and Sporadic vs. NIC (p=0.027)
Supplemental Figure 3. Heatmap expression of a 19 gene panel upregulated and a three gene panel downregulated in our study cohort compared to a previous report (VHL samples in yellow box) shows similar expression patterns (31), B. FLT12 mRNA overexpression in VHL vs. MEN1 (p=0.05), VHL vs. Sporadic (p=0.017) and VHL vs. NIC (p=0.09), C. VIM mRNA overexpression in VHL vs. MEN1 (p=0.05), VHL vs. Sporadic (p=0.13) and VHL vs. NIC (p=0.06)
Supplemental Figure 4. A. RT-qPCR expression analysis showing overexpression of CDK4 in the MEN1 vs. MEN1/Sporadic NFPanNETs (p=0.07) and MEN1 vs. Sporadic NFPanNETs subgroups (p=0.04). B. RT-qPCR expression analysis showing overexpression of CDK6 in the MEN1 vs. MEN1/Sporadic NFPanNETs (p=0.08) and MEN1 vs. Sporadic NFPanNETs subgroups (p=0.14).
Acknowledgments
Funding sources: This study was supported by the intramural program of the Center for Cancer Research, National Cancer Institute (1ZIABCO11275-7)
Footnotes
Author Contributions:
XK, SK, SG, MB, SA, MQ: experiment design and performance
RF, MC: statistical analysis
XM, RH, NN, MC, EK: Manuscript drafting and editing
Conflict of interest: There are no conflict of interest disclosures from any authors.
References
- 1.Halfdanarson TR, Rubin J, Farnell MB, Grant CS, Petersen GM. Pancreatic endocrine neoplasms: epidemiology and prognosis of pancreatic endocrine tumors. Endocrine-related cancer. 2008;15(2):409–27. doi: 10.1677/ERC-07-0221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bilimoria KY, Tomlinson JS, Merkow RP, Stewart AK, Ko CY, Talamonti MS, et al. Clinicopathologic features and treatment trends of pancreatic neuroendocrine tumors: analysis of 9,821 patients. Journal of gastrointestinal surgery : official journal of the Society for Surgery of the Alimentary Tract. 2007;11(11):1460–7. doi: 10.1007/s11605-007-0263-3. discussion 7-9. [DOI] [PubMed] [Google Scholar]
- 3.Horton WA, Wong V, Eldridge R. Von Hippel-Lindau disease: clinical and pathological manifestations in nine families with 50 affected members. Archives of internal medicine. 1976;136(7):769–77. doi: 10.1001/archinte.136.7.769. [DOI] [PubMed] [Google Scholar]
- 4.Keutgen XM, Hammel P, Choyke PL, Libutti SK, Jonasch E, Kebebew E. Evaluation and management of pancreatic lesions in patients with von Hippel-Lindau disease. Nature reviews Clinical oncology. 2016;13(9):537–49. doi: 10.1038/nrclinonc.2016.37. [DOI] [PubMed] [Google Scholar]
- 5.Kloppel G, Perren A, Heitz PU. The gastroenteropancreatic neuroendocrine cell system and its tumors: the WHO classification. Annals of the New York Academy of Sciences. 2004;1014:13–27. doi: 10.1196/annals.1294.002. [DOI] [PubMed] [Google Scholar]
- 6.Neychev V, Sadowski SM, Zhu J, Allgaeuer M, Kilian K, Meltzer P, et al. Neuroendocrine Tumor of the Pancreas as a Manifestation of Cowden Syndrome: A Case Report. The Journal of clinical endocrinology and metabolism. 2016;101(2):353–8. doi: 10.1210/jc.2015-3684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sadowski SM, Triponez F. Management of pancreatic neuroendocrine tumors in patients with MEN 1. Gland surgery. 2015;4(1):63–8. doi: 10.3978/j.issn.2227-684X.2014.12.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Mestier L, Gaujoux S, Cros J, Hentic O, Vullierme MP, Couvelard A, et al. Long-term Prognosis of Resected Pancreatic Neuroendocrine Tumors in von Hippel-Lindau Disease Is Favorable and Not Influenced by Small Tumors Left in Place. Annals of surgery. 2015;262(2):384–8. doi: 10.1097/SLA.0000000000000856. [DOI] [PubMed] [Google Scholar]
- 9.Blansfield JA, Choyke L, Morita SY, Choyke PL, Pingpank JF, Alexander HR, et al. Clinical, genetic and radiographic analysis of 108 patients with von Hippel-Lindau disease (VHL) manifested by pancreatic neuroendocrine neoplasms (PNETs) Surgery. 2007;142(6):814–8. doi: 10.1016/j.surg.2007.09.012. discussion 8 e1-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Charlesworth M, Verbeke CS, Falk GA, Walsh M, Smith AM, Morris-Stiff G. Pancreatic lesions in von Hippel-Lindau disease? A systematic review and meta-synthesis of the literature. Journal of gastrointestinal surgery : official journal of the Society for Surgery of the Alimentary Tract. 2012;16(7):1422–8. doi: 10.1007/s11605-012-1847-0. [DOI] [PubMed] [Google Scholar]
- 11.Hammel PR, Vilgrain V, Terris B, Penfornis A, Sauvanet A, Correas JM, et al. Pancreatic involvement in von Hippel-Lindau disease. The Groupe Francophone d’Etude de la Maladie de von Hippel-Lindau. Gastroenterology. 2000;119(4):1087–95. doi: 10.1053/gast.2000.18143. [DOI] [PubMed] [Google Scholar]
- 12.Libutti SK, Choyke PL, Bartlett DL, Vargas H, Walther M, Lubensky I, et al. Pancreatic neuroendocrine tumors associated with von Hippel Lindau disease: diagnostic and management recommendations. Surgery. 1998;124(6):1153–9. doi: 10.1067/msy.1998.91823. [DOI] [PubMed] [Google Scholar]
- 13.Jiao Y, Shi C, Edil BH, de Wilde RF, Klimstra DS, Maitra A, et al. DAXX/ATRX, MEN1, and mTOR pathway genes are frequently altered in pancreatic neuroendocrine tumors. Science. 2011;331(6021):1199–203. doi: 10.1126/science.1200609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dong M, Phan AT, Yao JC. New strategies for advanced neuroendocrine tumors in the era of targeted therapy. Clinical cancer research : an official journal of the American Association for Cancer Research. 2012;18(7):1830–6. doi: 10.1158/1078-0432.CCR-11-2105. [DOI] [PubMed] [Google Scholar]
- 15.Schmitt AM, Schmid S, Rudolph T, Anlauf M, Prinz C, Kloppel G, et al. VHL inactivation is an important pathway for the development of malignant sporadic pancreatic endocrine tumors. Endocrine-related cancer. 2009;16(4):1219–27. doi: 10.1677/ERC-08-0297. [DOI] [PubMed] [Google Scholar]
- 16.Zhang J, Francois R, Iyer R, Seshadri M, Zajac-Kaye M, Hochwald SN. Current understanding of the molecular biology of pancreatic neuroendocrine tumors. Journal of the National Cancer Institute. 2013;105(14):1005–17. doi: 10.1093/jnci/djt135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lonser RR, Glenn GM, Walther M, Chew EY, Libutti SK, Linehan WM, et al. von Hippel-Lindau disease. Lancet. 2003;361(9374):2059–67. doi: 10.1016/S0140-6736(03)13643-4. [DOI] [PubMed] [Google Scholar]
- 18.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic acids research. 2015;43(7):e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wilkerson MD, Hayes DN. ConsensusClusterPlus: a class discovery tool with confidence assessments and item tracking. Bioinformatics. 2010;26(12):1572–3. doi: 10.1093/bioinformatics/btq170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Larsson C, Calender A, Grimmond S, Giraud S, Hayward NK, Teh B, et al. Molecular tools for presymptomatic testing in multiple endocrine neoplasia type 1. Journal of internal medicine. 1995;238(3):239–44. doi: 10.1111/j.1365-2796.1995.tb00929.x. [DOI] [PubMed] [Google Scholar]
- 21.Manickam P, Guru SC, Debelenko LV, Agarwal SK, Olufemi SE, Weisemann JM, et al. Eighteen new polymorphic markers in the multiple endocrine neoplasia type 1 (MEN1) region. Human genetics. 1997;101(1):102–8. doi: 10.1007/s004390050595. [DOI] [PubMed] [Google Scholar]
- 22.Godin-Heymann N, Brabetz S, Murillo MM, Saponaro M, Santos CR, Lobley A, et al. Tumour-suppression function of KLF12 through regulation of anoikis. Oncogene. 2016;35(25):3324–34. doi: 10.1038/onc.2015.394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou HQ, Chen QC, Qiu ZT, Tan WL, Mo CQ, Gao SW. Integrative microRNA-mRNA and protein-protein interaction analysis in pancreatic neuroendocrine tumors. European review for medical and pharmacological sciences. 2016;20(13):2842–52. [PubMed] [Google Scholar]
- 24.Merrill MJ, Edwards NA, Lonser RR. Notch receptor and effector expression in von Hippel-Lindau disease-associated central nervous system hemangioblastomas. Journal of neurosurgery. 2011;115(3):512–7. doi: 10.3171/2011.5.JNS11271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van Adrichem RC, de Herder WW, Kamp K, Brugts MP, de Krijger RR, Sprij-Mooij DM, et al. Effects of Somatostatin Analogs and Dopamine Agonists on Insulin-Like Growth Factor 2-Induced Insulin Receptor Isoform A Activation by Gastroenteropancreatic Neuroendocrine Tumor Cells. Neuroendocrinology. 2016;103(6):815–25. doi: 10.1159/000444280. [DOI] [PubMed] [Google Scholar]
- 26.Johnson JD, Kuang S, Misler S, Polonsky KS. Ryanodine receptors in human pancreatic beta cells: localization and effects on insulin secretion. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2004;18(7):878–80. doi: 10.1096/fj.03-1280fje. [DOI] [PubMed] [Google Scholar]
- 27.Missiaglia E, Dalai I, Barbi S, Beghelli S, Falconi M, della Peruta M, et al. Pancreatic endocrine tumors: expression profiling evidences a role for AKT-mTOR pathway. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2010;28(2):245–55. doi: 10.1200/JCO.2008.21.5988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Erlic Z, Ploeckinger U, Cascon A, Hoffmann MM, von Duecker L, Winter A, et al. Systematic comparison of sporadic and syndromic pancreatic islet cell tumors. Endocrine-related cancer. 2010;17(4):875–83. doi: 10.1677/ERC-10-0037. [DOI] [PubMed] [Google Scholar]
- 29.Wang DD, Liu ZW, Han MM, Zhu ZM, Tu YL, Dou CQ, et al. Microarray based analysis of gene expression patterns in pancreatic neuroendocrine tumors. European review for medical and pharmacological sciences. 2015;19(18):3367–74. [PubMed] [Google Scholar]
- 30.Lopez-Avalos MD, Duvivier-Kali VF, Xu G, Bonner-Weir S, Sharma A, Weir GC. Evidence for a role of the ubiquitin-proteasome pathway in pancreatic islets. Diabetes. 2006;55(5):1223–31. doi: 10.2337/db05-0450. [DOI] [PubMed] [Google Scholar]
- 31.Speisky D, Duces A, Bieche I, Rebours V, Hammel P, Sauvanet A, et al. Molecular profiling of pancreatic neuroendocrine tumors in sporadic and Von Hippel-Lindau patients. Clinical cancer research : an official journal of the American Association for Cancer Research. 2012;18(10):2838–49. doi: 10.1158/1078-0432.CCR-11-2759. [DOI] [PubMed] [Google Scholar]
- 32.Yoshihara K, Shahmoradgoli M, Martinez E, Vegesna R, Kim H, Torres-Garcia W, et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nature communications. 2013;4:2612. doi: 10.1038/ncomms3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sadanandam A, Wullschleger S, Lyssiotis CA, Grotzinger C, Barbi S, Bersani S, et al. A Cross-Species Analysis in Pancreatic Neuroendocrine Tumors Reveals Molecular Subtypes with Distinctive Clinical, Metastatic, Developmental, and Metabolic Characteristics. Cancer discovery. 2015;5(12):1296–313. doi: 10.1158/2159-8290.CD-15-0068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bloomston M, Durkin A, Yang I, Rojiani M, Rosemurgy AS, Enkmann S, et al. Identification of molecular markers specific for pancreatic neuroendocrine tumors by genetic profiling of core biopsies. Annals of surgical oncology. 2004;11(4):413–9. doi: 10.1245/ASO.2004.03.077. [DOI] [PubMed] [Google Scholar]
- 35.Duerr EM, Mizukami Y, Ng A, Xavier RJ, Kikuchi H, Deshpande V, et al. Defining molecular classifications and targets in gastroenteropancreatic neuroendocrine tumors through DNA microarray analysis. Endocrine-related cancer. 2008;15(1):243–56. doi: 10.1677/ERC-07-0194. [DOI] [PubMed] [Google Scholar]
- 36.Walmsley SR, McGovern NN, Whyte MK, Chilvers ER. The HIF/VHL pathway: from oxygen sensing to innate immunity. American journal of respiratory cell and molecular biology. 2008;38(3):251–5. doi: 10.1165/rcmb.2007-0331TR. [DOI] [PubMed] [Google Scholar]
- 37.Ning H, Shao QQ, Ding KJ, Gao DX, Lu QL, Cao QW, et al. Tumor-infiltrating regulatory T cells are positively correlated with angiogenic status in renal cell carcinoma. Chinese medical journal. 2012;125(12):2120–5. [PubMed] [Google Scholar]
- 38.Tartour E, Pere H, Maillere B, Terme M, Merillon N, Taieb J, et al. Angiogenesis and immunity: a bidirectional link potentially relevant for the monitoring of antiangiogenic therapy and the development of novel therapeutic combination with immunotherapy. Cancer metastasis reviews. 2011;30(1):83–95. doi: 10.1007/s10555-011-9281-4. [DOI] [PubMed] [Google Scholar]
- 39.Mediavilla-Varela M, Luddy K, Noyes D, Khalil FK, Neuger AM, Soliman H, et al. Antagonism of adenosine A2A receptor expressed by lung adenocarcinoma tumor cells and cancer associated fibroblasts inhibits their growth. Cancer biology & therapy. 2013;14(9):860–8. doi: 10.4161/cbt.25643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cuperlovic-Culf M, Cormier K, Touaibia M, Reyjal J, Robichaud S, Belbraouet M, et al. (1)H NMR metabolomics analysis of renal cell carcinoma cells: Effect of VHL inactivation on metabolism. International journal of cancer. 2016;138(10):2439–49. doi: 10.1002/ijc.29947. [DOI] [PubMed] [Google Scholar]
- 41.Fjallskog ML, Hessman O, Eriksson B, Janson ET. Upregulated expression of PDGF receptor beta in endocrine pancreatic tumors and metastases compared to normal endocrine pancreas. Acta oncologica. 2007;46(6):741–6. doi: 10.1080/02841860601048388. [DOI] [PubMed] [Google Scholar]
- 42.McGary EC, Weber K, Mills L, Doucet M, Lewis V, Lev DC, et al. Inhibition of platelet-derived growth factor-mediated proliferation of osteosarcoma cells by the novel tyrosine kinase inhibitor STI571. Clinical cancer research : an official journal of the American Association for Cancer Research. 2002;8(11):3584–91. [PubMed] [Google Scholar]
- 43.Pietras K, Stumm M, Hubert M, Buchdunger E, Rubin K, Heldin CH, et al. STI571 enhances the therapeutic index of epothilone B by a tumor-selective increase of drug uptake. Clinical cancer research : an official journal of the American Association for Cancer Research. 2003;9(10 Pt 1):3779–87. [PubMed] [Google Scholar]
- 44.Fjallskog ML, Lejonklou MH, Oberg KE, Eriksson BK, Janson ET. Expression of molecular targets for tyrosine kinase receptor antagonists in malignant endocrine pancreatic tumors. Clinical cancer research : an official journal of the American Association for Cancer Research. 2003;9(4):1469–73. [PubMed] [Google Scholar]
- 45.Singhi AD, Lilo M, Hruban RH, Cressman KL, Fuhrer K, Seethala RR. Overexpression of lymphoid enhancer-binding factor 1 (LEF1) in solid-pseudopapillary neoplasms of the pancreas. Modern pathology : an official journal of the United States and Canadian Academy of Pathology, Inc. 2014;27(10):1355–63. doi: 10.1038/modpathol.2014.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gandhirajan RK, Staib PA, Minke K, Gehrke I, Plickert G, Schlosser A, et al. Small molecule inhibitors of Wnt/beta-catenin/lef-1 signaling induces apoptosis in chronic lymphocytic leukemia cells in vitro and in vivo. Neoplasia. 2010;12(4):326–35. doi: 10.1593/neo.91972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chu IM, Hengst L, Slingerland JM. The Cdk inhibitor p27 in human cancer: prognostic potential and relevance to anticancer therapy. Nature reviews Cancer. 2008;8(4):253–67. doi: 10.1038/nrc2347. [DOI] [PubMed] [Google Scholar]
- 48.Tang LH, Contractor T, Clausen R, Klimstra DS, Du YC, Allen PJ, et al. Attenuation of the retinoblastoma pathway in pancreatic neuroendocrine tumors due to increased cdk4/cdk6. Clinical cancer research : an official journal of the American Association for Cancer Research. 2012;18(17):4612–20. doi: 10.1158/1078-0432.CCR-11-3264. [DOI] [PubMed] [Google Scholar]
- 49.Li B, He H, Tao BB, Zhao ZY, Hu GH, Luo C, et al. Knockdown of CDK6 enhances glioma sensitivity to chemotherapy. Oncology reports. 2012;28(3):909–14. doi: 10.3892/or.2012.1884. [DOI] [PubMed] [Google Scholar]
- 50.Peixoto RD, Noonan KL, Pavlovich P, Kennecke HF, Lim HJ. Outcomes of patients treated with capecitabine and temozolamide for advanced pancreatic neuroendocrine tumors (PNETs) and non-PNETs. Journal of gastrointestinal oncology. 2014;5(4):247–52. doi: 10.3978/j.issn.2078-6891.2014.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li J, Klughammer J, Farlik M, Penz T, Spittler A, Barbieux C, et al. Single-cell transcriptomes reveal characteristic features of human pancreatic islet cell types. EMBO reports. 2016;17(2):178–87. doi: 10.15252/embr.201540946. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1: Gene expression profile (10 most upregulated genes) according to the VHL-, MEN1-, Sporadic-, MEN1/Sporadic NFPanNETs and Normal Islet cells groups shows distinctive genotype-specific transcriptional alterations (yellow boxes).
Supplemental Figure 2. A. KLF12 mRNA overexpression in VHL vs. NIC (p=0.021), MEN1 vs. NIC (p=0.027) and Sporadic vs. NIC (p=0.007). B. CCNDBP1 mRNA overexpression in VHL vs. NIC (p=0.047), MEN1 vs. NIC (p=0.13) and Sporadic vs. NIC (p=0.05). C. MMP1 mRNA underexpression in VHL vs. NIC (p=0.04), MEN1 vs. NIC (p=0.04) and Sporadic vs. NIC (p=0.027)
Supplemental Figure 3. Heatmap expression of a 19 gene panel upregulated and a three gene panel downregulated in our study cohort compared to a previous report (VHL samples in yellow box) shows similar expression patterns (31), B. FLT12 mRNA overexpression in VHL vs. MEN1 (p=0.05), VHL vs. Sporadic (p=0.017) and VHL vs. NIC (p=0.09), C. VIM mRNA overexpression in VHL vs. MEN1 (p=0.05), VHL vs. Sporadic (p=0.13) and VHL vs. NIC (p=0.06)
Supplemental Figure 4. A. RT-qPCR expression analysis showing overexpression of CDK4 in the MEN1 vs. MEN1/Sporadic NFPanNETs (p=0.07) and MEN1 vs. Sporadic NFPanNETs subgroups (p=0.04). B. RT-qPCR expression analysis showing overexpression of CDK6 in the MEN1 vs. MEN1/Sporadic NFPanNETs (p=0.08) and MEN1 vs. Sporadic NFPanNETs subgroups (p=0.14).


