Abstract
OBJECTIVE
We aimed to develop a prediction model for lymph node metastasis using a gene expression signature in patients with endometrioid-type endometrial cancer.
METHODS
Newly diagnosed endometrioid-type endometrial cancer cases in which the patients had undergone lymphadenectomy during a surgical staging procedure were identified from a national dataset (N = 330). Clinical and pathologic data were extracted from patient medical records, and gene expression datasets of their tumors were used to create a 12-gene predictive model for lymph node metastasis. We used principal components analysis on a training set (n = 110) to develop multivariate logistic models to predict low-risk patients having a probability of lymph node metastasis of less than 4%. The model with the highest prediction performance was selected for an evaluation set (n = 112), which, in turn, was validated in an independent validation set (n = 108).
RESULTS
The model applied to the evaluation set showed 100% sensitivity (90% confidence interval [CI], 74%–100%) and 42% specificity (90% CI, 34%–51%), which resulted in 100% negative predictive value (90% CI, 89%–100%). In the validation set, we confirmed that the model consistently showed 100% sensitivity (90% CI, 88%–100%), 42% specificity (90% CI, 32%–50%), and 100% negative predictive value (90% CI, 88%–100%).
CONCLUSIONS
Our 12-gene signature model is a useful tool for the identification of patients with endometrioid-type endometrial cancer at low risk of lymph node metastasis, particularly given that it can be used to analyze histologic tissue prior to surgery and used to tailor surgical options.
Keywords: Cancer genomics, endometrial cancer, personalized medicine, diagnosis and staging
1. Introduction
Endometrial cancer is the fifth most common female cancer in the world(1) and the most common gynecological cancer in the United States(2). The prognosis and survival of patients with endometrial cancer are largely predicted by histology and a staging procedure that includes a lymphadenectomy(3). However, the performance of lymphadenectomy during surgery is controversial, owing to risks of serious morbidity and deteriorated quality of life for some patients(4, 5). Currently, many guidelines do not advocate routine lymphadenectomy, allowing for its omission in low-risk patients, especially in those with apparently early-stage endometrial cancer(6, 7). To identify low-risk patients, many proposed models assess the risk of lymph node metastasis using well-known clinical risk factors, such as depth of invasion, tumor grade, or tumor size(8–10).
Advances in molecular profiling have provided important insights into the biologic nature of the tumor. These advances allow researchers to study the possible use of gene expression signatures as predictive tools for clinical outcomes, including metastasis(11, 12). Therefore, we hypothesized that a gene expression signature could predict the risk of lymph node metastasis in endometrial cancer and be used to tailor surgery. To test the hypothesis, we developed a risk prediction model using gene expression profiling and validated its reliability.
2. Materials and Methods
Subsequent to gaining approvals from the Scientific Review Board of H. Lee Moffitt Cancer Center and the Institutional Review Board of the University of South Florida, we identified cases from the Total Cancer Care (TCC) consortium network dataset of newly diagnosed endometrioid-type endometrial cancer in which the patients had undergone lymphadenectomy during a surgical staging procedure. TCC is a cohort of patients who have consented to the collection of their tumor specimens and clinical data at Florida hospitals (including H. Lee Moffitt Cancer Center) and 8 other national sites(13). Patients with a histologic type other than endometrioid-type endometrial cancer were excluded from this study. Clinical and pathologic data were extracted from patient medical records. Of the 562 patients in the TCC cohort, 330 fulfilled eligibility criteria for this study, with patient and tumor characteristics summarized in Table 1. Tumors from patients under the TCC protocol were arrayed on Affymetrix HuRSTA–2z520709 GeneChips (Affymetrix, Santa Clara, CA), which contain approximately 60 000 probesets representing approximately 25 037 unique genes (Affymetrix HuRSTA-2a520709, Gene Expression Omnibus database: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GPL10379).
Table 1.
Clinical Characteristics of Study Patients (N=330)
| Training Set (n = 110) |
Evaluation Set (n = 112) |
Validation Set (n = 108) |
Total (n = 330) |
|
|---|---|---|---|---|
| Age, y | ||||
| Median (range) | 63 (29–86) | 61 (36–90) | 63 (32–87) | 63 (29–90) |
| Stage, No. (%) | ||||
| IA | 55 (50.0) | 62 (55.4) | 63 (58.3) | 180 (54.5) |
| IB | 18 (16.4) | 24 (21.4) | 20 (18.5) | 62 (18.8) |
| II–IV | 37 (33.6) | 26 (23.2) | 25 (23.2) | 88 (26.7) |
| Histologic type, No. (%) | ||||
| Endometrioid | 110 (100.0) | 112 (100.0) | 108 (100.0) | 330 (100.0) |
| Histologic grade, No. (%) | ||||
| 1 | 39 (35.5) | 41 (36.6) | 43 (39.8) | 123 (37.3) |
| 2 | 49 (44.6) | 57 (50.9) | 50 (46.3) | 156 (47.3) |
| 3 | 22 (20.0) | 14 (12.5) | 15 (13.9) | 51 (15.4) |
| Depth of myometrial invasion, No. (%) | ||||
| Less than 50% | 66 (60.0) | 74 (66.1) | 76 (70.4) | 216 (65.4) |
| 50% or more | 44 (40.0) | 38 (33.9) | 32 (29.6) | 114 (34.6) |
| Tumor size, cm | ||||
| Median(range) | 3.7 (0.5–12) | 3.5 (0.1–13) | 3.5 (0.1–10) | 3.5 (0.1–13) |
| Lymph node metastasis, No. (%) | ||||
| Negative | 95 (86.4) | 97 (86.6) | 93 (86.1) | 285 (86.4) |
| Positive | 15 (13.6) | 15 (13.4) | 15 (13.9) | 45 (13.6) |
2.1 Statistical Analyses
To develop and validate a biomarker signature that provides a consistent prediction for stratifying the nodal status of patients in the heterogeneous population of endometrial cancer patients, we divided our patient data into 3 unique sets to be used for biomarker discovery, modeling/evaluation, and independent validation(14). Specifically, the patients were evenly split into the 3 sets according to the dates on which they underwent surgery. This strategy was designed to ensure that the final model, having been consistently validated from the 3 sets, would be relatively independent of the potentially different clinical settings involved. The first set was used as the training set, the second as the evaluation set, and the third as the validation set.
To effectively identify genes with P values of <0.0001 under the null assumption, we used a 2-sample t-test to compare probesets that were differentially expressed between lymph node-positive and lymph node-negative patients in the training and evaluation sets. Probesets with P values <0.01 were pre-selected on both sets. We adopted this statistical strategy with 2 independent patient sets for gene discovery and multivariable prediction model training to avoid the pitfalls that could arise from making multiple comparisons of a large number of candidate genes and models.
To obtain more biologically relevant gene predictors, we eliminated all non-annotated probes and selected probes from among those differentially expressed probesets that matched to genes showing copy number alterations or mutations of more than 5% in The Cancer Genome Atlas (TCGA) data. In particular, we used the cBioPortal tool for Cancer Genomics (http://cbioportal.org/) on the Uterine Corpus Endometrial Carcinoma provisional dataset generated by the TCGA Research Network (http://cancergenome.nih.gov/). After eliminating the probesets that were less biologically relevant, we generated models using multivariable logistic regression and principal components analysis; PCA was used to reduce data dimension, with the principal components explaining >60% of the variation in the training set. We ranked the probesets by their P values and considered the top 10 in the multivariable logistic regression models to stratify the lymph node status of patients in the training set. We then evaluated these competing models by performing 5-fold cross validation, using logistic regression with lymph node-positive as the outcome. Accuracy, sensitivity, specificity, positive predictive value, negative predictive value, and Youden index were calculated. On the basis of the literature, a false negative rate of 4% was clinically determined to be the cut-off point(15, 16). The optimal number of probesets was then determined by comparing multiple models of varying numbers of selected probesets based on their consistent statistical significance in both the training and evaluation sets, ie, by their mean P values. We then independently validated an objective model performance with the validation set, which was completely set aside from the above training and modeling steps, for its accuracy, sensitivity, specificity, positive predictive value, and negative predictive value.
3. Results
Datasets from the 330 patients with histologically proven endometrioid endometrial carcinoma were split into 3 sets in chronological order. One hundred and ten, 112, and 108 patients were assigned to a training set, an evaluation set, and a validation set, respectively. The median age of patients was 63 years (range, 29–90 years). The median number of retrieved lymph nodes was 28 (range, 1–38), and para-aortic lymph node dissection was performed in 118 of the 330 patients (35.8%). Lymph node metastasis was found in 45 of the 330 patients (13.6%). The additional clinical characteristics, including clinical risk factors for lymph node metastasis, are summarized in Table 1, and the model development process is illustrated by the flowchart in Figure 1.
Figure 1.
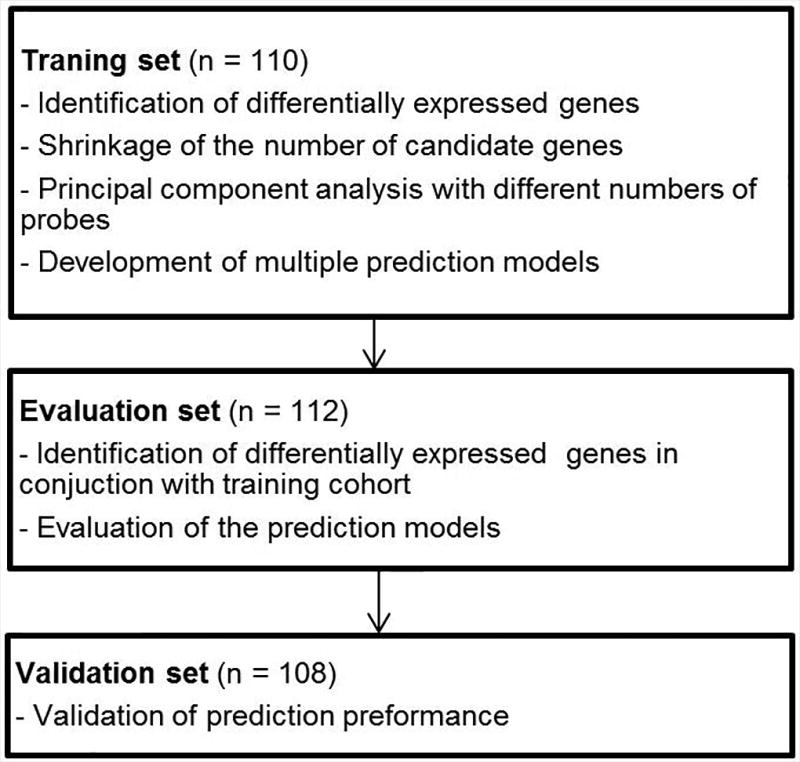
Study Flowchart. The training, evaluation, and validation sets were chronologically independent.
Using both the training and evaluation datasets, we developed a 12-gene signature predicting lymph node metastasis. Those genes were GREM2, FMO2, TMEM212, ESR1, RPTN, PRR9, TCHHL1, CPB1, CLCN2, ITLN2, PKHD1L1, and SLC9C2. The probe information and corresponding genes are summarized in Table 2. Those genes that were differentially expressed between the lymph node-positive and lymph node-negative groups, but did not show frequent genetic alteration in TCGA’s uterine endometrial cancer database, were discarded. After generating multiple prediction models using multivariate logistic regression and principal components analysis, we selected the model with the highest predictive performance by comparing Youden index values. Finally, the model with 18 probes corresponding to 12 genes was selected, because of a high Youden index in the training and evaluation sets, and subjected to further validation.
Table 2.
List of Selected Genes and Matching Probes
| Gene Symbol | Gene Name | Probe Identity | Location |
|---|---|---|---|
| TCHHL1 | Trichohyalin-like 1 | merck-NM_001008536_at | 1q21.3 |
|
| |||
| RPTN | Repetin | merck-ENST00000316073_at | 1q21.3 |
|
| |||
| PRR9 | Proline rich 9 | merck-ENST00000368744_at | 1q21.3 |
|
| |||
| ITLN2 | Intelectin 2 | merck-NM_080878_at | 1q22–q23 |
|
| |||
| FMO2 | Flavin containing monooxygenase 2 | merck2-AL833218_at Sense | 1q24.3 |
| merck-BC005894_a_at | |||
| merck-ENST00000209929_a_at | |||
| merck-NM_001460_at | |||
|
| |||
| SLC9C2 | Solute carrier family 9, member C2 | merck-NM_178527_at | 1q25.1 |
|
| |||
| GREM2 | Gremlin 2, DAN family BMP antagonist | merck-AK024848_a_at | 1q43 |
| merck-NM_001871_at | |||
| merck-BC046632_a_at | |||
|
| |||
| CPB1 | Carboxypeptidase B1 | merck-NM_001871_at | 3q24 |
|
| |||
| TMEM212 | Transmembrane protein 212 | merck-AK026825_x_at | 3q26.31 |
|
| |||
| CLCN2 | Chloride channel, voltage-sensitive 2 | merck-NM_004366_at | 3q27.1 |
| merck2-BC072004_at | |||
|
| |||
| ESR1 | Estrogen receptor 1 | merck-ENST00000347491_s_at | 6q25.1 |
| merck-BM544900_at | |||
| merck-NM_000125_at | |||
| merck2-NM_000125_at | |||
| merck2-AL050116_at | |||
|
| |||
| PKHD1L1 | Polycystic kidney and hepatic disease 1 (autosomal recessive)-like 1 | merck-NM_177531_s_at | 8q23 |
| merck2-AY219181_at | |||
| merck-AA443594_s_at | |||
When we allowed the pre-determined false negative rate of 4% for lymph node metastasis, the current model showed sensitivity of 100% (90% confidence interval [CI], 74%–100%) and specificity of 41% (90% CI, 34%–50%) in the validation set. The negative predictive value was 100% (90% CI, 89%–100%), while the positive predictive value was 21%. The sensitivity, specificity, and positive and negative predictive values for the training and evaluation sets are summarized in Table 3. In particular, in the multivariate logistic regression analysis that included well-known clinical risk factors (deep myometrial invasion and grade 3 histology), we found the linear estimates of our 12-gene signature model to be significant risk factors, independent of other competing clinical risk factors (P = 0.005).
Table 3.
The Diagnostic Performance of the 12-Gene Model for Lymph Node Metastasisa
| Sensitivity (90% CI) |
Specificity (90% CI) |
PPV (90% CI) |
NPV (90% CI) |
|
|---|---|---|---|---|
| Training set (n = 110) | 100% (74%–100%) | 42% (34%–51%) | 21% (14%–31%) | 100% (89%–100%) |
| Evaluation set (n = 112) | 100% (74%–100%) | 42% (34%–51%) | 21% (13%–31%) | 100% (89%–100%) |
| Validation set (n = 108) | 100% (74%–100%) | 41% (32%–50%) | 21% (14%–31%) | 100% (88%–100%) |
Abbreviations: CI, confidence interval; PPV, positive predictive value; NPV, negative predictive value.
On the basis of clinical consensus, we predetermined a cut-off probability of 0.04.
From the pooled analysis of 330 patients, our 12-gene signature model showed sensitivity of 100% (90% CI, 94%–100%) and specificity of 42% (90% CI, 32%–51%). The receiver operating characteristics curve area was 0.72 (90% CI, 0.69–0.75). The negative predictive value was 100% (90% CI, 98%–100%). Among the 330 patients, 137 patients had tumors showing deep myometrial invasion, grade 3 histology, or both, indicating that they could not be classified as a low-risk group. Of the 137 patients who were not clinically determined to be low-risk, our model classified 36 as low-risk with no false negative, which resulted in a sensitivity of 100% (90% CI, 92%–100%) and a negative predictive value of 100% (90% CI, 92%–100%).
4. Conclusions
In the current study, using a 12-gene expression signature, we developed a prediction model for identifying patients at low risk of lymph node metastasis in endometrioid-type endometrial cancer. Because the clinical usefulness of this model depends on the ability to reliably predict prognoses for patients with low risk of lymph node metastasis, we focused our analyses of sensitivity and negative predictive values on a low-risk group. The model showed consistently high sensitivity and negative predictive values in the training, evaluation, and independent validation sets. This high negative predictive value implies that patients classified as a low-risk group by this model may forgo systemic lymphadenectomy, which may be associated with serious morbidity or deteriorated quality of life for some patients.
Notably, our 12-gene model classified one-fourth of clinically high-risk patients as low-risk patients. As the model consistently showed high sensitivity and negative predictive values, it could provide a useful diagnostic tool for tailoring lymphadenectomy, even when clinical risk factors indicate a high risk for certain patients. Although the ability to exclude certain high-risk patients from a lymphadenectomy is useful, complete clinical risk information may not be readily available at the time of surgery. A genetic signature such as ours has real-world utility in that it can be effectively performed on histologic tissue prior to surgery, as demonstrated by the negative predictive value in the total data set. Despite the results from the 2 randomized trials(4, 5), lymphadenectomy is still widely recommended when patients have clinical risk factors, such as deep myometrial invasion or high tumor grade. Thus, although more than 50% to 60% of endometrial cancer patients may be exempt from having lymphadenectomy, 40% to 50% of patients are still subject to the procedure, which may deteriorate their quality of life. If our 12-gene prediction model could identify patients without lymph node metastasis even among high-risk patients classified by clinical risk factors, it could be of great utility in the decision of whether to proceed with lymphadenectomy (Figure 2).
Figure 2.
Principal Components (PC) Analysis of Selected Gene Probes in the Training (A) and Evaluation (B) Datasets.
The current study has several limitations. First, although we successfully validated the 12-gene signature using chronologically independent datasets, further validation of the current 12-gene model in independent datasets from different clinical settings and centers may be necessary to fully confirm its performance as a personalized surgical decision support tool. To help address this limitation, we currently plan to evaluate the performance of our model using multi-institutional patient data from the Oncology Research Information Exchange Network. Second, it is still unclear how the functional profile of the 12 genes in our model contributes to the progression of invasive phenotype of endometrial cancer. Although the functional aspect of the current model has yet to be identified, better insight may be revealed when the genetic alteration or protein expression of those genes are compared between endometrioid and papillary serous subtypes. When we reviewed the TCGA endometrial cancer dataset, we found that gene amplifications of those 12 genes were increased in the serous or papillary serous types of endometrial cancers by comparison with the endometrioid type. Therefore, we can speculate that our 12-gene expression profile may identify histologically endometrioid-type but biologically serous or serous papillary-type endometrial cancer. Third, we did not include clinical variables such as grade or myometrial invasion to develop a prediction model, given that those variables are only attainable as postoperative data.
The current study indicates that our 12-gene signature could be useful in the identification of endometrial cancer patients who have a very low risk of lymph node metastasis. In particular, the model may help patients with high clinical risk factors to avoid unnecessary lymphadenectomy. Further validation studies will be required to determine whether the 12-gene signature model can show such clinical benefits in endometrial cancer patients.
Figure 3.
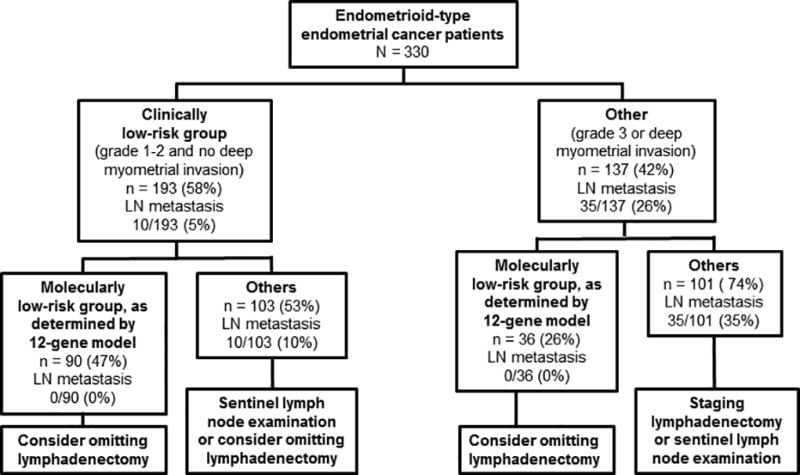
Possible Use of the Current 12-Gene Model in Precision Medicine to Treat Endometrial Cancer.
Acknowledgments
Funding/Support: This work has been supported, in part, by the Biostatistics Core Facility at the H. Lee Moffitt Cancer Center & Research Institute, an NCI designated Comprehensive Cancer Center (P30-CA076292). Dr. Chon is the recipient of a Wilma Williams Education and Clinical Research Award for Endometrial Cancer from the Foundation for Gynecologic Oncology.
Footnotes
Conflict of Interest Disclosures: None reported.
Additional Contributions: Manuscript editing assistance was provided by Sonya Smyk and Rasa Hamilton of the Moffitt Cancer Center. Neither received compensation outside their usual salaries.
References
- 1.Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359–86. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 3.Pecorelli S. Revised FIGO staging for carcinoma of the vulva, cervix, and endometrium. Int J Gynaecol Obstet. 2009;105:103–4. doi: 10.1016/j.ijgo.2009.02.012. [DOI] [PubMed] [Google Scholar]
- 4.Astec Study Group. Kitchener H, Swart AM, et al. Efficacy of systematic pelvic lymphadenectomy in endometrial cancer (MRC ASTEC trial): a randomised study. Lancet. 2009;373:125–36. doi: 10.1016/S0140-6736(08)61766-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Benedetti Panici P, Basile S, Maneschi F, et al. Systematic pelvic lymphadenectomy vs. no lymphadenectomy in early-stage endometrial carcinoma: randomized clinical trial. J Natl Cancer Inst. 2008;100:1707–16. doi: 10.1093/jnci/djn397. [DOI] [PubMed] [Google Scholar]
- 6.Colombo N, Creutzberg C, Amant F, et al. ESMO-ESGO-ESTRO Consensus Conference on Endometrial Cancer: diagnosis, treatment and follow-up. Ann Oncol. 2016;27:16–41. doi: 10.1093/annonc/mdv484. [DOI] [PubMed] [Google Scholar]
- 7.Querleu D, Planchamp F, Narducci F, et al. Clinical practice guidelines for the management of patients with endometrial cancer in France: recommendations of the Institut National du Cancer and the Societe Francaise d'Oncologie Gynecologique. Int J Gynecol Cancer. 2011;21:945–50. doi: 10.1097/IGC.0b013e31821bd473. [DOI] [PubMed] [Google Scholar]
- 8.Todo Y, Okamoto K, Hayashi M, et al. A validation study of a scoring system to estimate the risk of lymph node metastasis for patients with endometrial cancer for tailoring the indication of lymphadenectomy. Gynecol Oncol. 2007;104:623–8. doi: 10.1016/j.ygyno.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 9.Mariani A, Dowdy SC, Cliby WA, et al. Prospective assessment of lymphatic dissemination in endometrial cancer: a paradigm shift in surgical staging. Gynecol Oncol. 2008;109:11–8. doi: 10.1016/j.ygyno.2008.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kang S, Kang WD, Chung HH, et al. Preoperative identification of a low-risk group for lymph node metastasis in endometrial cancer: a Korean gynecologic oncology group study. J Clin Oncol. 2012;30:1329–34. doi: 10.1200/JCO.2011.38.2416. [DOI] [PubMed] [Google Scholar]
- 11.Ramaswamy S, Ross KN, Lander ES, Golub TR. A molecular signature of metastasis in primary solid tumors. Nat Genet. 2003;33:49–54. doi: 10.1038/ng1060. [DOI] [PubMed] [Google Scholar]
- 12.Blok EJ, van de Velde CJ, Smit VT. 70-Gene Signature in Early-Stage Breast Cancer. N Engl J Med. 2016;375:2199. doi: 10.1056/NEJMc1612048. [DOI] [PubMed] [Google Scholar]
- 13.Fenstermacher DA, Wenham RM, Rollison DE, Dalton WS. Implementing personalized medicine in a cancer center. Cancer J. 2011;17:528–36. doi: 10.1097/PPO.0b013e318238216e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ambroise C, McLachlan GJ. Selection bias in gene extraction on the basis of microarray gene-expression data. Proc Natl Acad Sci U S A. 2002;99:6562–6. doi: 10.1073/pnas.102102699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boronow RC. Surgical staging of endometrial cancer: evolution, evaluation, and responsible challenge--a personal perspective. Gynecol Oncol. 1997;66:179–89. doi: 10.1006/gyno.1997.4732. [DOI] [PubMed] [Google Scholar]
- 16.Sakuragi N. Emerging concept of tailored lymphadenectomy in endometrial cancer. J Gynecol Oncol. 2012;23:210–2. doi: 10.3802/jgo.2012.23.4.210. [DOI] [PMC free article] [PubMed] [Google Scholar]


