Abstract
Antibiotic exposure in children has been associated with the risk of Inflammatory Bowel Disease (IBD). Since antibiotic use in children or in their pregnant mother can affect how the intestinal microbiome develops, we asked whether the transfer of an antibiotic-perturbed microbiota from mothers to their children could affect their risk of developing IBD. Here we demonstrate that germ-free adult pregnant mice inoculated with a gut microbial community shaped by antibiotic exposure transmitted their perturbed microbiota to their offspring with high fidelity. Without any direct or continued exposure to antibiotics, this dysbiotic microbiota in the offspring remained distinct from controls for at least 21 weeks. By using both IL-10-deficient and wild type mothers, we showed that both inoculum and genotype shape the microbiota populations in the offspring. Since IL10−/− mice are genetically susceptible to colitis, we could assess the risk due to maternal transmission of an antibiotic-perturbed microbiota. We found that the IL10−/− offspring that had received the perturbed gut microbiota developed markedly increased colitis. Taken together, our findings indicate that antibiotic exposure shaping the maternal gut microbiota has effects that extend to their offspring with both ecological and long-term disease consequences.
Introduction
Antibiotics have long-lasting consequences on the gut microbiota with the potential to impact host physiology and health.1–6 Antibiotic exposure has been associated with increased IBD risk5,7,8- notable since antibiotics are massively used worldwide9,10. Antibiotic use is extensive during pregnancy11,12, with reported rates of 42% in France, 40% in the US, 33% in the UK, and 24% in Finland.12–15 β-lactam antibiotics are the most widely used in pregnancy because of their presumed safety, with penicillins being the antibiotics most frequently used. However, little is known about the transgenerational impact of an antibiotic-perturbed microbiota. Because of the potential importance of intergenerational microbiota transmission16–18, we hypothesized that a maternally inherited antibiotic-perturbed microbiota affects the risk of experimental colitis in offspring.
We tested this hypothesis in IL-10-deficient (IL10−/−) mice, which spontaneously develop colitis. The colitis that IL10−/− mice develop requires microbial exposure;19 in germ-free conditions they are protected from disease.20 The IL10−/− mouse has been a well-recognized model for chronic, immune-mediated colitis in humans since its first use in 1993.19 It has been extensively studied because of its relevance to human IBD and dependence on resident intestinal bacteria to stimulate effector TH1 and TH17 immune responses.20,21 Many of the features of the model, including pancolitis, small intestinal involvement in certain housing conditions, transmural inflammation, crypt hyperplasia, goblet cell depletion, infiltration of the lamina propria with activated T and B cells, a TH1/ Th17 cytokine profile, development of dysbiosis and a chronic progressive course, mimic clinical, pathologic, microbial and immunologic changes that occur in human ulcerative colitis and Crohn’s disease.22,23 On this basis, we performed studies to determine the consequences of maternal transmission of a perturbed microbiota in this model. By introducing antibiotic-perturbed or control microbiota to germ-free pregnant mice, we could ensure that we were examining the effects of the founding microbiota on properties in the offspring, without the interference of antibiotic treatment of their mothers. In this model system, we asked two major questions: (i) what was the fidelity of the transfer from the inocula to the dam, and then to the pups over an extended period of time; and (ii) did the microbiota in the antibiotic-perturbed inoculum confer enhanced risk of colitis in the pups more than 5 months after birth?
Results
Characterizing the inocula and microbiota changes with passage in the dams
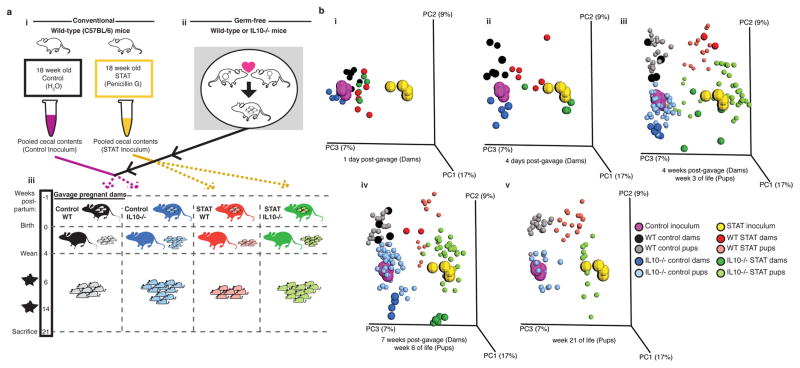
To investigate the role of an antibiotic-altered microbiome on colitis development, we sought to expose both wild type and IL10−/− mice to antibiotic-altered microbiota as early in life as possible; with both IL10−/− and wild type (WT) dams and two inocula, four groups of mice were studied (Figure 1a). We began the experiment with germ-free pregnant mice, conventionalizing them ~1 week before delivery with the specified inocula, facilitating transfer to their pups at birth. The inocula had been obtained at sacrifice from female C57BL/6 mice that had prolonged exposure to subtherapeutic levels of penicillin (STAT), or not (Control), which had been generated previously by pooling 3 donors for each inoculum2 (Figure 1ai). Pooling allowed us to create one inoculum for each treatment group and generate a large study cohort, though it may have limited the variability that is likely seen in humans. To maintain dietary consistency with the donor mice and thus reduce stress on the microbial community, all mice were maintained on a 45% high fat diet (HFD) for the entirety of the experiment.
Figure 1. Microbiome transfer to pregnant germ-free mice colonized 23 dams and 112 pups.
(a)(i) STAT and Control gut microbiota inocula were taken from a previous experiment.2 (ii) Germ-free (GF) mice were mated to yield pregnant GF dams. This procedure was done in C57BL/6 mice of wild type (WT) or IL10-deficient (IL10−/−) genotype. (iii) Approximately 1 week before delivery, the pregnant GF dams were gavaged with either the STAT or the Control inoculum, to conventionalize them. Pups were weaned at 4 weeks of age and sacrificed at 21 weeks of age. For the IL10−/− mice, there also were sacrifices at age 6 and 14 weeks. (b) Principal coordinate plots of unweighted UniFrac distances. The inocula used to colonize the dams before delivery are shown together with dam and pup fecal samples from the indicated time points. n=4 replicate aliquots for each inoculum: All numbers of mice/group are shown in Supplementary Table 1, along with P values from Adonis testing.
The STAT inoculum had fewer OTUs and a remodeled community composition and structure (Supplementary Figure 1a–b), with relative taxa abundance differences including decreased Erysipelotrichi and increased Verrucomicrobiae, consistent with prior observations2 (Supplementary Figure 1c). The microbial communities 1-day-post-gavage in the dams were similar (Figure 1bi), but by 4-days-post-gavage, each group became distinct (Figure 1bii; Supplementary Table 1), remaining so through pregnancy, nursing, and post-weaning (Figure 1bi–v). Over time, the communities shifted in parallel toward the original inocula (seen along PC1 in PCoA analysis; Supplementary Figure 1b), but both the founding populations (along PC3) and host genotype (along PC2) significantly determined the bacterial community structure (Supplementary Figure 2; Supplementary Figure 3a; Supplementary Table 2). Thus, we show that both host genotype and the composition of the founding microbiota determine microbial community composition and structure over time in animals that have inherited their microbiota.
Effects of mouse passage on the community dynamics
For pups, the first fecal samples (age 3-weeks) showed close resemblance to their dams (Figure 1biii), indicating that gavage of pregnant germ-free dams is effective for introducing particular inocula to pups at birth. The four groups determined by inoculum and host genotype were distinct, remaining consistently so to week 21 (Figure 1biii–v, Supplementary Table 1).
Since reduced microbial diversity is associated with IBD24 and other illnesses, we examined the effects of inoculum, host genotype, and mouse generation on community richness (α-diversity). After gavage, α-diversity dropped markedly in all groups with partial recovery within 4 days, and with stabilization by 7 days (Supplementary Figure 3b–d; Supplementary Table 3). Although the STAT and Control inocula were similarly diverse, α-diversity became lower in those dams receiving the STAT than the Control inoculum, continuing throughout the experiment (Supplementary Figure 3b–d; Supplementary Table 3). These trends were due to rapid community succession, as a few opportunists (Lactococcus, Turicibacter, and Staphylococcus) bloomed immediately after transfer, then were outcompeted by the usual resident microbiota (Supplementary Figure 1c, Supplementary Discussion).
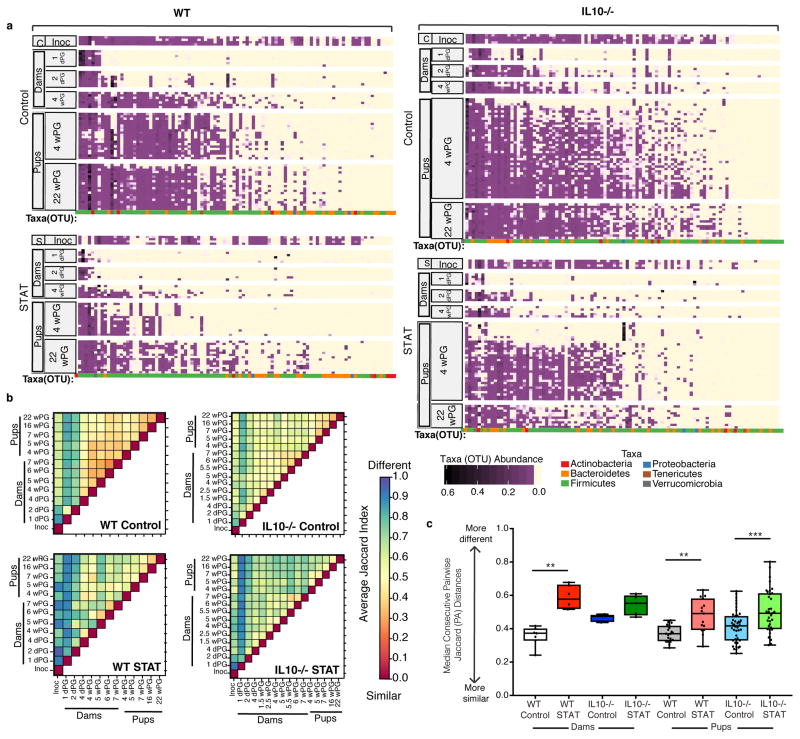
Next, we assessed the effects of the transfers on community stability. We found that OTU loss was greater in the intergenerational transfer from the dams that had received STAT microbiota than in those that received Control microbiota, and the response varied by genotype (Figure 2a, Supplementary Figure 4a, Supplementary Table 7). Mouse genotype played a role; transfer was more complete in IL10−/− mice (Figure 2a, Supplementary Figure 4a, Supplementary Figure 5, Supplementary Table 7, Supplementary Table 10, Supplementary Notes). We hypothesize this more complete transfer may be due to there being more extensive niches for colonization in a host with a defective immune system, although ultimately it is not clear why IL10−/− mice had richer microbial colonization from the inoculum. By Jaccard analysis of pairs of samples, there was instability in the dam microbiota for the first days post-gavage (highest Jaccard indices), mostly stabilizing by 4-days-post-gavage (Figure 2b). By 4-weeks-post-gavage, both dam and pup microbiota stabilized; yet the STAT pups had greater variation over time than Controls (Figure 2b–c, Supplementary Table 4). Thus, after immediate short-term community instability in the dams, the dams and pups were colonized with a mostly stable microbiota, highly resembling the inoculum. We show that the inheritance of a perturbed microbiota leads to increased instability of community composition in a later generation, with loss of richness.
Figure 2. Intergenerational microbiota transfer efficiency and stability over time.
(a) Heat map of relative abundance of OTUs from Control inoculum (C-Inoc), STAT inoculum (S-Inoc), and from dams and pups from all 4 groups of mice at indicated time points (dPG = days post-gavage, wPG = weeks post-gavage). Each row represents an individual mouse microbiota or in the case of the inoculum, a replicate sample. Each column represents an OTU. Phylum level taxonomic assignment of each OTU is indicated by color at the bottom of each column per mouse group. Rows and columns are sorted within each group by OTU prevalence and abundance. n=4 replicates for each inoculum. See Supplementary Table 1 for numbers of mice/group. (b) Average of pairwise Jaccard index values for each pair of samples for each group, as indicated. Pairwise Jaccard distances were generated between pup, dam and inoculum samples for each pup. Summarized heatmaps were created for each group by averaging individuals. (c) Median consecutive pairwise Jaccard distance of dam and pup samples calculated by comparing each animal’s sample to its previous sample, over time from the first time point (1 dPG for dams, 4 wPG for pups) to the last time point (7 wPG for dams, 22 wPG for pups). Boxplots show median values with interquartile range and bars ranging from minimum to maximum values, ** p<0.01, *** p<0.001 one-way ANOVA with Sidak’s multiple comparison test. See Supplementary Table 4 for all P values.
Specific taxa were nearly identically represented in the Control WT pups and their dams (Supplementary Figure 4b); however, for both inocula, Akkermansia was more abundant in dams than their pups (Supplementary Figure 4c, Supplementary Table 8). Populations in Control pups were more similar to their inoculum than were STAT pups, respectively, by both Jaccard Index (Supplementary Figure 4d, Supplementary Table 9), and as a percent of shared OTUs (Supplementary Figure 4a, Supplementary Table 7), indicative of higher transfer fidelity. In total, mouse genotype influences community structure after inoculation, with specific taxa differing in propensity for intergenerational transfer.
Effects of treatment and host genotype on the metagenome
To assess the differential selection associated with founding inoculum and host genotype, fecal samples from 3-week-old pups underwent shotgun sequencing to identify metagenomic effects. Analysis of Bray-Curtis distances of the metagenomic data showed significantly separated groups, except amongst the WT mice (Supplementary Figure 6a). Metagenomic analysis also permitted identification and fine scale resolution of specific taxa that were differentially present in the Control and STAT pups (Supplementary Figure 6b). There were many parallel changes in KEGG pathways in the IL10−/− compared to WT mice for the recipients of the STAT and Control microbiota (Supplementary Figure 6c); such conservation suggests genotype-specific selection for function. In particular, the pathways over-represented in WT STAT mice include ones involved with branched chain amino acids (BCAA), similar to an earlier observation25, whereas in the IL10−/− mice, many differences were identified that we have not seen reported elsewhere (Supplementary Figure 6d–e). This is potentially significant because BCAA have been associated with differential risk for auto-immune and metabolic diseases including type 1 diabetes as well as type 2 diabetes and obesity.26–29 The abundance of antibiotic-resistance genes was similar in the inocula from the Control and STAT mice (Supplementary Figure 6f). However, in the pups, abundance of resistance genes was lower in the recipients of the STAT microbiota than the Control microbiota, in both of the mouse backgrounds, and significant (p< 0.01) in the wild type mice. These findings suggest that without further antibiotic pressure, selection in STAT mice favored the growth of transferred organisms without resistance determinants (i.e. greater fitness in the absence of selection). While unexpected, there are several possible explanations for this finding (See Supplementary Discussion). The metagenomic analysis also revealed a significant enrichment of genes involved in the Sulfate Reduction I pathway in IL10−/− STAT pups, which has been associated with both Akkermansia muciniphila and colonic dysfunction (Supplementary Figure 7, Supplementary Notes).30,31 In total, both the maternal inocula and the host genotype led to differential metagenomic gene and pathway effects.
Effect of inoculum on characteristics of the pups
Finally, we examined the effects on host properties of receiving the transferred microbiota. Confirming prior reports32, bone mineral content and density were decreased in IL10−/− compared to WT mice (Supplementary Figure 8a–b, Supplementary Table 11). However, body composition was not affected by inoculum, differing from prior studies2,33, which might be related to dietary differences between the studies.
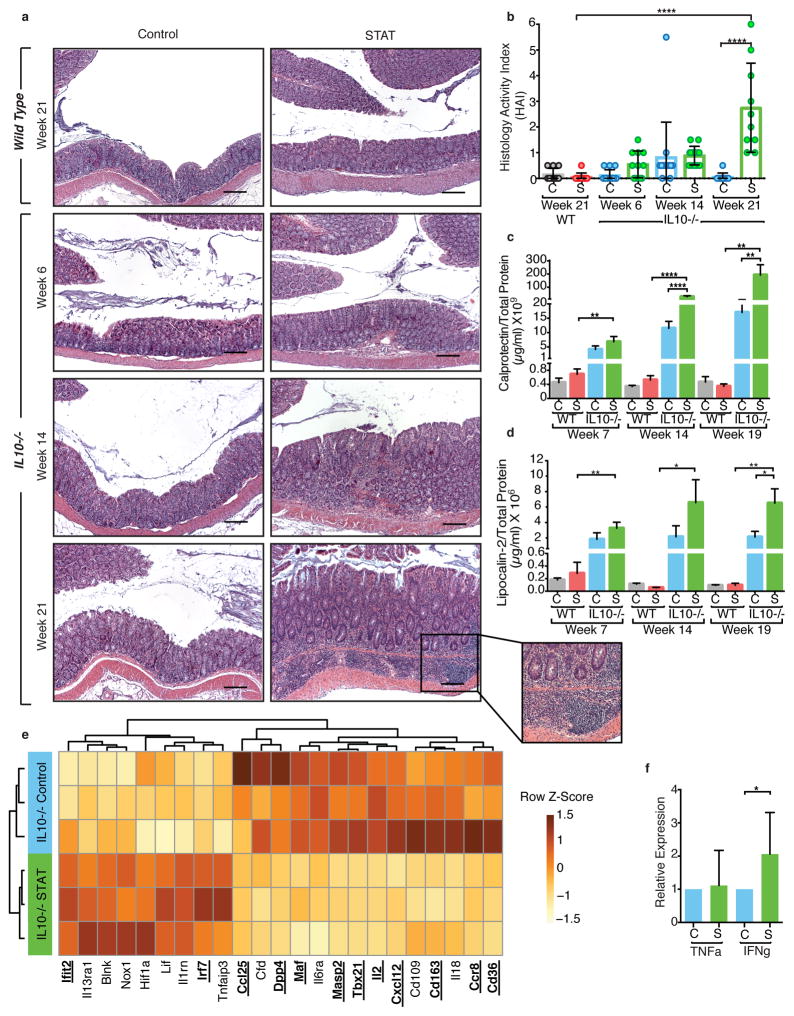
To determine the role of the antibiotic-altered microbiota on IBD development, we collected intestinal samples before the onset of severe weight loss, rectal prolapse, or bloody stool which often appear by week 26.34 The highest incidence of UC and CD occurs in 15–30 year-old young adults, but can occur at any age; 26-week old mice are adult.23 IL10−/− mice in the C57BL/6J background develop less aggressive IBD than in other genetic backgrounds,35 consistent with the low IBD histology activity index (HAI) we observed for the IL10−/− Control pups. However, even without apparent illness, the IL10−/− STAT pups had IBD lesions (colonic inflammation with lamina propria mononuclear cell infiltration, mucosal hyperplasia, dysplasia) at week 6 of life, with progressively increasing severity; as expected, the WT mice had no lesions (Figure 3a). In the 21-week-old IL10−/− mice, the mean IBD HAI was 55-fold higher (p<0.0001) in pups inheriting the STAT vs. Control microbiota (Figure 3b, Supplementary Table 5). Across all ages, based on HAI, the antibiotic-altered microbiota led to more severe inflammatory disease than the Control microbiota (Odds ratio 20.5 (CI 6.5 – 64.1) in IL10−/− pups) (Figure 3b, Supplementary Figure 9a, Supplementary Table 5). Using calprotectin and lipocalin-2, biomarkers of colonic inflammation in both human and murine IBD,36,37 temporal patterns of colitis development paralleled the observed histopathology (Figure 3c–d, Supplementary Table 5), with differences between the recipients of the two inocula most pronounced at week 21 (Figure 3a inset). We show that inheritance of an antibiotic-perturbed microbiota is not only faithfully recapitulated in the next generation, but that it confers added disease risk.
Figure 3. STAT microbiota in IL10−/− mouse increases colonic inflammation.
(a) Representative H&E-stained colon sections from WT (top panels, week 21) and IL10−/− mice (bottom 3 rows of panels, weeks 6, 14, and 21) colonized with Control microbiota (left) or STAT microbiota (right). Inset showing inflammation in IL10−/− STAT mice at week 21 at higher magnification on bottom right. Images are representative from a single experiment. Scale bar = 200 μm. (b) Mean histology activity index (HAI) +/− SD for WT and IL10−/− mice colonized with Control or STAT microbiota. ****p<0.0001, Kruskall-Wallis with Dunn’s post-test. (a–b) WT: n = 10 per group; IL10−/−: Control n = 13, 14, 10 and STAT n = 11, 13, 10 at weeks 6, 14, and 21 respectively. (c–d) ELISA results of calprotectin and lipocalin-2 normalized to total protein levels from fecal supernatants. WT: n = 9 per group; IL10−/−: Control n = 10, 11, 10 and STAT n = 9, 10, 9 at weeks 7, 14, and 19 respectively. Mean +/− SEM, * p<0.05, **p<0.01, ***p<0.001, ****p<0.0001 one way ANOVA with Sidak’s multiple comparison test. (e) Expression of colonic genes that were significantly altered by inoculum in IL10−/− pups (n = 3 each) at 21 weeks, measured by the Nanostring nCounter Mouse Immunology panel. Genes shown in bold and underlined have FDR-adjusted p value < 0.05, by t-test. Genes shown with normal font have FDR-adjusted p value between 0.05 and 0.10. (f) Relative expression of TNF-α and IFN-γ in colonic tissue from week 21 IL10−/− pups, measured by RT-qPCR (STAT TNF-α: n=12; all other groups: n=8), mean +/− SEM *p<0.05 t-test. (a–f) See Supplementary Tables 5–6 for detailed statistics.
In analyses using a panel of 547 mouse genes related to immunity and inflammation, we found that the IL10−/− mice showed increased colonic expression of Granzyme A, MHC Class II, Tnf, Ccl2, and Cxcl10 and decreased CD36, CD163, and IL15 compared to WT (Supplementary Figure 9b, Supplementary Table 12), consistent with prior studies38. Amongst the IL10−/− mice, the Control and STAT groups showed differential intestinal gene expression; 10 of the 12 significantly differential immune genes (Figure 3e, Supplementary Table 6) were decreased in the STAT pups, consistent with our prior studies.2 The IL10−/− STAT mice also showed elevated colonic IFN-γ expression (Figure 3f, Supplementary Table 5), a hallmark of IL10−/− colitis.19 These histologic, immunologic, and gene expression findings indicate that conventionalization of the IL10−/− mothers with the antibiotic-perturbed microbiota led to enhanced colitis in their offspring during adulthood.
Relationship between taxa and host phenotypes
We next used LEfSe and Random Forest modeling to determine which taxa could be important in the phenotypes we have characterized. Although these bioinformatic tools are insufficient to determine the significance of taxa without further experimental validation, they can identify candidates for further study.39 By LEfSe analyses, only two genera (Bacteroides and Odoribacter) more abundant in the STAT than the Control inoculum (Supplementary Figure 10ai) also were more abundant in the pre-inflammation pup samples. Five other taxa (Supplementary Figure 10aii) including Blautia producta, Staphylococcus, and Lachnospiraceae, were more abundant in the pre-colitis IL10−/− STAT pups, and might be considered as disease-promoting candidates.
Nine taxa were more abundant in the Control than the STAT inoculum (Supplementary Figure 10aiii); for seven (including Lactobacillus reuteri and Allobaculum), differences persisted in the pups. Nine other taxa had consistently increased abundance in the IL10−/− Control pups (including Bilophila, Roseburia, and Ruminococcus) (Supplementary Figure 10aiv). These 16 taxa, more abundant in Controls, might have disease-protecting roles.
Similarly, we used supervised learning to identify taxa predictive of high level HAI in the IL10−/− mice at week 21 (Supplementary Figure 10b). From samples obtained ≥5 weeks post-gavage, Random Forest modeling identified five OTUs (within Bilophila, Lachnospiraceae, Lactobacillus reuteri, Ruminococcus, and Bacteroidales S24-7) that highly predicted HAI scores. Four other OTUs (Rikenellaceae, Lactobacillus, Allobaculum, and Roseburia) were predictive mostly before weaning. Thus, while LEfSe and Random Forest modeling use different approaches, both analyses indicated that Bilophila, Lactobacillus reuteri, Roseburia, Ruminococcus, and Allobaculum were associated with protection against IBD onset, with the opposite for Odoribacter, Bacteroides, Lachnospiraceae, and Rikenellaceae.
Discussion
These results show that exposing pregnant germ-free mice to specified inocula allows the high fidelity transmission of the taxa to the dams over a period of weeks and to their pups over months. Fidelity was maintained despite housing the mice in a conventional facility in which mouse-adapted strains are circulating and which could invade the test mice. Nevertheless, it is clear that the control inoculum was better maintained in the recipients than was the STAT inoculum, which showed both loss of taxa and lower stability in Jaccard analyses. We also show that host genotype matters, with the IL10−/− (immunodeficient) mice more permissive for the transmission of the original inocula. A very recently published study, using a different study design in the IL10−/− model in which broad spectrum antibiotics were directly given to pregnant dams, provides evidence similar to our findings40, and the results of these studies together can be used to benchmark future studies of transfers to pregnant germ-free recipients.
By transferring microbes rather than treating mothers with antibiotics, we have shown in this model system that the antibiotic-perturbed microbiota, not the antibiotic itself, was responsible for the accelerated disease onset, measured during development and at sacrifice. We show that the biological effects in the next generation are exclusively due to the effects of antibiotic perturbation of the microbiota in a prior generation. These findings are consistent with associations between early-life antibiotic exposures and enhanced IBD risk in human children5, but move the exposure to the prior generation, mirroring allergy and asthma studies in Finnish children.15,41 Further, since the early-life microbiota may be critical for proper immunological development,42,43 perturbation may have important health consequences. We note that these studies are in a model system in mice with many differences from human biology, but the simplicity of the study design with only two variables—inoculum and genotype—with a single test diet and mouse strain background, and appropriate inoculum and genotype controls permits robust hypothesis testing. In further studies it will be important to test whether the accelerated colitis onset phenotype in mice with the STAT microbiota can be rescued with a microbiota transplant. Multiple variables will need to be considered in these studies including timing of the microbiota transplantation and the source and composition of the donor microbiota.Studies of present-day human children,44–46 and across hominid evolution,16 provide strong evidence of vertical transmission of intestinal taxa. Given the importance of founding bacterial populations that we show, the high-fidelity transmission that we observed becomes a useful tool for understanding inheritance in IBD and other inflammatory conditions. The extent of the inherited susceptibility to IBD that is due to human genes versus the intergenerational transfer of microbes (and their genes) should now be more closely examined. Our findings also are consistent with a multi-generational model of microbiome perturbation due to nutritional stress.18 As such, this model can be used to identify specific taxa as potentially disease-promoting or disease-protecting in future studies. Understanding these relationships is a biologically complex problem, involving multiple time points and multiple taxa. We have developed a first approximation of important taxa by identifying several in which the results of the analyses at different time points and across models have yielded consistent data. These might be considered to be especially strong candidates for playing causal roles. As noted in the results, these include Odoribacter, Bacteroides, Lachnospiraceae, Blautia, and Rikenellaceae species. These taxa have been identified in prior studies in models of IBD development and in human subjects.40,47,48 The metagenomic studies also point to a potentially critical injurious role of one taxon Akkermansia mucinophila, and an important metabolic pathway, leading to H2S production. However, it may be unlikely that any single taxon will play a crucial role. It is also possible that the major factor in the development of disease is the loss of protective organisms, such as Roseburia, Ruminococcus, and Bilophila, a role for which also is supported by the literature.49,50 Of course, an important caveat of human IBD studies is they typically examine patients with existing IBD and their respective controls. In those studies, it is not possible to determine causal relationships since differences observed could be causal, an effect of the disease, and/or an effect of the treatments. The taxa that we identified as differential in our studies must be considered only as preliminary candidates, whose importance must be tested in definitive experiments.
In total, these findings provide evidence that the founding inoculum, host genotype, and intergenerational transfer all may affect intestinal microbial community structure and composition in interacting ways. That antibiotic impact on the gut microbiota could potentially affect disease susceptibility for more than one generation is possibly relevant to the large proportion of women receiving antibiotics before and during pregnancy.11,12
Methods
Animals
All animal experiments were conducted according to protocols approved by the New York University School of Medicine Institutional Animal Care and Use Committee (IACUC). After mice were received from the National Gnotobiotic Rodent Resource Center (NGRRC), University of North Carolina, Chapel Hill, they were maintained on irradiated high fat diet (HFD, Rodent Diet D12451i, Research Diets, 45% kcal%).
Microbiota Inoculum Preparation
Wild type C57BL/6 mice were raised to 18 weeks of age with sub-therapeutic levels of penicillin in their drinking water (STAT) or not (Control). Cecal contents were collected from 3 Control and 3 STAT penicillin female mice that were fed a high fat diet (HFD, Rodent Diet D12451i, Research Diets, 45% kcal%), as has been described.2 Following euthanasia, 1 set of flame-sterilized instruments were used to open the visceral wall, and a second set of flame-sterilized instruments were used to collect 1/3 of the cecal content from each mouse which were immediately placed in 1 mL pre-reduced liquid dental anaerobic transport medium (Anaerobe Systems, Morgan Hill, CA). Contents were homogenized in an anaerobic environment, tissue removed, pooled according to experimental group (total 3 ml) and diluted with 5 mL of pre-reduced anaerobically sterilized saline (Anaerobe Systems, Morgan Hill, CA). This inoculum was stored in 1 mL aliquots at −80°C until use.
We used a pool rather than individual mice because we could only realistically perform one transfer, since the experiment involved 4 groups of mice (2 genotypes, 2 inocula) representing 16 litters and 135 mice studied in all (and 1137 samples). Also, importantly, the intestinal microbiota shows individual variation, even in genetically identical mice that are housed in the same cage. In order to generate a representative microbiota from our control and antibiotic-treated mice, we selected the three mice with the weights closest to the median.
Microbiota Transfer into Pregnant Germ-Free Mice
Pregnant germ-free adult mice ≥8 weeks of age (wild-type C57BL/6J and IL10−/− C57BL/6J), received from the NGRRC ~14 days after mating plugs were detected, acclimated to the NYU animal facility overnight in their sterilized shipping containers. Control and STAT murine inocula, which had been frozen at −80°C2, were allowed to thaw, and the mice were briefly restrained while 250 μl of Control or STAT inoculum was placed into their stomach by oral gavage. Recipient mice were chosen randomly and inoculations alternated between Control and STAT with glove changes between each. Immediately after gavage, a fecal sample was collected from each mouse. There was no amplification of the 16S rRNA gene in any of these samples, confirming their germ-free status. Mice were then housed individually in autoclaved cages. We aimed to have 5 litters per group to produce at least 15 mice per group for statistical significance. All mice were maintained on a 45% high fat diet (HFD, Research Diets D12451i) as the donor mice were receiving this diet before they were sacrificed to create the cecal inocula.2 Dams gave birth 6.8 ± 2.3 days after gavage. Five IL10−/− dams were gavaged with each inoculum and all 10 litters survived. Of 12 WT dams gavaged, in six cases, the mothers ate their pups shortly after birth (3/7 given the Control inoculum, and 3/5 receiving the STAT inoculum). Fecal pellets and scale weights were collected daily from the dams for the first week post-gavage.
Maintenance of the Pups
The dams were left undisturbed after giving birth until their pups were 21 days old, except for the IL10−/− dams, which were weighed and fecal pellets obtained on days 4 and 11 post-partum. After the pups reached 21 days, dams and pups were weighed and fecal pellets obtained 1–5 times per week. After weaning, pups were housed 4 per cage with other pups from the same inoculum and genotype group (WT Control with WT Control, WT STAT with WT STAT, etc). Both sexes were studied. Cage assignments were random within each inoculum and genotype group to minimize litter/cage differences on the microbiome. Dams were sacrificed 2–3 weeks after pups were weaned. All 35 WT pups were sacrificed at 21 weeks of age. Subgroups of IL10−/− pups were sacrificed at 6 (n=24), 14 (n=27), and 21 (n=26) weeks of age.
Histopathology
At sacrifice, tissues were collected as described,51 fixed with 10% neutral-buffered formalin, routinely processed, and paraffin-embedded. Colon samples were sectioned and stained with hemotoxylin and eosin (H&E), coded, and scored by a board-certified veterinary pathologist (A.B.R.) who was blinded to experimental status. These samples were scored according to a previously established histopathologic grading scheme for experimental colitis51 with scores of 0–4 given for each of three categories – inflammation, hyperplasia, and dysplasia. A total histology activity index (HAI) was calculated by combining these three separate values. Sections from 10–14 mice were scored in each group, means were calculated and tested for statistical significance using Kruskall-Wallis with Dunn’s post-test.
Odds Ratio Calculation
The severity score of the disease status of the gut of the mice was scored on a scale of 0–6 in 11 response categories (Supplementary Figure 9a). A Proportional Odds model52 was used to fit a common-slopes cumulative model, which is a parallel lines regression model based on the cumulative probabilities of the response categories rather than on their individual probabilities. The cumulative model has the form: g(Pr(Y ≤ I | x)) = αi + β’x, i=k,…,k where g is the logit link function, αi,…, αk are k intercept parameters, and β is the vector of slope parameters. Based on this model, treatment effects can be represented as the Odds Ratio (OR) of going to the next level in the disease score of treatment versus Control.
Microbial Community Analysis
DNA was extracted from frozen fecal, cecal, and ileal samples using the MO BIO PowerSoil DNA Isolation Kit (MO BIO, Carlsbad, CA) in the 96-well format. The 16S rRNA gene was amplified with barcoded fusion primers targeting the V4 region, as described.53 Amplicon pools were sequenced on the Illumina Miseq platform with 150 bp paired-end reads. The QIIME pipeline54 was used for downstream analysis.
Bioinformatic Analysis
Paired end reads were joined using fastq-join from EA-utils55 with a minimum overlap of 30 base pairs. Only reads that were perfectly matched in this overlapping region were kept. Sequences were demultiplexed and quality filtered with QIIME. If a read had more than 3 consecutive low quality bases (phred q score ≤ 20) it would be truncated, and any read that ended up being <75% of its original length was discarded. Sequences were assigned taxonomy with the open reference method in QIIME using UCLUST56 and the Green Genes 2013 May database release as a reference.57 Samples with less than 5000 reads were removed from the dataset. This dataset was then used to generate relative abundance plots, calculate alpha diversity, and measure UniFrac distances to generate PCoA plots. Linear discriminant analysis effect size (LEfSe)39 was used to detect significantly differential taxa between various pairings of groups. To reduce the number of features the dataset was filtered to exclude any taxa that accounted for 0.01% or less of the total observation count. The average sequencing depth for this study was 15,564. As such, 0.01% abundance represents only about 1–2 reads/sample. Although there are low abundance taxa that could be relevant, they were considered to be below the detection threshold of the current study, since single reads may represent sequencing artifacts. Significance thresholds were retained as the default LEfSe settings.
Whole genome sequencing
Fecal samples from 4 female pups from each group at 3 weeks of life were chosen for metagenomic study, along with each inoculum. Extracted genomic DNA (2 ng) from each sample was used for library preparation and subsequent WGS using the Illumina HiSeq 2500 platform. Samples were sequenced over 4 flow cell lanes as 100-bp paired-end reads. The metagenomes were pre-processed for quality metrics by removing nucleotides with a Phred score < 20 and reads shorter than 60bp using Trimmomatic.58 Next sequences were aligned to the mouse (mm10) genome to reduce host-contaminated sequences using KneadData. After filtering low quality and contaminated sequences, we performed functional profiling to detect microbial genes and pathways using HUMAnN2 v0.9.5,59 with default settings and screened against the EC-filtered UniRef90 database. All metagenomes have been deposited in the European Nucleotide Archive (ENA) and are available at the European Bioinformatics Institute (EBI), under the project ID PRJEB18627.
Intergenerational transfer heatmap
Relative abundances of filtered OTUs were plotted in a heatmap grouped by mouse genotype and treatment over selected timepoints. Heatmap was generated using custom R scripts and the following packages: biom, phyloseq, qiimer.60–62
Jaccard Index Analyses
Pairwise Jaccard distances were generated between each pair of samples for each pup, dam and inoculum sample, using the vegan R-package63 and the parameter binary = T. Plots were created using the levelplot function found within the lattice R-package. Summarized heat maps were generated using the median distance across all sample groups for each group. Boxplots were generated using the median consecutive pairwise Jaccard distance, by comparing each pup sample to its previous sample, over time. For each mouse, the median Jaccard values were calculated by comparing between dam, pup and inoculum groups. The distribution of values was assessed by the pairwise Wilcoxon test with Holm correction for statistical analysis.
Supervised classification of histopathology results
Random forest classification models were built to predict high (1+) vs. low (<1) HAI histology scores (class) as a function of the microbial community OTU relative abundances (features), for each examined week of pup life, as described.33 Models were built by growing 1,000 trees per forest and model error was calculated using a 10-fold cross validation approach. All samples were randomly subsampled at 5000 OTU/sample prior to analysis to avoid bias from uneven sampling efforts. Subsampling and analysis was performed in 10 independent trials that were then used to calculate mean model error and OTU importance. At all time points, the model had substantially better than random (error ratio >2) predictive power. The same strategy was used to predict inoculum (class) as a function of microbial community (features).
Quantitation of fecal lipocalin-2 and calprotectin by ELISA
Frozen fecal samples were vortexed with PBS to obtain a homogenous suspension. Samples were then centrifuged for 10 min at 9,500 g, at 4°C. Supernatants were collected and stored at −80°C until analysis. Lipocalin-2 levels were estimated in the supernatants using Duoset murine lipocalin-2 ELISA kit (R&D Systems, Minneapolis, MN). Calprotectin levels were estimated in the supernatants using the S100A9/Calprotectin mouse ELISA kit (Hycult Biotech, Plymouth Meeting, PA).37 Both were standardized to total protein, as determined using measured with the Pierce BCA Protein Assay Kit (Thermo Fisher Scientific, Waltham, MA).
Gene Expression Studies
Colon, cecum and small intestine tissues were obtained at sacrifice, rinsed, and stored in RNA later for 24 hours at 4°C. Tissues were then placed in a new tube and stored at −80 until RNA could be extracted using the RNeasy Mini Kit (QIAGEN, Valencia, CA).
Nanostring
Colonic expression of a panel of immune genes was measured in 21-week-old WT and IL10−/− mice whose mothers had received either the STAT or Control inoculum (4 groups, n=3/group). Assays were performed using the nCounter Gene Expression Mouse Immunology Kit (NanoString Technologies, Seattle WA).
QPCR
RNA was reverse transcribed to cDNA with the Verso cDNA Synthesis kit (Thermo Scientific). QPCR was performed on a LightCycler 480 ii (Roche, Indianapolis, IN) using LightCycler 480 SYBR Green I Master. Specific mouse oligonucleotides were used to measure IFN-γ (5′-CGGCACAGTCATTGAAAGCC-3′ and 5′-TGTCACCATCCTTTTGCCAGT-3′) and TNF-α (5′-GTTCTGCAAAGGGAGAGTGG-3′ and 5′-GCACCTCAGGGAAGAGTCT-3′). Relative expression of these genes was calibrated to the 18S rRNA endogenous Control gene (5′-CCATCCAATCGGTAGTAGCG-3′ and 5′-GTAACCCGTTGAACCCCATT-3′) and analyzed with the comparative CT (ΔΔCT) method to calculate the fold-differences in expression between Control and STAT IL10−/− pups.
Body Composition Measurements
Body composition was assessed at ~4 week intervals beginning when the pups were weaned at 4 weeks of age. Body composition was measured by dual energy x-ray absorptiometry (DEXA) with a Lunar PIXImus II mouse densitometer (GE Medical Systems, Waukesha WI) with anesthesia by isofluorane inhalation.
Statistical Analyses
Values were expressed as mean ± SEM or SD, as noted in figure legends. Analyses were performed using GraphPad Prism (GraphPad Software Inc., Ca, USA). A one-way ANOVA with Sidak’s multiple comparison test or unpaired two-tailed students t-test was used to analyze parametric data while nonparametric data were analyzed with a Mann-Whitney U test.
Data Availability
All 16S rRNA sequencing data have been deposited at QIITA [ID 11113 (https://qiita.ucsd.edu/study/description/11113)] and EBI (accession number ERP104982). NanoString gene expression data have been deposited in NCBI’s Gene Expression Omnibus and are accessible through GEO Series accession number GSE92623 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE92623). Whole metagenome shotgun sequencing data have been deposited in the European Nucleotide Archive (ENA) under the project PRJEB18627 (http://www.ebi.ac.uk/ena/data/view/PRJEB18627).
Supplementary Material
Acknowledgments
We acknowledge Maureen Bower and the National Gnotobiotic Rodent Resource Center, University of North Carolina, Chapel Hill for supplying us with mice, NYUMC Genome Technology Center for help with sequencing (partially supported by the Cancer Center Support Grant, P30CA016087, at the Laura and Isaac Perlmutter Cancer Center) and the NYUMC Histology Core for assistance preparing samples for histology. These studies were supported by NIH grants DK090989, OD010995, DK034987 and the Crohn’s and Colitis Foundation of America, and the Ziff Family, Knapp Family, and C & D funds, and the Diane Belfer Program for Human Microbial Ecology.
Footnotes
Author Contributions
A.F.S., R.B.S., and M.J.B designed experiments and interpreted data. A.F.S. performed experiments and participated in analysis of the data. Y.A. and V.E.R. contributed to interpretation of immunologic data. Y.A. and M.H. performed protein expression assays. T.B. advised on microbiome analytical methods and performed data analyses. L.B. performed odds ratio calculations and other statistical tests. S.R, T.W, and D.K. contributed to microbiota stability analysis. A.B.R. performed histological analyses. L.M.C. and R.B.S. provided essential reagents and procedural advice. A.F.S. and M.J.B. were responsible for writing this manuscript, as reviewed and edited by all authors.
Competing Interests Statement
The authors declare no competing financial interests.
References
- 1.Jernberg C, Löfmark S, Edlund C, Jansson JK. Long-term ecological impacts of antibiotic administration on the human intestinal microbiota. ISME J. 2007;1:56–66. doi: 10.1038/ismej.2007.3. [DOI] [PubMed] [Google Scholar]
- 2.Cox LM, et al. Altering the intestinal microbiota during a critical developmental window has lasting metabolic consequences. Cell. 2014;158:705–721. doi: 10.1016/j.cell.2014.05.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Azad MB, Bridgman SL, Becker AB, Kozyrskyj AL. Infant antibiotic exposure and the development of childhood overweight and central adiposity. Int J Obes. 2014;38:1290–1298. doi: 10.1038/ijo.2014.119. [DOI] [PubMed] [Google Scholar]
- 4.Arrieta M-C, et al. Early infancy microbial and metabolic alterations affect risk of childhood asthma. Sci Transl Med. 2015;7:307ra152–307ra152. doi: 10.1126/scitranslmed.aab2271. [DOI] [PubMed] [Google Scholar]
- 5.Hviid A, Svanstrom H, Frisch M. Antibiotic use and inflammatory bowel diseases in childhood. Gut. 2011;60:49–54. doi: 10.1136/gut.2010.219683. [DOI] [PubMed] [Google Scholar]
- 6.Shaw SY, Blanchard JF, Bernstein CN. Association Between the Use of Antibiotics and New Diagnoses of Crohn’s Disease and Ulcerative Colitis. Am J Gastroenterol. 2011;106:2133–2142. doi: 10.1038/ajg.2011.304. [DOI] [PubMed] [Google Scholar]
- 7.Ng SC, et al. Geographical variability and environmental risk factors in inflammatory bowel disease. Gut. 2013;62:630–649. doi: 10.1136/gutjnl-2012-303661. [DOI] [PubMed] [Google Scholar]
- 8.Kronman MP, Zaoutis TE, Haynes K, Feng R, Coffin SE. Antibiotic Exposure and IBD Development Among Children: A Population-Based Cohort Study. Pediatrics. 2012;130:e794–e803. doi: 10.1542/peds.2011-3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Van Boeckel TP, et al. Global antibiotic consumption 2000 to 2010: An analysis of national pharmaceutical sales data. Lancet Infect Dis. 2014;14:742–750. doi: 10.1016/S1473-3099(14)70780-7. [DOI] [PubMed] [Google Scholar]
- 10.Hicks LA, et al. US outpatient antibiotic prescribing variation according to geography, patient population, and provider specialty in 2011. Clin Infect Dis. 2015;60:1308–1316. doi: 10.1093/cid/civ076. [DOI] [PubMed] [Google Scholar]
- 11.Stokholm J, et al. Prevalence and predictors of antibiotic administration during pregnancy and birth. PLoS One. 2013;8:1–7. doi: 10.1371/journal.pone.0082932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Andrade SE, et al. Prescription drug use in pregnancy. Am J Obstet Gynecol. 2004;191:398–407. doi: 10.1016/j.ajog.2004.04.025. [DOI] [PubMed] [Google Scholar]
- 13.Lacroix I, et al. Prescription of drugs during pregnancy: A study using EFEMERIS, the new French database. Eur J Clin Pharmacol. 2009;65:839–846. doi: 10.1007/s00228-009-0647-2. [DOI] [PubMed] [Google Scholar]
- 14.Petersen I, Gilbert R, Evans S, Ridolfi A, Nazareth I. Oral antibiotic prescribing during pregnancy in primary care: UK population-based study. J Antimicrob Chemother. 2010;65:2238–2246. doi: 10.1093/jac/dkq307. [DOI] [PubMed] [Google Scholar]
- 15.Metsälä J, et al. Mother’s and Offspring’s Use of Antibiotics and Infant Allergy to Cow’s Milk. Epidemiology. 2013;24:303–309. doi: 10.1097/EDE.0b013e31827f520f. [DOI] [PubMed] [Google Scholar]
- 16.Moeller AH, et al. Cospeciation of gut microbiota. Science (80−) 2016;353:380–382. doi: 10.1126/science.aaf3951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goodrich JK, et al. Human genetics shape the gut microbiome. Cell. 2014;159:789–799. doi: 10.1016/j.cell.2014.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sonnenburg ED, et al. Diet-induced extinctions in the gut microbiota compound over generations. Nature. 2016;529:212–215. doi: 10.1038/nature16504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kühn R, Löhler J, Rennick D, Rajewsky K, Müller W. Interleukin-10-deficient mice develop chronic enterocolitis. Cell. 1993;75:263–274. doi: 10.1016/0092-8674(93)80068-p. [DOI] [PubMed] [Google Scholar]
- 20.Sellon RK, et al. Resident enteric bacteria are necessary for development of spontaneous colitis and immune system activation in interleukin-10-deficient mice. Infect Immun. 1998;66:5224–5231. doi: 10.1128/iai.66.11.5224-5231.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Krause P, et al. IL-10-producing intestinal macrophages prevent excessive antibacterial innate immunity by limiting IL-23 synthesis. Nat Commun. 2015;6:7055. doi: 10.1038/ncomms8055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sartor RB, Wu GD. Roles for Intestinal Bacteria, Viruses, and Fungi in Pathogenesis of Inflammatory Bowel Diseases and Therapeutic Approaches. Gastroenterology. 2017;152:327–339.e4. doi: 10.1053/j.gastro.2016.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jones-Hall YL, Grisham MB. Immunopathological characterization of selected mouse models of inflammatory bowel disease: Comparison to human disease. Pathophysiology. 2014;21:267–288. doi: 10.1016/j.pathophys.2014.05.002. [DOI] [PubMed] [Google Scholar]
- 24.Manichanh C. Reduced diversity of faecal microbiota in Crohn’s disease revealed by a metagenomic approach. Gut. 2006;55:205–211. doi: 10.1136/gut.2005.073817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Livanos AE, et al. Antibiotic-mediated gut microbiome perturbation accelerates development of type 1 diabetes in mice. Nat Microbiol. 2016;1:16140. doi: 10.1038/nmicrobiol.2016.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pedersen HK, et al. Human gut microbes impact host serum metabolome and insulin sensitivity. Nature. 2016;535:376–381. doi: 10.1038/nature18646. [DOI] [PubMed] [Google Scholar]
- 27.Orešič M, et al. Dysregulation of lipid and amino acid metabolism precedes islet autoimmunity in children who later progress to type 1 diabetes. J Exp Med. 2008;205:2975–2984. doi: 10.1084/jem.20081800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nagata C, et al. Branched-chain Amino Acid Intake and the Risk of Diabetes in a Japanese Community: The Takayama Study. Am J Epidemiol. 2013;178:1226–1232. doi: 10.1093/aje/kwt112. [DOI] [PubMed] [Google Scholar]
- 29.Lynch CJ, Adams SH. Branched-chain amino acids in metabolic signalling and insulin resistance. Nat Rev Endocrinol. 2014;10:723–736. doi: 10.1038/nrendo.2014.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Attene-Ramos MS, et al. DNA damage and toxicogenomic analyses of hydrogen sulfide in human intestinal epithelial FHs 74 Int cells. Environ Mol Mutagen. 2010;51:304–314. doi: 10.1002/em.20546. [DOI] [PubMed] [Google Scholar]
- 31.van Passel MWJ, et al. The genome of Akkermansia muciniphila, a dedicated intestinal mucin degrader, and its use in exploring intestinal metagenomes. PLoS One. 2011;6 doi: 10.1371/journal.pone.0016876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cohen SL, Moore AM, Ward WE. Interleukin-10 knockout mouse: a model for studying bone metabolism during intestinal inflammation. Inflamm Bowel Dis. 2004;10:557–563. doi: 10.1097/00054725-200409000-00009. [DOI] [PubMed] [Google Scholar]
- 33.Mahana D, et al. Antibiotic perturbation of the murine gut microbiome enhances the adiposity, insulin resistance, and liver disease associated with high-fat diet. Genome Med. 2016;8:48. doi: 10.1186/s13073-016-0297-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Spencer DM, Veldman GM, Banerjee S, Willis J, Levine AD. Distinct inflammatory mechanisms mediate early versus late colitis in mice. Gastroenterology. 2002;122:94–105. doi: 10.1053/gast.2002.30308. [DOI] [PubMed] [Google Scholar]
- 35.Berg DJ, et al. Enterocolitis and colon cancer in interleukin-10-deficient mice are associated with aberrant cytokine production and CD4+ Th1-like responses. J Clin Invest. 1996;98:1010–1020. doi: 10.1172/JCI118861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lehmann FS, Burri E, Beglinger C. The role and utility of faecal markers in inflammatory bowel disease. Therap Adv Gastroenterol. 2015;8:23–36. doi: 10.1177/1756283X14553384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chassaing B, et al. Fecal Lipocalin 2, a Sensitive and Broadly Dynamic Non-Invasive Biomarker for Intestinal Inflammation. PLoS One. 2012;7:3–10. doi: 10.1371/journal.pone.0044328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hansen JJ, Holt L, Sartor RB. Gene expression patterns in experimental colitis in IL-10-deficient mice. Inflamm Bowel Dis. 2009;15:890–899. doi: 10.1002/ibd.20850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Segata N, et al. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12:R60. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miyoshi J, et al. Peripartum Antibiotics Promote Gut Dysbiosis, Loss of Immune Tolerance, and Inflammatory Bowel Disease in Genetically Prone Offspring. Cell Rep. 2017;20:491–504. doi: 10.1016/j.celrep.2017.06.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Metsälä J, et al. Prenatal and post-natal exposure to antibiotics and risk of asthma in childhood. Clin Exp Allergy. 2015;45:137–145. doi: 10.1111/cea.12356. [DOI] [PubMed] [Google Scholar]
- 42.Gensollen T, Iyer SS, Kasper DL, Blumberg RS. How colonization by microbiota in early life shapes the immune system. Science (80−) 2016;352:539–544. doi: 10.1126/science.aad9378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gomez de Aguero M, et al. The maternal microbiota drives early postnatal innate immune development. Science (80−) 2016;351:1296–1302. doi: 10.1126/science.aad2571. [DOI] [PubMed] [Google Scholar]
- 44.Bokulich NA, et al. Antibiotics, birth mode, and diet shape microbiome maturation during early life. Sci Transl Med. 2016;8:343ra82–343ra82. doi: 10.1126/scitranslmed.aad7121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Asnicar F, et al. Studying Vertical Microbiome Transmission from Mothers to Infants by Strain-Level Metagenomic Profiling. mSystems. 2017;2:e00164–16. doi: 10.1128/mSystems.00164-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nayfach S, Rodriguez-Mueller B, Garud N, Pollard KS. An integrated metagenomics pipeline for strain profiling reveals novel patterns of bacterial transmission and biogeography. Genome Res. 2016;26:1612–1625. doi: 10.1101/gr.201863.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Palm NW, et al. Immunoglobulin A coating identifies colitogenic bacteria in inflammatory bowel disease. Cell. 2014;158:1000–1010. doi: 10.1016/j.cell.2014.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pascal V, et al. A microbial signature for Crohn’s disease. Gut. 2017;66:813–822. doi: 10.1136/gutjnl-2016-313235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Halfvarson J, et al. Dynamics of the human gut microbiome in inflammatory bowel disease. 2017;2:17004. doi: 10.1038/nmicrobiol.2017.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gevers D, et al. The treatment-naive microbiome in new-onset Crohn’s disease. Cell Host Microbe. 2014;15:382–392. doi: 10.1016/j.chom.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
References for Methods
- 51.Rogers AB, Houghton J. Helicobacter -Based Mouse Models of Digestive System Carcinogenesis. 2009:267–295. doi: 10.1007/978-1-59745-447-6_11. [DOI] [PubMed] [Google Scholar]
- 52.Cox D, Snell EJ. Analysis of Binary Data. Chapman and Hall/CRC press; 1989. [Google Scholar]
- 53.Caporaso JG, et al. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci. 2011;108:4516–4522. doi: 10.1073/pnas.1000080107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Caporaso JG, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Publ Gr. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Aronesty E. Comparison of Sequencing Utility Programs. Open Bioinforma J. 2013;7:1–8. [Google Scholar]
- 56.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 57.McDonald D, et al. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 2012;6:610–618. doi: 10.1038/ismej.2011.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bolger AM, Lohse M, Usadel B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Abubucker S, et al. Metabolic reconstruction for metagenomic data and its application to the human microbiome. PLoS Comput Biol. 2012;8 doi: 10.1371/journal.pcbi.1002358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bittinger K. qiimer: Work with QIIME Output Files in R. 2015. [Google Scholar]
- 61.McMurdie PJ. biom: An interface package for the BIOM file format. 2014. [Google Scholar]
- 62.McMurdie PJ, Holmes S. Phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS One. 2013;8 doi: 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dixon P. VEGAN, a package of R functions for community ecology. J Veg Sci. 2003;14:927–930. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All 16S rRNA sequencing data have been deposited at QIITA [ID 11113 (https://qiita.ucsd.edu/study/description/11113)] and EBI (accession number ERP104982). NanoString gene expression data have been deposited in NCBI’s Gene Expression Omnibus and are accessible through GEO Series accession number GSE92623 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE92623). Whole metagenome shotgun sequencing data have been deposited in the European Nucleotide Archive (ENA) under the project PRJEB18627 (http://www.ebi.ac.uk/ena/data/view/PRJEB18627).