Abstract
Over the past sixty years, researchers have made outmost efforts to clarify the structural organization and functional regulation of the complexes that configure the mitochondrial respiratory chain. As a result, the entire composition of each individual complex is practically known and, aided by notable structural advances in mammals, it is now widely accepted that these complexes stablish interactions to form higher-order supramolecular structures called supercomplexes and respirasomes. The mechanistic models and players that regulate the function and biogenesis of such superstructures are still under intense debate, and represent one of the hottest topics of the mitochondrial research field at present. Noteworthy, understanding the pathways involved in the assembly and organization of respiratory chain complexes and supercomplexes is of high biomedical relevance because molecular alterations in these pathways frequently result in severe mitochondrial disorders. The purpose of this review is to update the structural, biogenetic and functional knowledge about the respiratory chain supercomplexes and assembly factors involved in their formation, with special emphasis on their implications in mitochondrial disease. Thanks to the integrated data resulting from recent structural, biochemical and genetic approaches in diverse biological systems, the regulation of the respiratory chain function arises at multiple levels of complexity.
Keywords: Respiratory Chain Function, Supercomplexes, Respirasome, Assembly Factors
Introduction
Recent investigations have shed light into the sophistication surrounding the biogenesis of the oxidative phosphorylation (OXPHOS) system. In mammals, the OXPHOS system is formed by five multiprotein enzyme complexes and two mobile electron carriers embedded in the inner mitochondrial membrane. The first four enzyme complexes (CI–CIV) comprise the mitochondrial respiratory chain (MRC), which facilitates electron transfer from reducing equivalents to molecular oxygen coupled to the generation of a proton gradient across the inner membrane that will be used by the ATP synthase (complex V) to drive ATP synthesis. In addition to their structural subunits, the biogenesis of these five hetero-oligomeric enzymatic complexes requires an extensive number of ancillary factors to coordinate subunits maturation, incorporation of prosthetic groups and assembly into the holoenzymes. The discovery that some MRC enzymes physically interact to form a variety of supramolecular structures called supercomplexes (SCs) and respirasomes, and the existence of SC-specific assembly factors involved in their assembly, has put the spotlight on the structural and functional properties of the SCs, and on the regulatory pathways involved in their biogenesis. It is currently debated whether SCs play a relevant functional role in cellular bioenergetics, in the formation of reactive intermediates, or in the stabilization of the individual MRC complexes. Since genetic alterations in MRC subunits and assembly factors often lead to severe encephalomyopathies and neurodegenerative disorders, a full understanding of the structural organization and biosynthetic regulation of the MRC is essential to understand the molecular mechanisms underlying mitochondrial pathology. In this review we will explore the current knowledge on mammalian respiratory SCs, integrating the historical perspective with the most recent structural findings, and putting this information in the context of mitochondrial disease.
1. Mitochondrial respiratory chain, a long-lasting journey
1.1. Composition and function of the respiratory chain
The production of adenosine 5’-triphosphate (ATP), the key energy source of the cell, through aerobic substrate oxidation is the main function of the mitochondrial metabolism. In the late 50s, several redox enzymes and prosthetic groups responsible for the classic mitochondrial electron transfer chain were defined [1] followed by their reconstitution in the early 60s [2]. The overall respiratory chain activity was postulated as a sequential transfer of electrons between four major multi-enzymatic complexes dispersed in the inner mitochondrial membrane (IMM): NADH dehydrogenase:ubiquinone oxidoreductase (complex I, CI), succinate:ubiquinone oxidoreductase (complex II, CII), ubiquinol:cytochrome c oxidoreductase or cytochrome bc1 complex (complex III, CIII), and cytochrome c oxidase (complex IV, CIV). In addition, the electron transfer was ensured by the diffusion of two mobile components acting as co-substrates: the lipophilic ubiquinone, also designated as coenzyme Q (CoQ), embedded in the membrane lipid bilayer, and the hydrophilic heme protein cytochrome c (cytc) located on the external surface of the IMM [3,4]. Altogether, these components form the mitochondrial respiratory chain (MRC) where cellular respiration takes place. Organic nutrients are catabolized into small electron donor molecules, NADH2 and FADH2, which transfer the electrons to CI and CII, respectively. CoQ uptakes the electrons from both sources, transferring them to dimeric CIII (CIII2), then to cytc and finally to CIV, that yields the electrons to molecular oxygen. This electron flux is coupled to a proton pump from the matrix to the intermembrane space through complexes I, III and IV, which generates an electrochemical gradient across the IMM that provides the necessary free energy for the ATP synthase (complex V, CV) to synthesize ATP through the mechanism known as oxidative phosphorylation [5].
1.2. Models for the structural organization of the respiratory chain
Despite the well-known functional relevance of the respiratory chain, the structural organization of its components remains unclear.
1.2.1. Solid-state Model
The pioneering spectrophotometric studies of Chance and Williams represented the MRC as a solid state assembly of prosthetic groups that carry out sequential redox reactions in a protein matrix [1]. The evidences in favour of this “rigid” or “solid-state model” were based on the isolation of CI–CIII and CII–CIII active units in a stoichiometric molar ratio of 1:1 during intermediate purification steps of the individual enzymes, that were interpreted as secondary enzymatically active complexes [6]; and on the reconstitution of a “repeating unit of electron transfer” containing all MRC complexes from bovine heart mitochondria [7]. This model implied the notion of permanently-bound CoQ to the MRC units. Accordingly, vesicle reconstitution experiments showed stoichiometric associations between CI and CIII2 at high protein concentrations with no exchange between free and bound ubiquinone, i.e., no 'CoQ-pool' behaviour [8]. Only when sufficient lipid was added the CoQ-pool behaviour was restored, but this was interpreted as the movement of complex-associated CoQ rather than of free CoQ [9]. Later studies in Saccharomyces cerevisiae provided evidence that CoQ and cytc only diffused freely along the membrane upon the addition of chaotropic agents, suggesting that the respiratory chain in yeast also behaves as one functional unit that at least comprises complexes III and IV with bound CoQ and cytc [10].
1.2.2. Liquid-state Model
The general vision gradually evolved into a “random collision”, “fluid” or “liquid-state” model, proposed by Hackenbrock and co-workers, that pictured all membrane proteins and redox components that catalyse electron transport and ATP synthesis in constant and independent diffusional motion, where electron transfer to place through diffusion-based collisions among the redox partners [11]. Evidence in favour of the “liquid-state model” arose from kinetic analyses proposing not only that electron transfer in the CoQ and cytc regions obeyed a pool behaviour in mammalian mitochondria, but also that it followed saturation kinetics with regards to CoQ and cytc concentrations [3,12]. Furthermore, enzymatic activities were retained upon isolation of the individual OXPHOS complexes [2] and the use of fluorescent antibodies against CIII2 and CIV caused independent aggregation of these complexes, suggesting that they diffuse laterally in the membrane plane independent of one another [13].
1.2.3. Dynamic aggregate or Plasticity Model
The two previous models were proposed for the way in which MRC components interact to accomplish a maximal-efficient electron transfer, representing extreme examples where intermediate modes of operation are feasible. Alternative studies by Ferguson-Miller and collaborators that analysed the role of lateral diffusion of cytc in electron transfer within native mitochondrial membranes, gave rise to the “dynamic aggregate” model [14]. In this model, a dynamic equilibrium exists between freely diffusing and associated-forms of the MRC components, all active in electron transfer. This new representation of the respiratory chain reconciled the two classical models, as it incorporated transient aggregates as well as free lateral diffusion of redox components to account for the electron transport rates. The reversible formation of specific MRC aggregates additionally offered a mechanism for localized proton flow and the possibility of regulating the direction and efficiency of electron transfer [15,16]. This idea was disregarded in favour of the “fluid” model until new data from two groups, based on the use of blue native polyacrylamide gel electrophoresis (BN-PAGE) developed by Schägger and collaborators, showed the co-existence of individual MRC complexes together with their inter-associations in supramolecular assemblies that were termed supercomplexes (SCs) [17,18]. Experimental evidences demonstrated that SCs are evolutionarily conserved stable structures in both prokaryotes and eukaryotes, and not random associations of MRC complexes [19,20], and recent high-resolution cryo-electron microscopy (cryo-EM) analyses showed the detailed structural architecture of mammalian respiratory SCs (Figure 1) [21–23]. The coexistence of individual MRC complexes and SCs thus supports the principles of the “dynamic aggregate” model, which was lately renamed as the “plasticity” model by Enríquez and co-workers [24,25]. In its current form, this model proposes that the switch between freely moving MRC complexes to fixed SCs would optimize electron flux from different substrates (NADH and FADH2), adapting the efficiency of the respiratory chain to changes in cellular metabolism via partitioned CoQ and cytc pools [26]. The catalytic relevance of this hypothesis, based on BN-PAGE and oxygen consumption analyses in mouse cells depleted of MRC complexes, was however questioned by more direct spectroscopic and kinetic experiments in yeast and bovine submitochondrial particles showing that: a) the metabolic pathways for NADH and succinate oxidation occur through a single and universally accessible CoQ-pool [27], and b) that cytc does not encounter major barriers to its free diffusion [28]. Moreover, this model was proposed on the basis of differences in the steady-state levels of respiratory chain structures on BN-PAGE gels, and therefore, the reversible and dynamic association/dissociation events –the so-called “plasticity”- between the individual complexes and the respirasomes remains to be proven. The prospective evolution of the “plasticity” model will thus rely on the extensive characterization of the structural, functional and kinetic properties of the SCs upon variations in physiological conditions [29,30].
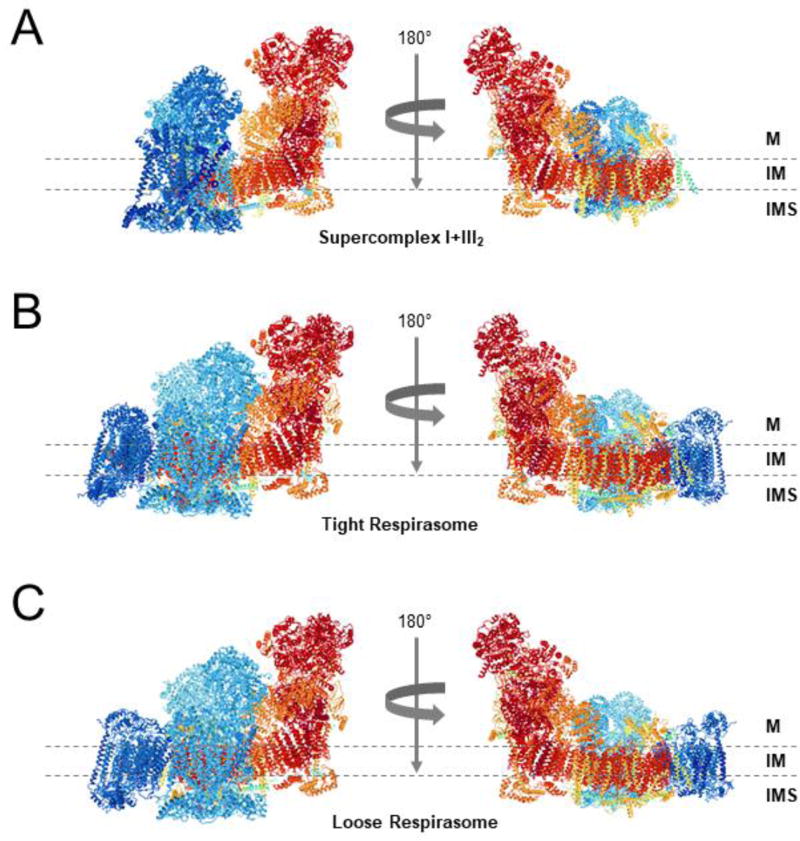
Figure 1. Architectures of the mammalian respirasomes.
Side views along the membrane from (A) Supercomplex I+III2, (B) the tight respirasome, and (C) the loose respirasome, according to the structures proposed by Letts et al. [22]. Images were obtained from the RCSB Protein Data Bank in combination with the NGL viewer. The structural models of CI, CIII2, and CIV are colored in red, turquoise, and navy blue, respectively. The transmembrane region is indicated by two dashed lines. M, matrix; IM, mitochondrial inner membrane; IMS, intermembrane space.
2. Structural architecture of respiratory chain supercomplexes
2.1. Types of respiratory chain supercomplexes
The notion of “supercomplexes” first appeared upon the observations of preferential associations between bacterial MRC complexes [31,32]. Respiratory chain SCs of different compositions and stoichiometries were later reported by means of BN-PAGE analyses of mitochondrial fractions solubilized with the mild detergent digitonin. This method allows the retention of labile supramolecular assemblies of membrane protein complexes that would otherwise be dissociated (Figure 2A). The solubilization of OXPHOS complexes from yeast and bovine heart mitochondria using varying digitonin-to-protein ratios and subsequent BN-PAGE allowed to separate assorted types of stoichiometric associations of complexes I, III and IV within the molecular mass range from ~750 to ~2100 kDa, in an overall 1:3:6 stoichiometry [33]. Additional BN-PAGE, single-particle electron microscopy (EM) and cryo-electron tomography studies, consistently reported specific associations between CI, CIII2 and CIV in a wide range of organisms [19]. It is worth mentioning that for a long time, many researchers attributed the appearance of SCs on BN-PAGE gels to protein aggregation as a consequence of detergent solubilisation. However, the migrations of these bands in the gels are consistently reproducible, as shown by many different laboratories in the last 17 years, and well-defined structures from these bands extracted from the gels are now available.
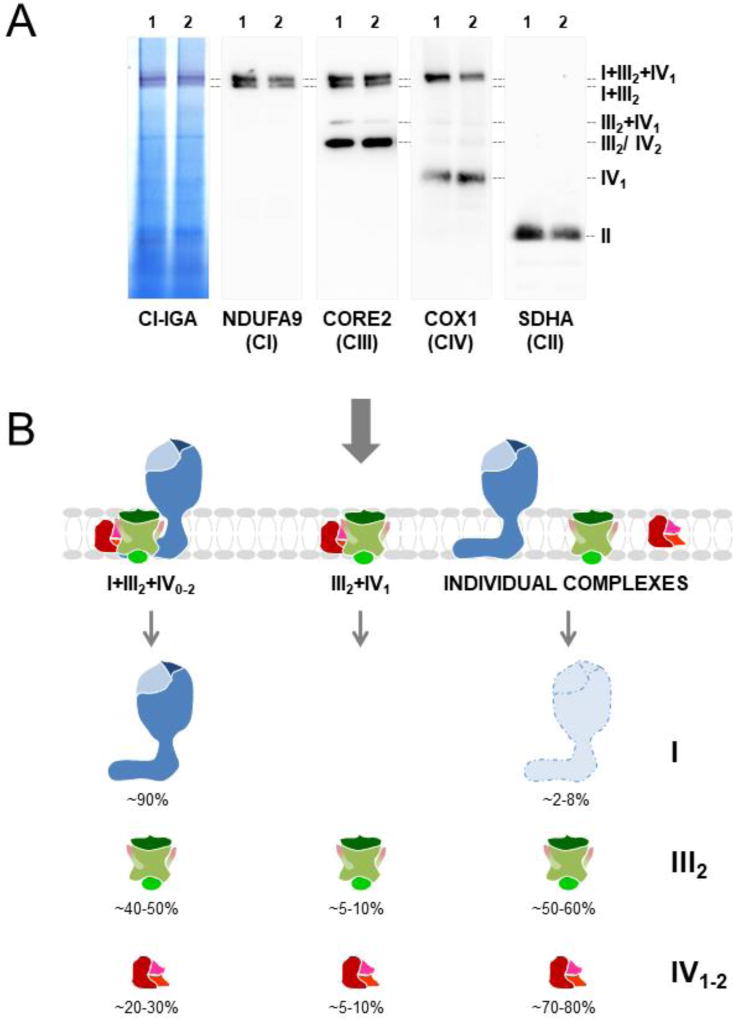
Figure 2. Distribution of human respiratory chain complexes in supercomplexes.
(A) Mitochondria isolated from cultured 143B cells (1) and cybrids (2) were analysed by BN-PAGE in combination with CI in-gel activity (IGA) assay and western blot with antibodies against the indicated MRC subunits. (B) Most CI (blue), ~half of CIII2 (green) and ~20–30% of CIV (red) are localized in the respirasome (I+III2+IV0–1). SC III2+IV1 represents ~5% of the total amount of MRC structures, as well as CIV dimers (IV2). CII (II) is not present in SCs. Free CI (light blue) requires to be associated in supercomplexes to minimize destabilization and ROS generation [29].
The composition and abundance of the respiratory chain SCs may vary among organisms and tissues depending on the metabolic and physiological conditions [17,26,34–40], as well as on the lipid content of the mitochondrial inner membrane [41–44]. In most CI-containing eukaryotes, CI primarily interacts with CIII2 and CIV to form the most abundant SC, I+III2+IV1, to which additional CIV monomers are added to form SCs I+III2+IV2–4. These structures are known as the respirasomes [17], since they contain all the components required to transfer electrons from NADH to molecular oxygen. Additionally, CI associates with CIII2 to form SC I+III2; and CIII2 binds one to two CIV monomers to assemble SCs III2+IV1–2. In mammals, most CI, ~40–50% of CIII2 and ~20–30% of CIV are localized in the largest SCs I+III2 and I+III2+IV1–4 (Figure 2B). CIII2 and CIV may also interact to form SC III2+IV1 that scarcely represents 5–10% of the total MRC structures [33,45]. In higher plants mitochondria, CIV-containing SCs are barely detectable, and SC I+III2 is the predominant macrostructure [46]. In S. cerevisiae, which lacks CI, two bands of ~750 and ~1000 kDa corresponding to SCs III2+IV1 and III2+IV2, respectively, are predominant [17,18]. These variations in the relative abundance of SCs may reflect different stoichiometries of CI, CIII2 and CIV among organisms. Regarding CII and CV, these enzymes form oligomers that do not interact with complexes I, III and IV under normal physiological conditions [17,33,45,47–50].
Based on the differential solubilisation of MRC complexes and SCs on BN-PAGE, a higher organization level of SCs in “respiratory strings” was proposed for mammals and yeast [51], where respirasomes would be interconnected by CIV tetramers at regular intervals, thus generating linear assemblies of respiratory SCs. The observation of CI dimers (CI2) in Yarrowia lipolytica [52] redefined this model as “respiratory patches” generated by the interactions between two CI monomers within adjacent respiratory strings. An alternative model for plants based on single-particle EM, proposed the repetition of I2III2IV2 units into a respiratory string mediated by the interaction of two neighbouring CIV through a dimeric interface [53], which differed from the previous model by the lower abundance of CIV copies. The structural and functional characterization of SCs among different species would benefit very much from in situ studies without disrupting the mitochondrial membranes with detergents; for example, using fluorescence life-time imaging, as recently published [54].
2.2. Structural properties of respiratory chain supercomplexes
2.2.1. Supercomplexes I+III2 and III2+IV1–2
The first structure of SC I+III2 from Arabidopsis thaliana mitochondria was determined at 18Å resolution [46], revealing the lateral association of CIII2 to the membrane-embedded distal part of CI. However, the lack of an atomic CI structure failed to decipher the precise subunit interactions within this SC. Recently, the architectures of the mammalian SC I+III2 were resolved at high resolution in ovine (Figure 1A) [22] and bovine [55]. In these structures, contacts between CI and CIII2 were similar to the ones detected for SC I+III2+IV1 (described in Section 3.2.2.).
A pseudo-atomic model at 15Å resolution of yeast SCs III2+IV1–2 [56] revealed that CIII2 is attached to the convex side of two CIV monomers, leaving the opposite interfaces open for CIV dimerization. Cardiolipin and phosphatidylethanolamine lipids were identified at the CIII2-CIV interface. This structure also revealed cytc bound to CIII2, which moves and rotates within a distance of 40Å (~4 nm) to mediate electron transfer between CIII2 and CIV. Later studies based on 3D-cryo-EM maps specified the distance between the cytc binding sites of CIII2 and CIV as ~6nm, considered to be sufficiently short to enable the channelling of cytc between these complexes [57], although this was previously refuted [28]. The yeast CIII2 subunits cytochrome b, cytochrome c1, Qcr6 (Hinge protein), Qcr7, Qcr8 and Qcr9 were identified at the CIII2-CIV interface, as well as the CIV subunits CoxI, CoxII, CoxIII, CoxIV, CoxVIc, CoxVIIa and CoxVIIc [56]. However, direct evidence supporting precise subunit interactions was missing. Due to its low relative abundance, SCs III2+IV1–2 from higher organisms await structural characterization.
2.2.2. Respirasome or supercomplex I+III2+IV1
In mammalian mitochondria, I+III2+IV1 is the most abundant SC. The first 3D maps of bovine heart SC I+III2+IV1 [47,50,58,59] revealed the lateral binding of CIII2 to the middle part of the CI membrane arm, with CIV positioned in the distal tip of CI while laterally interacting with CIII2, and cardiolipin molecules filling the gaps between the transmembrane domains at the interfaces between the individual complexes [47,59]. The recent characterization of the atomic structure of mammalian CI by cryo-EM [60,61] represented a major step forward that enabled to obtain high-resolution projection maps of the mammalian respirasomes [21–23,55]. Yang and collaborators solved the conformation of porcine SC I+III2+IV1 [21,23], where CIV would loosely bind CI and CIII2. In addition, Letts et al. [22] distinguished two architectures for the ovine respirasome, a major “tight” and a minor “loose” conformations, where CIV would contact both CI and CIII2 within the tight form, but only CI within the loose form (Figures 1B–1C). Sousa et al. also resolved two classes of SC I+III2+IV1 in bovine [55]. The tight form of the ovine respirasome was essentially identical to the bovine respirasome class 1. However, the bovine respirasome class 2 differed in the conformational flexibility of CIII2, as this rotates by 25° relative to CI while CIV remains unchanged. The bovine respirasome additionally showed clear density in one of the two membrane extrinsic iron-sulphur domains of CIII2, suggesting that only one CIII monomer would be active [55]. The heterogeneity among these structures deserves further consideration [30], as it could reflect the existence of independent structural entities resulting from the dynamic association and dissociation of the MRC complexes in response to, e.g., tissue-specific phospholipid environments or ROS levels.
The respirasome-bound CI is more compact than free CI due to its associations with CIII2 and CIV [60,61]. Although several interaction points exist between CI, CIII2 and CIV, the most extensive and stable interactions take place between CI supernumerary subunits (absent in bacteria) and CIII2 at two major points: CI subunits NDUFA11 (B14.7 in bovine) and NDUFB4 (B15) directly interact with CIII2 subunit UQCRQ at the matrix and inner membrane interface, and CI subunits NDUFB4 and NDUFB9 (B22) bind CIII2 subunits UQCRC1 and UQCRFS1 in the matrix [22,23]. Another important interaction occurs between CI subunit NDUFB7 (B18) and subunit UQCRH on CIII2. Both subunits contain disulphide bonds, suggesting that redox regulation might modulate the interactions between MRC complexes [22]. Since the long helix of NDUFB7 is poised at the interface of the three complexes, it may also interact with CIV through the COX7A and COX8B subunits at the intermembrane space and inner membrane interface [22,23]. CIV is less tightly bound to the respirasomes and major contacts differ among structures, reflecting its varying location. There is a close association of CIV subunit COX7C and ND5 on CI, as well as an interaction of COX7A on CIV with CIII2 subunits UQCR11, UQCRC1 and UQCRB at the matrix and inner membrane interface [22,23]. This interaction between CIII2 and IV seems to swing away in the loose respirasome form, where only COX7A would contact CI through subunit ND5 [22]. It must be noted that the differences that exist between the structural models presented from ovine/bovine/porcine probably rely on the species-specific protein sequences of the MRC subunits. Therefore, structural variations in the supercomplexes and respirasomes from other species, such as rodents and human, appear well possible depending on the degree of conservation of the specific protein domains and residues that promote the interactions within these structures.
2.3. Effect of cardiolipin on the stabilization of supercomplexes
Supporting the idea that phospholipids mediate protein–protein interactions in the inner mitochondrial membrane, cardiolipin molecules were detected within yeast SCs III2+IV1–2 [57], where they stabilize these structures [41,42]. Moreover, studies in lymphocytes from patients with Barth syndrome, a mitochondrial disorder in which cardiolipin levels are drastically reduced due to mutations affecting Tafazzin (an enzyme involved in cardiolipin maturation), revealed the specific destabilization of SC I+III2+IV1 [62]. A pluripotent stem cell model system of this disorder later confirmed the role of cardiolipin content for SC stabilization [63]. Consistent with these observations, the atomic structure of the respirasome revealed clear gaps between CI, CIII2 and CIV that were occupied by cardiolipin molecules to further stabilize the respirasome [23,59]. Although cardiolipin is considered to stabilize SCs, phosphatidylethanolamine, another phospholipid of the inner membrane, seems to exert the opposite effect [43]. Therefore, differences in the balance between phospholipid species may contribute to the specific reorganization of the MRC complexes and SCs.
3. Functional roles of the supercomplexes
The discovery of SCs represents a great progress in the study of the functional and structural properties of the MRC, and their existence should provide functional advantages that remain far to be fully-understood.
3.1. Catalytic enhancement of the electron flux through substrate channelling
The arrangement into SCs was initially proposed to maximize the efficiency of the electron flux across the MRC [17]. Indeed, spectrophotometric assays of the MRC activities of isolated SCs from bovine heart mitochondria showed that CI in SC I+III2 displays about half the activity of that in SC I+III2+IV1, suggesting that the full respirasome was the most active unit [50]. Substrate channelling was proposed as a possible mechanism to explain the increased rates of electron transfer within SCs based on flux control analyses of the MRC complexes in bovine heart mitochondria [64]. The authors suggested that CI and CIII2 behave as a single enzymatic unit, where electron transfer through CoQ is accomplished by channelling between the two redox enzymes without following a pool behaviour, in agreement with other reports in yeast [10] and bovine mitochondria [8]. Following studies that analysed the roles of CoQ and cytc in the attenuation of CIII2 and CIV pharmacological inhibition on the respiratory flux supported the dynamic compartmentalization of the respiratory substrates [65], as well as studies based in the competition of substrates for NADH and succinate oxidation [26]. The proposal that SCs may provide distinct electron translocation pathways through the partition of CoQ into different pools to mediate metabolic adaptation [26], was questioned by kinetic and flux control studies showing that the metabolic pathways for NADH and succinate oxidation comprise different CoQ redox steady states, but communicate and converge on a single non-partitioned CoQ pool [27,30]. It has also been argued that the cytc pool is equally compartmentalized [26], but studies monitoring the reduction potential of CIII upon addition of NADH, succinate or both [27], evidenced against cytc partitioning. In agreement, time-resolved spectroscopic analysis of cytc oxidation in intact yeast cells showed that cytc is not trapped within SCs and, therefore, there are no restrictions that limit its diffusion [28]. Moreover, the respirasome structures showed no evidence of a protein-mediated substrate channel connecting the CoQ binding sites of CI and CIII2 [22], since both active sites are open to the membrane and separated by 10 nm, as also evidenced for cytc, in agreement with previous studies that questioned substrate channelling based on the distances between the substrates binding sites on SCs [47]. Therefore, the function of a direct catalytic role for mitochondrial SCs remains questionable [30].
3.2. Assembly and stability of complex I
Experimental evidence accumulated on respiratory chain disease models, suggests that the formation of mammalian SCs confer stability to their individual components, and most particularly to CI. The first description of a patient with progressive exercise intolerance due to a nonsense mutation in the CIII2 subunit gene MT-CYB associated with a combined enzyme deficiency of complexes I and III [66], was followed by a more extensive study showing that genetic alterations leading to a loss of CIII2 prevented respirasome formation and led to the secondary loss of CI [67]. Further studies confirmed that not only the structural integrity of CIII2 [68], but also that of CIV [69–71], were essential to maintain the stability of mammalian CI. Despite these evidences, ostensible CI functional alterations are relatively infrequent to most patients presenting with CIII2 or CIV enzyme deficiencies [72,73], indicating that only severe structural alterations of these two complexes induce a parallel CI dysfunction. On the contrary, a dramatic decrease in CI levels do not generally lead to CIII2 and CIV functional defects in mammals [67]. In agreement, the depletion of 28 different CI accessory subunits in human HEK293T cells showed a loss of SCs I+III2 and I+III2+IV1 with no alterations in the steady-state levels of CIII2 or CIV [74].
The specific dependence of CI stability on the respirasome biosynthesis has important repercussions for our understanding of the respiratory chain disorders. In this regard, two main hypotheses are currently debated to explain the integration and stabilization of CI into SCs (Figures 3 and 4, see Section 5.1.). However, the fact that CI is purified and remains active in its free form adds to the debate whether CI stability may rely on additional molecular mechanisms, like alterations in the mitochondrial membrane potential [49] or ROS levels (see Section 4.3.). In this regard, the absence of respirasomes due to the lack of cytc [75], CIII2 or CIV induced reverse electron transport from reduced CoQ to CI, triggering local superoxide generation and CI degradation [76].
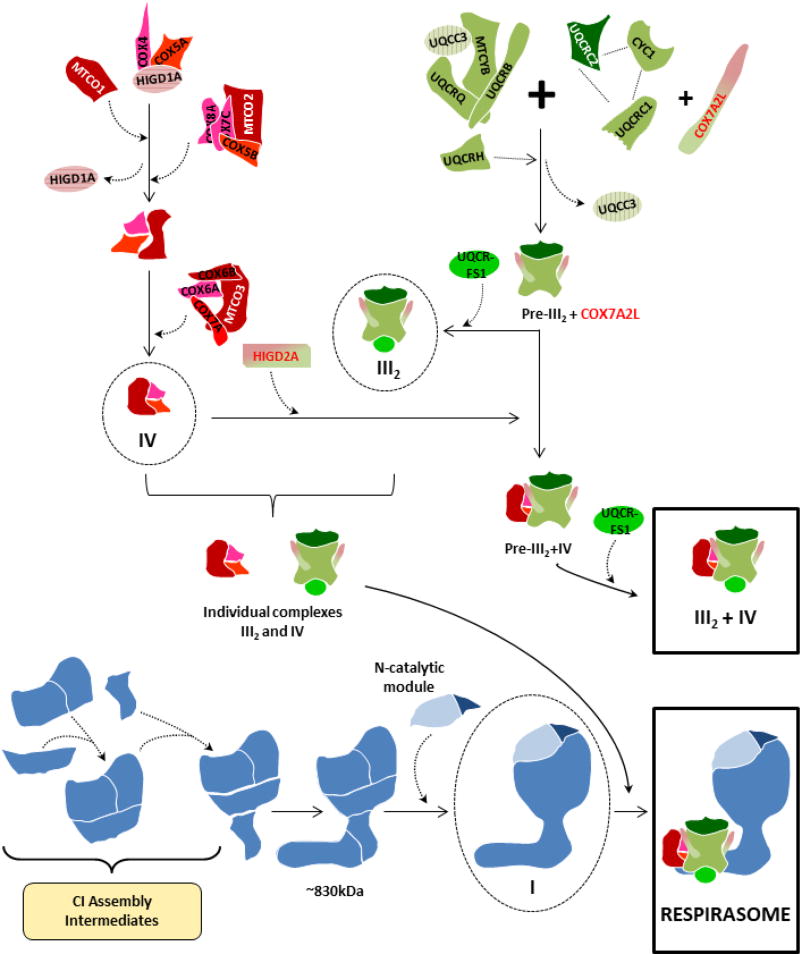
Figure 3. Respirasome biogenesis through the direct association of fully-assembled respiratory chain complexes.
This model [24,86] proposes that the mammalian respirasome (I+III2+IV1) originates by the direct association of single preassembled CI (blue), CIII2 (green) and CIV (red). The assembly pathways of the individual MRC complexes are depicted according to stablished models [72,86,108], and SC III2+IV1 is formed independently of the respirasomes [98]. Supercomplex assembly factors COX7A2L and HIGD2A are marked in green/red.
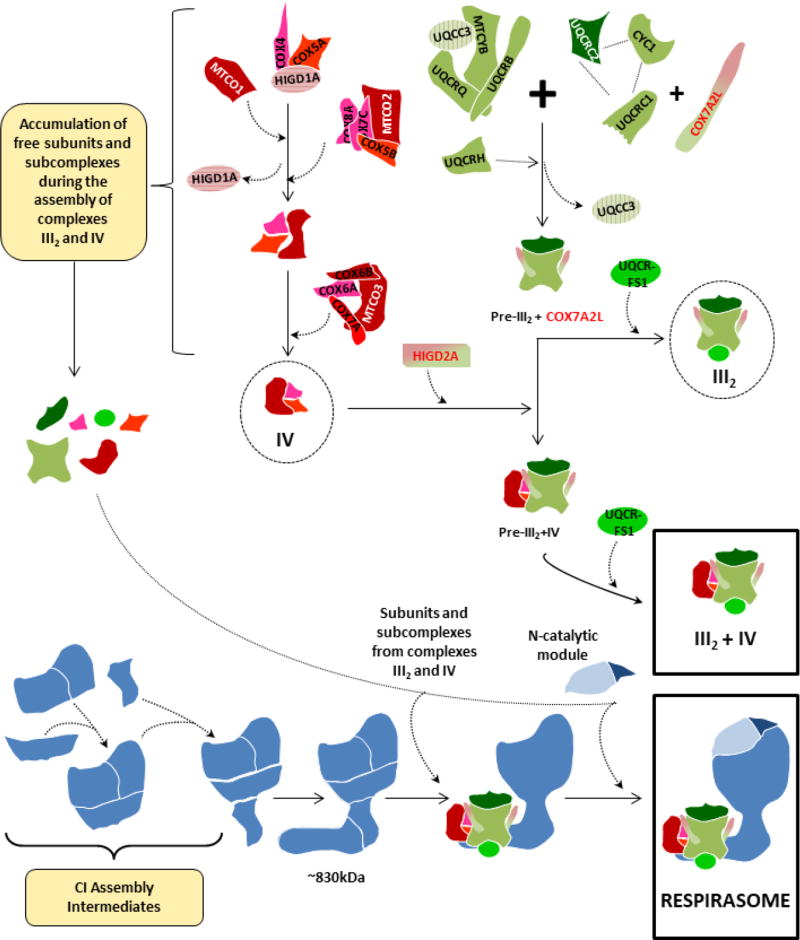
Figure 4. Respirasome biogenesis through the stepwise association of partially-assembled respiratory chain complexes and submodules.
This model [45] proposes the sequential and coordinated association of submodules and free subunits from CIII2 (green) and CIV (red) to a CI-scaffold (blue) that lacks the N catalytic module, which is incorporated at the latest assembly stage to ensure respirasome activation in the presence of all the necessary structural components. The assembly pathways of individual CIII2 and CIV are depicted according to stablished models [72,108], and SC III2+IV1 is formed independently of the respirasomes [98]. Supercomplex assembly factors COX7A2L and HIGD2A are marked in green/red.
3.3. Modulation of ROS production
Because CI and CIII2 constitute the main redox centres responsible for oxygen reduction to superoxide [77,78], it has been hypothesized that their arrangement into SCs could minimize ROS production. Measurements in bovine heart mitochondria provided the first demonstration that the disruption of SC I+III2 enhanced the generation of superoxide from CI [79], and a direct correlation between ROS levels and CI dissociated from SCs was established in neurons and astrocytes [80]. According to the ovine respirasome structure [22], the two CoQ binding cavities on CIII2 are arranged in such way that the symmetry of CIII2 is broken, and this asymmetry could limit ROS production at the expense of maximal activity.
Although decreased ROS production may be a functional consequence of SCs, oxygen levels can also modulate the assembly of these superstructures and the efficiency of mitochondrial respiration. For instance, during prolonged hypoxia, potato mitochondria showed a rearrangement of CI from SC I+III2 to its free form [36]. Or studies in mouse fibroblasts lacking the Rieske Fe-S protein of CIII2 showed that enhanced ROS disrupted SCs and produced a deleterious effect on the stability of complexes I and IV [81]. One way the cells adapt to hypoxic conditions is by building a more efficient respiratory chain through CIV, a key enzyme composed of oxygen-regulated subunit isoforms in yeast and mammals [82–84]. Variations in oxygen levels may thus affect the assembly state of CIV and its incorporation into respirasomes. Aging, a fundamental biological process that affects all eukaryotic lives, is generally attributed to increased oxidative damage induced by ROS, and this is usually accompanied by a decay of SCs levels [85]. Therefore, the interplay between oxygen levels, superoxide production, association and dissociation of SCs and impaired respiration, arises as a crucial regulatory process in health, disease and ageing.
4. Biogenesis of respiratory chain supercomplexes
4.1. The respirasome assembly pathway
The precise mechanisms that regulate the biosynthesis of mammalian mitochondrial SCs remain unsolved. Two models have been proposed to explain the association of CI with complexes III and IV to form the respirasome [45,86]. The first model (Figure 3) proposes that CI gets fully-assembled prior to its binding to SCs [86]. Time-course incorporation analyses of the 13 radiolabelled mitochondrial-encoded polypeptides into complexes and SCs showed the existence of a temporal gap between the formation of the complexes and their co-localization in SCs, suggesting that SCs originate by the direct association of single preassembled complexes [24]. Recent proteomics studies based on BN-PAGE and complexome profiling of CI intermediates upon mitochondrial translation inhibition with chloramphenicol, agreed that CI was independently assembled before SC formation [86]. In contrast, previous studies from the same group in Ndufs4-KO mice had shown that CI lacking its functional NADH-binding (N) module is associated with CIII2 in a partially-assembled SC [87], suggesting that the biogenesis of SCs does not necessarily require the preassembly of individual complexes. The possibility that the formation of the respirasome could be achieved instead through a coordinated association of submodules and free subunits was supported by the observation in Neurospora crassa that the assembly of SC I+III2 occurs before the individual CI is formed [88], and by studies showing that, in mitochondria from patients with chronically reduced CIV levels, newly-imported COX subunits preferentially integrate into SCs [89,90]. In accordance with this idea, we analysed the formation of SC assembly intermediates by reversibly depleting control cell lines of OXPHOS complexes by long treatment with doxycycline, a reversible inhibitor of mitochondrial translation. Results led us to propose a second model (Figure 4) that involves the sequential binding of subcomplexes from CIII2 and CIV to an almost complete CI scaffold that lacks the N catalytic module, which would be incorporated at the end of the assembly process to ensure the activation of the respirasome once all the essential structural components are present [45]. This model implies that the SCs constitute the structural units where CI gets fully-assembled and activated, providing a basis to explain the relevance of the respirasomes for CI stability [67] and why certain mutations in CIII2 or CIV-associated genes lead to a combined enzyme deficiency of CI [45]. The differences between both CI assembly models [45,86] could be attributed to technical differences related to the use of two different mitochondrial translation inhibitors, the length of the inhibition periods, or the time gaps between the collected samples, which probably affect the balances between the synthesis of individual MRC subunits and their kinetics of incorporation into the different MRC structures. Although the first model [86] supports the dynamic exchange of CI between its free form and a SC-associated form, the fact that “free CI” is underrepresented in mammalian tissues under mild purification conditions [33] mainly agrees with our statement that SCs provide a scaffold for the full-assembly and stability of CI. Additionally, our model supports the idea that, when “unable to associate to / or once dissociated from” SCs, free CI is prone to degradation by, i.e. locally-produced superoxide [76,79]; moreover, it does not exclude the dynamic exchange of CIII2 and CIV once the respirasome assembly has been completed (being compatible with the variable stoichiometry of CIV within SCs), and it allows for the dynamic replacement on site of specific CI reassembled modules to avoid futile continuous cycles of turnover and de novo synthesis of this large complex. In this regard, we agree with the previous reports where a CI intermediate lacking the N catalytic module is stably associated in SCs [55,87,91]. Extensive structural studies in cellular and animal models of respiratory chain disease are necessary to clarify the intriguing mechanisms that govern the stepwise biogenesis of the mitochondrial respirasome.
4.2. Supercomplex assembly factors
An important issue is the regulatory function that chaperones or assembly factors play on the assembly of SCs and respirasomes. Here we will discuss the roles of the two best-studied protein types defined as SC assembly factors: COX7A2L and the RCFs.
4.2.1. COX7A2L/COX7RP/SCAFI
An interesting debate concerns the regulatory role of the protein COX7A2L/COX7RP in the formation and stabilization of mitochondrial SCs. COX7A2L was first reported to be present in the respirasomes and SC III2+IV1 but not in CIII2 or free CIV, therefore constituting the first SC-specific assembly factor that was renamed SCAFI [26]. Enríquez and co-workers discovered that certain wild-type mouse strains widely used in biological research, e.g. C57BL/6J and BALB/c, were homozygous for a 6 bp deletion in the Cox7a2l gene and expressed a short, unstable COX7A2L isoform that failed to support CIV association into SCs, thereby promoting differences in mitochondrial respiration rates and ATP production. The authors proposed that COX7A2L is a SC-specific assembly factor that adapts respirasomes formation and mitochondrial function to metabolic variations [26,29]. This hypothesis was challenged by Larsson and co-workers, who demonstrated normal respirasome formation and respiratory chain function in liver and heart mitochondria from mice strains bearing the truncated COX7A2L isoform [92], which was conclusively supported by other studies [93–97]. A recent work from our laboratory showed that COX7A2L is neither essential for respirasome formation in human cell lines [98]. This protein is not uniquely found associated in SCs, since it independently interacts with both CIII2 and free CIV to promote the stabilization of SC III2+IV1, results later supported by other groups [84,96,99]. In the absence of CI and CIV, COX7A2L remains associated with CIII2-containing SCs, thereby showing a preferential association for CIII2 [98]. The fact that the formation/stabilization of mammalian SC III2+IV1 is regulated by a specific protein in a respirasome-independent manner implies the co-existence of independent regulatory mechanisms for the biogenesis and turnover of different SC structures that deserve further attention.
Variations observed among tissues in the relative distribution of BN-PAGE bands above the canonical respirasome (SC I+III2+IV1) in mice strains bearing the short COX7A2L isoform [94–96], opened the possibility that the assembly of specific SCs could be regulated in a tissue-specific manner. The high sequence similarity between COX7A2L and tissue-specific isoforms of the CIV subunit COX7A, led to speculate that COX7A2L may replace COX7A in SCs, acting as a bridge to stabilize the interaction between CIII2 and CIV [22]. Mass spectrometric analyses of SCs from CD1 mice mitochondria (that express the long COX7A2L isoform), revealed that COX7A2 was present almost exclusively in CIV1–2, whereas COX7A2L was present in SCs III2+IV and I+III2+IV1–4 [84]. These observations suggested that the homologous region of COX7A2L could displace COX7A from CIV to form a tight CIII2-CIV interface within SCs, while in the absence of COX7A2L, respirasomes would form with a weaker CIII2-CIV interface solely stabilized by interactions with CI. This possibility remains to be confirmed by higher resolution cryo-EM density maps of the “tight” and “loose” respirasome structures that allow identification of the individual residues [22].
4.2.2. Rcf 1–3
Three independent groups identified two respiratory complex factors (Rcf1 and Rcf2) that control the formation and stabilization of yeast SC III2+IV1–2 [100–102]. Rcf1 and Rcf2 belong to the conserved hypoxia induced gene 1 (Hig1) protein family, and both are CIV-binding proteins that may also interact with CIII2. Regarding Rcf1, deletion mutants (Rcf1Δ) showed an impaired incorporation of Rcf2, and subunits Cox13 and Cox12 (Cox6a and Cox6b in mammals, respectively) into CIV [101,102], resulting in defective CIV activity, decreased SC III2+IV1–2 levels and increased ROS production [100,102]. Remarkably, SC III2+IV was detectable in the Rcf1Δ/Rcf2Δ double mutants, indicating that the core of CIV could still bind CIII2 in the absence of these proteins [101]. These results, as well as recent kinetic and spectroscopic studies in Rcf1Δ and Rcf2Δ mutants [103], revealed the presence of at least two different enzymatically-active CIV forms, which differ in their protein composition and are independently incorporated into SCs, maybe to adapt the respiratory function to different physiological conditions. Rcf1 interacts with the Aac2 protein [101], a member of the ADP/ATP carrier protein family that facilitates the equimolar exchange of ATP for ADP across the inner membrane [104]; therefore, changes on the conformational state of the Aac2 protein could influence the binding of Rcf1 to CIV and the regulation of its activity [101]. Latest studies showed that Rcf1 interacts with CIV to regulate late stages of its assembly process [105], suggesting that it functions in the assembly of individual CIV rather than playing a direct role in SC III2+IV1–2. Rcf1 eukaryotic homologs include two variants, HIGD1A and HIGD2A, that display the broadest expression pattern in mammals [100]. Silencing of HIGD1A in mice C2C12 cells did not induce structural alterations on mammalian SCs [100]; instead, this protein was shown to bind CIV in early assembly stages [107], and to upregulate CIV activity under hypoxic cellular stress [106]. However, HIGD2A knockdown caused the depletion of SC III2+IV1 and of CIV-containing bands above the SC I+III2+IV1 without altering free CIV levels [100], suggestive of a true SC stabilizing role. Rcf2 is found in yeast species, and it is specifically required for CIV assembly [101,102]. Recent studies showed that Rcf2 is processed into a stable C-terminal fragment that remains associated with CIV and SCs, and a labile N-terminal fragment that displays high sequence similarity with another mitochondrial protein, Rcf3 [107]. Rcf2 and Rcf3 associate predominantly with CIV and CIV-containing SCs in the inner membrane, and in the absence of any of these proteins, CIV activity is increased. However, Rcf2Δ/Rcf3Δ double mutants displayed a decrease in CIV activity that resulted in the loss of yeast respiratory growth, indicating an overlapping function of both proteins. Whether the Rcf proteins are truly SC assembly factors is a matter of discussion, as they primarily assist CIV assembly rather than the biogenesis of SCs. However, the fact that all three Rcf proteins remain associated with CIII2 in the absence of a functional CIV [107], open new prospects regarding their functional implications, as well as those of their human orthologues, in the assembly of SCs.
5. Conclusions and Perspectives
The structural and functional organization of the respiratory chain has been long debated. Although the existence of specific associations of complexes I, III and IV into higher-order structures (i.e., respirasomes and intermediate SCs I+III2 and III2+IV1–2) is generally accepted, their functional relevance remain unsolved. Recent structural advances in the composition and organization of the mammalian respirasomes represent a significant step forward to understand the functional consequences of the dynamic rearrangements between individual MRC complexes and SCs, as well as their implications in the regulation of the respiratory chain function in different physiological conditions. This is of particular importance for the understanding of the molecular mechanisms underlying respiratory chain disorders and ageing, where a general deterioration of SCs formation is commonly observed. The following research excitingly point towards the decryption of the specific interactions that govern the bindings among the individual complexes in a variety of superstructures, and to unravelling the molecular mechanisms and players that regulate SCs formation.
HIGHLIGHTS.
The respirasome is the most abundant supercomplex in mammalian mitochondria.
The functional consequences of the supercomplexes remain questioned.
There are two current hypotheses for the biogenesis of the respirasome.
The biogenesis of SC III2+IV is independent from the respirasome.
The recent structures of the respirasomes represent an important novelty.
Acknowledgments
This work was funded by Instituto de Salud Carlos III-MINECO and European FEDER Funds (grant number PI14-00209), and by NIH-NIGMS (1R01GM105781-01).
ABBREVIATIONS
- MRC
mitochondrial respiratory chain
- CoQ
coenzyme Q or ubiquinone
- cytc
cytochrome c
- SC
supercomplex
- CI–CIV
respiratory chain complexes I to IV
- CIII2
dimeric CIII
- CV
ATP synthase
- BN-PAGE
blue native gel electrophoresis
- EM
electron microscopy
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Chance B, Williams GR. A method for the localization of sites for oxidative phosphorylation. Nature. 1955;176:250–254. doi: 10.1038/176250a0. [DOI] [PubMed] [Google Scholar]
- 2.Hatefi Y, Haavik AG, Fowler LR, Griffiths DE. Studies on the electron transfer system. XLII. Reconstitution of the electron transfer system. J. Biol. Chem. 1962;237:2661–2669. [PubMed] [Google Scholar]
- 3.Kröger A, Klingenberg M. The Kinetics of the Redox Reactions of Ubiquinone Related to the Electron-Transport Activity in the Respiratory Chain. Eur. J. Biochem. 1973;34:358–368. doi: 10.1111/j.1432-1033.1973.tb02767.x. [DOI] [PubMed] [Google Scholar]
- 4.Margoliash E, Ferguson-Miller S, Tulloss J, Kang CH, Feinberg B, Brautigan DL, et al. Separate intramolecular pathways for reduction and oxidation of cytochrome c in electron transport chain reactions. Proc. Natl. Acad. Sci. USA. 1973;70:3245–3249. doi: 10.1073/pnas.70.11.3245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reid RA, Moyle J, Mitchell P. Synthesis of adenosine triphosphate by a protonmotive force in rat liver mitochondria. Nature. 1966;212:257–258. doi: 10.1038/212257a0. [DOI] [PubMed] [Google Scholar]
- 6.Fowler LR, Hatefi Y. Reconstitution of the electron transport system III. Reconstitution of DPNH oxidase, succinic oxidase, and DPNH, succinic oxidase. Biochem. Biophys. Res. Commun. 1961;5:203–208. doi: 10.1016/0006-291x(61)90110-3. [DOI] [PubMed] [Google Scholar]
- 7.Blair PV. Preparation and properties of repeating units of electron transfer. Methods Enzymol. 1967;10:208–212. [Google Scholar]
- 8.Ragan CI, Heron C. The interaction between mitochondrial NADH-ubiquinone oxidoreductase and ubiquinol-cytochrome c oxidoreductase. Evidence for stoicheiometric association. Biochem. J. 1978;174:783–790. doi: 10.1042/bj1740783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heron C, Ragan CI, Trumpower BL. The interaction between mitochondrial NADH-ubiquinone oxidoreductase and ubiquinol-cytochrome c oxidoreductase. Restoration of ubiquinone-pool behaviour. Biochem. J. 1978;174:791–800. doi: 10.1042/bj1740791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boumans H, Grivell LA, Berden JA. The respiratory chain in yeast behaves as a single functional unit. J. Biol. Chem. 1998;273:4872–4877. doi: 10.1074/jbc.273.9.4872. [DOI] [PubMed] [Google Scholar]
- 11.Höchli M, Hackenbrock CR. Fluidity in mitochondrial membranes: thermotropic lateral translational motion of intramembrane particles. Proc. Natl. Acad. Sci. USA. 1976;73:1636–1640. doi: 10.1073/pnas.73.5.1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gupte SS, Hackenbrock CR. The role of cytochrome c diffusion in mitochondrial electron transport. J. Biol. Chem. 1988;263:5248–5253. [PubMed] [Google Scholar]
- 13.Höchli M, Höchli L, Hackenbrock CR. Independent lateral diffusion of cytochrome bc1 complex and cytochrome oxidase in the mitochondrial inner membrane. Eur. J. Cell Biol. 1985;38:1–5. [PubMed] [Google Scholar]
- 14.Hochman JH, Schindler M, Lee JG, Ferguson-Miller S. Lateral mobility of cytochrome c on intact mitochondrial membranes as determined by fluorescence redistribution after photobleaching. Proc. Natl. Acad. Sci. USA. 1982;79:6866–6870. doi: 10.1073/pnas.79.22.6866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hochman J, Ferguson-Miller S, Schindler M. Mobility in the mitochondrial electron transport chain. Biochemistry. 1985;24:2509–2516. doi: 10.1021/bi00331a017. [DOI] [PubMed] [Google Scholar]
- 16.Ferguson-Miller S, Hochman J, Schindler M. Aggregation and diffusion in the mitochondrial electron-transfer chain: role in electron flow and energy transfer. Biochem. Soc. Trans. 1986;14:822–824. doi: 10.1042/bst0140822. [DOI] [PubMed] [Google Scholar]
- 17.Schägger H, Pfeiffer K. Supercomplexes in the respiratory chains of yeast and mammalian mitochondria. EMBO J. 2000;19:1777–1783. doi: 10.1093/emboj/19.8.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cruciat CM, Brunner S, Baumann F, Neupert W, Stuart RA. The cytochrome bc1 and cytochrome c oxidase complexes associate to form a single supracomplex in yeast mitochondria. J. Biol. Chem. 2000;275:18093–18098. doi: 10.1074/jbc.M001901200. [DOI] [PubMed] [Google Scholar]
- 19.Chaban Y, Boekema EJ, Dudkina NV. Structures of mitochondrial oxidative phosphorylation supercomplexes and mechanisms for their stabilisation. Biochim. Biophys. Acta. 2014;1837:418–426. doi: 10.1016/j.bbabio.2013.10.004. [DOI] [PubMed] [Google Scholar]
- 20.Melo AMP, Teixeira M. Supramolecular organization of bacterial aerobic respiratory chains: From cells and back. Biochim. Biophys. Acta. 2016;1857:190–197. doi: 10.1016/j.bbabio.2015.11.001. [DOI] [PubMed] [Google Scholar]
- 21.Gu J, Wu M, Guo R, Yan K, Lei J, Gao N, et al. The architecture of the mammalian respirasome. Nature. 2016;537:639–643. doi: 10.1038/nature19359. [DOI] [PubMed] [Google Scholar]
- 22.Letts JA, Fiedorczuk K, Sazanov LA. The architecture of respiratory supercomplexes. Nature. 2016;537:644–648. doi: 10.1038/nature19774. [DOI] [PubMed] [Google Scholar]
- 23.Wu M, Gu J, Guo R, Huang Y, Yang M. Structure of Mammalian Respiratory Supercomplex I1III2IV1. Cell. 2016;167:1598–1609. doi: 10.1016/j.cell.2016.11.012. [DOI] [PubMed] [Google Scholar]
- 24.Acín-Pérez R, Fernández-Silva P, Peleato ML, Pérez-Martos A, Enriquez JA. Respiratory active mitochondrial supercomplexes. Mol. Cell. 2008;32:529–539. doi: 10.1016/j.molcel.2008.10.021. [DOI] [PubMed] [Google Scholar]
- 25.Acin-Perez R, Enriquez JA. The function of the respiratory supercomplexes: The plasticity model. Biochim. Biophys. Acta. 2014;1837:444–450. doi: 10.1016/j.bbabio.2013.12.009. [DOI] [PubMed] [Google Scholar]
- 26.Lapuente-Brun E, Moreno-Loshuertos R, Acin-Perez R, Latorre-Pellicer A, Colas C, Balsa E E, et al. Supercomplex Assembly Determines Electron Flux in the Mitochondrial Electron Transport Chain. Science. 2013;340:1567–1570. doi: 10.1126/science.1230381. [DOI] [PubMed] [Google Scholar]
- 27.Blaza JN, Serreli R, Jones AJY, Mohammed K, Hirst J. Kinetic evidence against partitioning of the ubiquinone pool and the catalytic relevance of respiratory-chain supercomplexes. Proc. Natl. Acad. Sci. USA. 2014;111:15735–15740. doi: 10.1073/pnas.1413855111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Trouillard M, Meunier B, Rappaport F. Questioning the functional relevance of mitochondrial supercomplexes by time-resolved analysis of the respiratory chain. Proc. Natl. Acad. Sci. USA. 2011;108:E1027–1034. doi: 10.1073/pnas.1109510108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barrientos A, Ugalde C. I Function, therefore I am: Overcoming skepticism about mitochondrial supercomplexes. Cell Metab. 2013;18:147–149. doi: 10.1016/j.cmet.2013.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Milenkovic D, Blaza JN, Larrson N-G, Hirst J. The enigma of the respiratory chain supercomplex. Cell Metab. 2017:765–776. doi: 10.1016/j.cmet.2017.03.009. [DOI] [PubMed] [Google Scholar]
- 31.Sone N, Sekimachi M, Kutoh E. Identification and properties of a quinol oxidase super-complex composed of a bc1 complex and cytochrome oxidase in the thermophilic bacterium PS3. J. Biol. Chem. 1987;262:15386–15391. [PubMed] [Google Scholar]
- 32.Berry EA, Trumpower BL. Isolation of ubiquinol oxidase from Paracoccus denitrificans and resolution into cytochrome bc1 and cytochrome c-aa3 complexes. J. Biol. Chem. 1985;260:2458–2467. [PubMed] [Google Scholar]
- 33.Schägger H, Pfeiffer K. The Ratio of Oxidative Phosphorylation Complexes I–V in Bovine Heart Mitochondria and the Composition of Respiratory Chain Supercomplexes. J. Biol. Chem. 2001;276:37861–37867. doi: 10.1074/jbc.M106474200. [DOI] [PubMed] [Google Scholar]
- 34.Dencher NA, Frenzel M, Reifschneider NH, Sugawa M, Krause F. Proteome Alterations in Rat Mitochondria Caused by Aging. Ann. N. Y. Acad. Sci. 2007;1100:291–298. doi: 10.1196/annals.1395.030. [DOI] [PubMed] [Google Scholar]
- 35.Helbig AO, De Groot MJL, van Gestel RA, Mohammed S, de Hulster EAF, Luttik MAH, et al. A three-way proteomics strategy allows differential analysis of yeast mitochondrial membrane protein complexes under anaerobic and aerobic conditions. Proteomics. 2009;9:4787–4798. doi: 10.1002/pmic.200800951. [DOI] [PubMed] [Google Scholar]
- 36.Ramírez-Aguilar SJ, Keuthe M, Rocha M, Fedyaev VV, Kramp K, Gupta KJ, et al. The composition of plant mitochondrial supercomplexes changes with oxygen availability. J. Biol. Chem. 2011;286:43045–43053. doi: 10.1074/jbc.M111.252544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cogliati S, Frezza C, Soriano ME, Varanita T, Quintana-Cabrera R, Corrado M, et al. Mitochondrial Cristae Shape Determines Respiratory Chain Supercomplexes Assembly and Respiratory Efficiency. Cell. 2013;155:160–171. doi: 10.1016/j.cell.2013.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Greggio C, Jha P, Kulkarni SS, Lagarrigue S, Broskey NT, Boutant M, et al. Enhanced Respiratory Chain Supercomplex Formation in Response to Exercise in Human Skeletal Muscle. Cell Metab. 2017;25:1–11. doi: 10.1016/j.cmet.2016.11.004. [DOI] [PubMed] [Google Scholar]
- 39.Jang S, Lewis TS, Powers C, Khuchua Z, Baines CP, Wipf P, et al. Elucidating mitochondrial ETC supercomplexes in the heart during ischemia-reperfusion. Antioxid. Redox Signal. 2017;27:57–69. doi: 10.1089/ars.2016.6635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rosca MG, Vazquez EJ, Kerner J, Parland W, Chandler MP, Stanley W, et al. Cardiac mitochondria in heart failure: Decrease in respirasomes and oxidative phosphorylation. Cardiovasc. Res. 2008;80:30–39. doi: 10.1093/cvr/cvn184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang M, Mileykovskaya E, Dowhan W. Gluing the respiratory chain together: Cardiolipin is required for supercomplex formation in the inner mitochondrial membrane. J. Biol. Chem. 2002;277:43553–43556. doi: 10.1074/jbc.C200551200. [DOI] [PubMed] [Google Scholar]
- 42.Pfeiffer K, Gohil V, Stuart RA, Hunte C, Brandt U, Greenberg ML, et al. Cardiolipin Stabilizes Respiratory Chain Supercomplexes. J. Biol. Chem. 2003;278:52873–52880. doi: 10.1074/jbc.M308366200. [DOI] [PubMed] [Google Scholar]
- 43.Böttinger L, Horvath SE, Kleinschroth T, Hunte C, Daum G, Pfanner N, et al. Phosphatidylethanolamine and cardiolipin differentially affect the stability of mitochondrial respiratory chain supercomplexes. J. Mol. Biol. 2012;423:677–686. doi: 10.1016/j.jmb.2012.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tasseva G, Bai HD, Davidescu M, Haromy A, Michelakis E, Vance JE. Phosphatidylethanolamine Deficiency in Mammalian Mitochondria Impairs Oxidative Phosphorylation and Alters Mitochondrial Morphology. J. Biol. Chem. 2013;288:4158–4173. doi: 10.1074/jbc.M112.434183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moreno-Lastres D, Fontanesi F, García-Consuegra I, Martín MA, Arenas J, Barrientos A, et al. Mitochondrial complex I plays an essential role in human respirasome assembly. Cell Metab. 2012;15:324–335. doi: 10.1016/j.cmet.2012.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dudkina NV, Eubel H, Keegstra W, Boekema EJ, Braun H-P. Structure of a mitochondrial supercomplex formed by respiratory-chain complexes I and III. Proc. Natl. Acad. Sci. U. S. A. 2005;102:3225–3229. doi: 10.1073/pnas.0408870102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dudkina NV, Kudryashev M, Stahlberg H, Boekema EJ. Interaction of complexes I, III, and IV within the bovine respirasome by single particle cryoelectron tomography. Proc. Natl. Acad. Sci. USA. 2011;108:15196–15200. doi: 10.1073/pnas.1107819108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Muster B, Kohl W, Wittig I, Strecker V, Joos F, Haase W, et al. Respiratory chain complexes in dynamic mitochondria display a patchy distribution in life cells. PLoS One. 2010;5 doi: 10.1371/journal.pone.0011910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Quarato G, Piccoli C, Scrima R, Capitanio N. Variation of flux control coefficient of cytochrome c oxidase and of the other respiratory chain complexes at different values of protonmotive force occurs by a threshold mechanism. Biochim. Biophys. Acta. 2011;1807:1114–1124. doi: 10.1016/j.bbabio.2011.04.001. [DOI] [PubMed] [Google Scholar]
- 50.Schäfer E, Seelert H, Reifschneider NH, Krause F, Dencher NA, Vonck J. Architecture of active mammalian respiratory chain supercomplexes. J. Biol. Chem. 2006;281:15370–15375. doi: 10.1074/jbc.M513525200. [DOI] [PubMed] [Google Scholar]
- 51.Wittig I, Carrozzo R, Santorelli FM, Schägger H. Supercomplexes and subcomplexes of mitochondrial oxidative phosphorylation. Biochim. Biophys. Acta. 2006;1757:1066–1072. doi: 10.1016/j.bbabio.2006.05.006. [DOI] [PubMed] [Google Scholar]
- 52.Nubel E, Wittig I, Kerscher S, Brandt U, Schägger H. Two-dimensional native electrophoretic analysis of respiratory supercomplexes from Yarrowia lipolytica. Proteomics. 2009;9:2408–2418. doi: 10.1002/pmic.200800632. [DOI] [PubMed] [Google Scholar]
- 53.Bultema JB, Braun HP, Boekema EJ, Kouřil R. Megacomplex organization of the oxidative phosphorylation system by structural analysis of respiratory supercomplexes from potato. Biochim. Biophys. Acta. 2009;1787:60–67. doi: 10.1016/j.bbabio.2008.10.010. [DOI] [PubMed] [Google Scholar]
- 54.Rieger B, Shalaeva DN, Söhnel AC, Kohl W, Duwe P, Mulkidjanian AY, Busch KB. Lifetime imaging of GFP at CoxVIIIa reports respiratory supercomplex assembly in live cells. Sci. Rep. 2017;7:46055. doi: 10.1038/srep46055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sousa JS, Mills DJ, Vonck J, Kühlbrandt W. Functional asymmetry and electron flow in the bovine respirasome. Elife. 2016;5:1–17. doi: 10.7554/eLife.21290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Heinemeyer J, Braun HP, Boekema EJ, Kouril R, Kouřil R. A structural model of the cytochrome C reductase/oxidase supercomplex from yeast mitochondria. J. Biol. Chem. 2007;282:12240–12248. doi: 10.1074/jbc.M610545200. [DOI] [PubMed] [Google Scholar]
- 57.Mileykovskaya E, Penczek PA, Fang J, Mallampalli VKPS, Sparagna GC, Dowhan W. Arrangement of the respiratory chain complexes in Saccharomyces cerevisiae supercomplex III2IV2 revealed by single particle cryo-electron microscopy. J. Biol. Chem. 2012;287:23095–23103. doi: 10.1074/jbc.M112.367888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schäfer E, Dencher Na, Vonck J, Parcej DN. Three-dimensional structure of the respiratory chain supercomplex I1III2IV1 from bovine heart mitochondria. Biochemistry. 2007;46:12579–12585. doi: 10.1021/bi700983h. [DOI] [PubMed] [Google Scholar]
- 59.Althoff T, Mills DJ, Popot J-L, Kühlbrandt W. Arrangement of electron transport chain components in bovine mitochondrial supercomplex I1III2IV1. EMBO J. 2011;30:4652–4664. doi: 10.1038/emboj.2011.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhu J, Vinothkumar KR, Hirst J. Structure of mammalian respiratory complex I. Nature. 2016;536:354–358. doi: 10.1038/nature19095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fiedorczuk K, Letts JA, Degliesposti G, Kaszuba K, Skehel M, Sazanov LA. Atomic structure of the entire mammalian mitochondrial complex I. Nature. 2016:1–21. doi: 10.1038/nature19794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.McKenzie M, Lazarou M, Thorburn DR, Ryan MT. Mitochondrial Respiratory Chain Supercomplexes Are Destabilized in Barth Syndrome Patients. J. Mol. Biol. 2006;361:462–469. doi: 10.1016/j.jmb.2006.06.057. [DOI] [PubMed] [Google Scholar]
- 63.Dudek J, Cheng IF, Balleininger M, Vaz FM, Streckfuss-Bömeke K, Hübscher D, et al. Cardiolipin deficiency affects respiratory chain function and organization in an induced pluripotent stem cell model of Barth syndrome. Stem Cell Res. 2013;11:806–819. doi: 10.1016/j.scr.2013.05.005. [DOI] [PubMed] [Google Scholar]
- 64.Blanchi C, Genova ML, Castelli GP, Lenaz G. The mitochondrial respiratory chain is partially organized in a supercomplex assembly: Kinetic evidence using flux control analysis. J. Biol. Chem. 2004;279:36562–36569. doi: 10.1074/jbc.M405135200. [DOI] [PubMed] [Google Scholar]
- 65.Benard G, Faustin B, Galinier A, Rocher C, Bellance N, Smolkova L K, et al. Functional dynamic compartmentalization of respiratory chain intermediate substrates: Implications for the control of energy production and mitochondrial diseases. Int. J. Biochem., Cell Biol. 2008;40:1543–1554. doi: 10.1016/j.biocel.2007.11.023. [DOI] [PubMed] [Google Scholar]
- 66.Lamantea E, Carrara F, Mariotti C, Morandi L, Tiranti V, Zeviani M. A novel nonsense mutation (Q352X) in the mitochondrial cytochrome b gene associated with a combined deficiency of complexes I and III. Neuromuscul. Disord. 2002;12:49–52. doi: 10.1016/s0960-8966(01)00244-9. [DOI] [PubMed] [Google Scholar]
- 67.Schägger H, de Coo R, Bauer MF, Hofmann S, Godinot C, Brandt U. Significance of respirasomes for the assembly/stability of human respiratory chain complex I. J. Biol. Chem. 2004;279:36349–36353. doi: 10.1074/jbc.M404033200. [DOI] [PubMed] [Google Scholar]
- 68.Acín-Pérez R, Bayona-Bafaluy MP, Fernández-Silva P, Moreno-Loshuertos R, Pérez-Martos A, Bruno C, et al. Respiratory complex III is required to maintain complex I in mammalian mitochondria. Mol. Cell. 2004;13:805–815. doi: 10.1016/s1097-2765(04)00124-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li Y, D’Aurelio M, Deng JH, Park JS, Manfredi G, Hu P, Lu J, Bai Y. An assembled complex IV maintains the stability and activity of complex I in mammalian mitochondria. J. Biol. Chem. 2007;282:17557–17562. doi: 10.1074/jbc.M701056200. [DOI] [PubMed] [Google Scholar]
- 70.Hornig-Do H-T, Tatsuta T, Buckermann A, Bust M, Kollberg G, Rötig A, et al. Nonsense mutations in the COX1 subunit impair the stability of respiratory chain complexes rather than their assembly. EMBO J. 2012;31:1293–1307. doi: 10.1038/emboj.2011.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Diaz F, Fukui H, Garcia S, Moraes CT. Cytochrome c oxidase is required for the assembly/stability of respiratory complex I in mouse fibroblasts. Mol. Cell. Biol. 2006;26:4872–4881. doi: 10.1128/MCB.01767-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Fernández-Vizarra E, Zeviani M. Nuclear gene mutations as the cause of mitochondrial complex III deficiency. Front. Genet. 2015;6:134. doi: 10.3389/fgene.2015.00134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rak M, Benit P, Chretien D, Bouchereau J, Schiff M, El-Khoury R, et al. Mitochondrial cytochrome c oxidase deficiency. Clin. Sci. 2016;130:393–407. doi: 10.1042/CS20150707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Stroud DA, Surgenor EE, Formosa LE, Reljic B, Frazier AE, Dibley MG, et al. Accessory subunits are integral for assembly and function of human mitochondrial complex I. Nature. 2016:1–17. doi: 10.1038/nature19754. [DOI] [PubMed] [Google Scholar]
- 75.Vempati UD, Han X, Moraes CT. Lack of cytochrome c in mouse fibroblasts disrupts assembly/stability of respiratory complexes I and IV. J. Biol. Chem. 2009;284:4383–4391. doi: 10.1074/jbc.M805972200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Guarás A, Perales-Clemente E, Calvo E, Acín-Pérez R, Loureiro-Lopez M, Pujol C, et al. The CoQH2/CoQ Ratio Serves as a Sensor of Respiratory Chain Efficiency. Cell Rep. 2016;15:197–209. doi: 10.1016/j.celrep.2016.03.009. [DOI] [PubMed] [Google Scholar]
- 77.Genova ML, Ventura B, Giuliano G, Bovina C, Formiggini G, Parenti Castelli G, et al. The site of production of superoxide radical in mitochondrial Complex I is not a bound ubisemiquinone but presumably iron-sulfur cluster N2. FEBS Lett. 2001;505:364–368. doi: 10.1016/s0014-5793(01)02850-2. [DOI] [PubMed] [Google Scholar]
- 78.Sun J, Trumpower BL. Superoxide anion generation by the cytochrome bc1 complex. Arch. Biochem. Biophys. 2003;419:198–206. doi: 10.1016/j.abb.2003.08.028. [DOI] [PubMed] [Google Scholar]
- 79.Maranzana E, Barbero G, Falasca AI, Lenaz G, Genova ML. Mitochondrial Respiratory Supercomplex Association Limits Production of Reactive Oxygen Species from Complex I. Antioxidants & Redox Signal. 2013;19:1469–1480. doi: 10.1089/ars.2012.4845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lopez-Fabuel I, Le Douce J, Logan A, James AM, Bonvento G, Murphy MP, et al. Complex I assembly into supercomplexes determines differential mitochondrial ROS production in neurons and astrocytes. Proc. Natl. Acad. Sci. USA. 2016;113:13063–13068. doi: 10.1073/pnas.1613701113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Diaz F, Enríquez JA, Moraes CT. Cells lacking Rieske iron-sulfur protein have a reactive oxygen species-associated decrease in respiratory complexes I and IV. Mol. Cell. Biol. 2012;32:415–429. doi: 10.1128/MCB.06051-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Cogliati S, Calvo E, Loureiro M, Guaras AM, Nieto-Arellano R, Garcia-Poyatos C, et al. Mechanism of super-assembly of respiratory complexes III and IV. Nature. 2016;539:579–582. doi: 10.1038/nature20157. [DOI] [PubMed] [Google Scholar]
- 83.Aras S, Pak O, Sommer N, Finley R, Hüttemann M, Weissmann N, et al. Oxygen-dependent expression of cytochrome c oxidase subunit 4-2 gene expression is mediated by transcription factors RBPJ, CXXC5 and CHCHD2. Nucleic Acids Res. 2013;41:2255–2266. doi: 10.1093/nar/gks1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Liu J, Barrientos A. Transcriptional Regulation of Yeast Oxidative Phosphorylation Hypoxic Genes by Oxidative Stress. Antioxidants & Redox Signal. 2012;19:1916–1927. doi: 10.1089/ars.2012.4589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Frenzel M, Rommelspacher H, Sugawa MD, Dencher NA. Ageing alters the supramolecular architecture of OxPhos complexes in rat brain cortex. Exp. Gerontol. 2010;45:563–572. doi: 10.1016/j.exger.2010.02.003. [DOI] [PubMed] [Google Scholar]
- 86.Guerrero-Castillo S, Baertling F, Kownatzki D, Wessels HJ, Arnold S, Brandt U, et al. The Assembly Pathway of Mitochondrial Respiratory Chain Complex I. Cell Metab. 2016:1–12. doi: 10.1016/j.cmet.2016.09.002. [DOI] [PubMed] [Google Scholar]
- 87.Calvaruso MA, Willems P, den brand M, Valsecchi F, Kruse S, Palmiter R, et al. Mitochondrial complex III stabilizes complex I in the absence of NDUFS4 to provide partial activity. Hum. Mol. Genet. 2012;21:115–120. doi: 10.1093/hmg/ddr446. [DOI] [PubMed] [Google Scholar]
- 88.Marques I, Dencher NA, Videira A, Krause F. Supramolecular organization of the respiratory chain in Neurospora crassa mitochondria. Eukaryot. Cell. 2007;6:2391–2405. doi: 10.1128/EC.00149-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kovářová N, Čížková Vrbacká A, Pecina P, Stránecký V, Pronicka E, Kmoch S, et al. Adaptation of respiratory chain biogenesis to cytochrome c oxidase deficiency caused by SURF1 gene mutations. Biochim. Biophys. Acta. 2012;1822:1114–1124. doi: 10.1016/j.bbadis.2012.03.007. [DOI] [PubMed] [Google Scholar]
- 90.Lazarou M, Smith SM, Thorburn DR, Ryan MT, McKenzie M. Assembly of nuclear DNA-encoded subunits into mitochondrial complex IV, and their preferential integration into supercomplex forms in patient mitochondria. FEBS J. 2009;276:6701–6713. doi: 10.1111/j.1742-4658.2009.07384.x. [DOI] [PubMed] [Google Scholar]
- 91.Lazarou M, McKenzie M, Ohtake A, Thorburn DR, Ryan MT. Analysis of the assembly profiles for mitochondrial- and nuclear-DNA-encoded subunits into complex I. Mol. Cell. Biol. 2007;27:4228–4237. doi: 10.1128/MCB.00074-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Mourier A, Matic S, Ruzzenente B, Larsson NG, Milenkovic D. The respiratory Chain supercomplex organization is independent of COX7A2L isoforms. Cell Metab. 2014;20:1069–1075. doi: 10.1016/j.cmet.2014.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ikeda K, Shiba S, Horie-Inoue K, Shimokata K, Inoue S. A stabilizing factor for mitochondrial respiratory supercomplex assembly regulates energy metabolism in muscle. Nat. Commun. 2013;4:2147. doi: 10.1038/ncomms3147. [DOI] [PubMed] [Google Scholar]
- 94.Davoudi M, Kotarsky H, Hansson E, Kallijärvi J, Fellman V. COX7A2L/SCAFI and Pre-Complex III Modify Respiratory Chain Supercomplex Formation in Different Mouse Strains with a Bcs1l Mutation. PLoS One. 2016;11:e0168774. doi: 10.1371/journal.pone.0168774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Jha P, Wang X, Auwerx J. Curr. Protoc. Mouse Biol. John Wiley & Sons Inc.; Hoboken, NJ, USA: 2016. Analysis of Mitochondrial Respiratory Chain Supercomplexes Using Blue Native Polyacrylamide Gel Electrophoresis (BN-PAGE) pp. 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Williams EG, Wu Y, Jha P, Dubuis S, Blattmann P, Argmann CA, et al. Systems proteomics of liver mitochondria function. Science. 2016;352:aad0189–aad018914. doi: 10.1126/science.aad0189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Sun D, Li B, Qiu R, Fang H, Lyu J. Cell type-specific modulation of respiratory chain supercomplex organization. Int. J. Mol. Sci. 2016;17:6–11. doi: 10.3390/ijms17060926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Perez-Perez R, Lobo-Jarne T, Milenkovic D, Mourier A, Bratic A, Garcia-Bartolome A, et al. COX7A2L Is a Mitochondrial Complex III Binding Protein that Stabilizes the III2+IV Supercomplex without Affecting Respirasome Formation. Cell Rep. 2016;16:2387–2398. doi: 10.1016/j.celrep.2016.07.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zhang K, Wang G, Zhang X, Hüttemann PP, Qiu Y, Liu J, et al. COX7AR is a Stress-inducible Mitochondrial COX Subunit that Promotes Breast Cancer Malignancy. Sci. Rep. 2016;6:31742. doi: 10.1038/srep31742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Chen YC, Taylor EB, Dephoure N, Heo JM, Tonhato A, Papandreou I, et al. Identification of a protein mediating respiratory supercomplex stability. Cell Metab. 2012;15:348–360. doi: 10.1016/j.cmet.2012.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Strogolova V, Furness A, Robb-McGrath M, Garlich J, Stuart RA. Rcf1 and Rcf2, members of the hypoxia-induced gene 1 protein family, are critical components of the mitochondrial cytochrome bc1-cytochrome c oxidase supercomplex. Mol. Cell. Biol. 2012;32:1363–1373. doi: 10.1128/MCB.06369-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Vukotic M, Oeljeklaus S, Wiese S, Vögtle FN, Meisinger C, Meyer A HE, et al. Rcf1 mediates cytochrome oxidase assembly and respirasome formation, revealing heterogeneity of the enzyme complex. Cell Metab. 2012;15:336–347. doi: 10.1016/j.cmet.2012.01.016. [DOI] [PubMed] [Google Scholar]
- 103.Rydström Lundin C, von Ballmoos C, Ott M, Ädelroth P, Brzezinski P. Regulatory role of the respiratory supercomplex factors in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA. 2016:E4475–E4485. doi: 10.1073/pnas.1601196113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Klingenberg M. Molecular aspects of the adenine nucleotide carrier from mitochondria. Arch. Biochem. Biophys. 1989;270:1–14. doi: 10.1016/0003-9861(89)90001-5. [DOI] [PubMed] [Google Scholar]
- 105.Garlich J, Strecker V, Wittig I, Stuart RA. Mutational Analysis of the QRRQ Motif in the Yeast Hig1-type 2 Protein, Rcf1, Reveals a Regulatory Role for the Cytochrome c Oxidase Complex. J. Biol. Chem. 2017;292:5216–5226. doi: 10.1074/jbc.M116.758045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Hayashi T, Asano Y, Shintani Y, Aoyama H, Kioka H, Tsukamoto O, et al. Higd1a is a positive regulator of cytochrome c oxidase. Proc. Natl. Acad. Sci. USA. 2015;112:1553–1558. doi: 10.1073/pnas.1419767112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Römpler K, Müller T, Juris L, Wissel M, Vukotic M, Hofmann K, et al. Overlapping Role of Respiratory Supercomplex Factor Rcf2 and Its N-terminal Homolog Rcf3 in Saccharomyces cerevisiae. J. Biol. Chem. 2016;291:23769–23778. doi: 10.1074/jbc.M116.734665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Vidoni S, Harbour ME, Guerrero-Castillo S, Signes A, Ding S, Fearnley IM, et al. MR-1S Interacts with PET100 and PET117 in Module-Based Assembly of Human Cytochrome c Oxidase. Cell Rep. 2017;18:1727–1738. doi: 10.1016/j.celrep.2017.01.044. [DOI] [PubMed] [Google Scholar]