Abstract
Chloropyrene, one of the major chlorinated polycyclic aromatic hydrocarbon contaminants, was incubated with human cytochrome P450 (P450 or CYP) enzymes including CYP1A1, 1A2, 1B1, 2A6, 2A13, 2B6, 2C9, 2D6, 2E1, 3A4, and 3A5. Catalytic differences in 1-chloropyrene oxidation by polymorphic two CYP1B1 and five CYP2A13 allelic variants were also examined.
CYP1A1 oxidized 1-chloropyrene at the 6- and 8-positions more actively than at the 3-position, while both CYP1B1.1 and 1B1.3 preferentially catalyzed 6-hydroxylation.
Five CYP2A13 allelic variants oxidized 8-hydroxylation much more than 6- and 3-hydroxylation, and the variant CYP2A13.3 was found to slowly catalyze these reactions with a lower kcat value than other CYP2A13.1 variants.
CYP2A6 catalyzed 1-chloropyrene 6-hydroxylation at a higher rate than the CYP2A13 enzymes, but the rate was lower than the CYP1A1 and 1B1 variants. Other human P450 enzymes had low activities towards 1-chloropyrene.
Molecular docking analysis suggested differences in the interaction of 1-chloropyrene with active sites of CYP1 and 2A enzymes. In addition, a naturally occuring Thr134 insertion in CYP2A13.3 was found to affect the orientation of Asn297 in the I-helix in interacting with 1-chloropyrene (and also 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone, NNK) and caused changes in the active site of CYP2A13.3 as compared with CYP2A13.1.
Keywords: P450 1B1, P450 2A13, P450 2A6, docking, chloropyrene
Introduction
Chlorinated polycyclic aromatic hydrocarbons (chlorinated PAHs) have been detected in tap water, air, soil, and foodstuffs (Shiraishi et al., 1985; Sugiyama et al., 1999; Ishaq et al., 2003; Ohura et al., 2004; 2005; 2008; Ding et al., 2012; 2013). 1-Chloropyrene and 6-chlorobenzo[a]pyrene are the most commonly occurring chlorinated PAHs in the environment, and approximately 15% of chlorinated PAHs are in the nanoparticle phase (<0.1 µm) in the air (Ohura et al., 2008; Kakimoto et al., 2014; Kakimoto et al., 2017;Ma et al., 2013). Ohura et al. (2016) reported that photochlorination of pyrene under acidic conditions in the presence of both UV and visible light causes formation of 1-chloropyrene as the main product, whereas 6-chlorobenzo[a]pyrene is formed through irradiation of benzo[a]pyrene by visible light.
Some chlorinated PAHs show mutagenicity in Salmonella typhimurium TA98 and TA100 (Colmsjo et al., 1984; Bhatia et al., 1987) and induce aryl hydrocarbon receptor-mediated activities in yeast assay systems (Ohura et al., 2007). However, little is known about the levels of exposue of these chlorinated PAHs in humans and if these chemicals induce adverse effects in humans and other living organisms. Recently, 1-chloropyrene has been shown to be oxidized by human CYP1A1 and 1B1 to form at least three oxidative metabolites, namely the 3-, 6-, and 8-hydroxylated products (Kakimoto et al., 2015); however, little is known about roles of other human P450 enzymes in the oxidation of 1-chloropyrene. Our previous studies have shown that pyrene, 1-hydroxypyrene, 1-nitropyrene, and 1-acetylpyrene—which are structurally similar to 1-chloropyrene—are oxidized by CYP2A13 (and 2A6) at similar or much higher rates than CYP1A1 and 1B1 (Shimada et al., 2016a), and it is of interest to determine whether 1-chloropyrene is oxidized by these or other human P450 enzymes.
We examined the oxidation of 1-chloropyrene by different recombinant human P450 enzymes including CYP1A1, 1A2, 1B1, 2A6, 2A13, 2B6, 2C9, 2D6, 2E1, 3A4, and 3A5; the enzymes had been co-expressed with NADPH-P450 reductase in Escherichia coli (Shimada et al., 2015, 2016a, 2016b, 2016c) or in microsomes of Trichoplusia ni cells (GENTEST, Corning Life Sciences, Woburn, MA). Two polymorphic CYP1B1 variants (1B1.1 and 1B1.3) and five polymorphic variants CYP2A13 (CYP2A13.1, 2A13.2, 2A13.3, 2A13.10A, 2A13.10B) were also studied; these P450 enzymes have been expressed in our laboratories to study roles of these CYP1B1 and 2A13 variants in catalytic roles of oxidation of various xenobiotic chemicals (Kim et al., unpublished results) and we expanded to study the roles of these enzymes in 1-chloropyrene and pyrene oxidations in this study. Molecular docking analysis of interaction of 1-chloropyrene with the active sites of these P450 enzymes was done, and the ligand-interaction energies (U values) were used for the analysis of structure-function relationships of chemicals in interacting with active sites of these two P450 enzymes.
Materials and Methods
Chemicals
1-Chloropyrene was obtained from Frinton Laboratories (Hainesport, NJ, USA) and purified by HPLC; its oxidation metabolites, 3-, 6-, and 8-hydroxy-1-chloropyrene were synthesized and purified as described previously (Kakimoto et al., 2015). Pyrene and 1-hydroxypyrene were obtained from Sigma-Aldrich (St. Louis, MO, USA) or Wako Pure Chemicals (Osaka, Japan). Other chemicals and reagents used in this study were obtained from the sources described previously or were of the highest quality commercially available (Shimada et al., 2013; 2015; 2016a; 2016b: 2016c).
Enzymes
Expression in E. coli and purification of human P450 enzymes have been described previously (Parikh et al., 1997; Shimada et al., 2009; 2011; 2013; Han et al., 2012). Bacterial bicistronic CYP1A2, 1B1.1, 1B1.3, 2A6, 2A13.1, 2A13.2, 2A13.3, 2A13.10A, 2A13.10B, 2C9, and 3A4 membranes in which human NADPH-P450 reductase was co-expressed were prepared, and the E. coli membranes were suspended in 10 mM Tris-HCl buffer (pH 7.4) containing 1.0 mM EDTA and 20% glycerol (v/v) as described (Sandhu et al., 1993; 1994; Guengerich, 2015; Han et al., 2012).
Two polymorphic CYP1B1.1 (R48A119L432N453) and 1B1.3 RAVN (R48A119V432N453) variants and six polymorphic CYP2A13.1 (wild-type), 2A13.2 (R257C), 2A13.3 (133_134InsThr; D158E). CYP2A13.10A (I331T), 2A13.10B (R257C, I331T) were also expressed in E. coli (together with human NADPH-P450 reductase) by the methods essentially as described above (Lim and Kim, unpublished results; Han et al., 2012).
CYP1A1, 2A6, 1B1.1, 1B1.3, 2A6, 2A13.1, 2A13.2, 2A13.3, 2A13.10A, 2A13.10B, 2C9, 3A4, NADPH-P450 reductase, and cytochrome b5 (b5) were purified from membranes of recombinant E. coli as described elsewhere (Parikh et al., 1997; Sandhu et al., 1993; 1994; Han et al., 2012).
Recombinant CYP2B6, 2E1, 2D6, and 3A5 expressed in microsomes of T. ni cells, infected with baculovirus containing human P450 and NADPH-P450 reductase cDNA inserts, were obtained from GENTEST (Corning Life Sciences, Woburn, MA, USA). P450 contents in these microsomes were accepted from the values in the data sheets provided by the manufacturer.
Spectral Binding Titrations
Purified P450 enzymes were diluted to 1.0 µM in 0.10 M potassium phosphate buffer (pH 7.4) containing 20% glycerol (v/v), and binding spectra were recorded with subsequent additions of 1-chloropyrene and pyrene in a JASCO V-550 or an OLIS-Aminco DW2a spectrophotometer (On-Line Instrument Systems, Bogart, GA, USA) as described previously (Shimada et al., 2009; 2011; 2013; 2016a). Binding intensities (ΔAmax values) were estimated by subtracting absorbance at the trough at 418 nm from the absorbance at the peak at 386 nm in the difference spectra in the presence and absence of chemicals. Spectral dissociation constants (Ks) were estimated using GraphPad Prism software (GraphPad Software, San Diego, CA, USA), either using hyperbolic plots or quadratic fits for tight binding.
Oxidation of 1-Chloropyrene by P450 Enzymes
Oxidative metabolism of 1-chloropyrene by P450 enzymes was determined in a standard incubation mixture (0.25 ml) containing bicistronic P450s (50 pmol of P450 in E. coli membranes or 10 pmol of P450s in microsomes of T. ni cells (co-expressing human NADPH-P450 reductase), 10 µM 1-chloropyrene, and an NADPH-generating system consisting of 0.5 mM NADP+, 5 mM glucose 6-phosphate, and 0.5 unit of yeast glucose 6-phosphate dehydrogenase/ml (Shimada et al., 2015; 2016a). 1-Chloropyrene and other chemicals were dissolved in (CH3)2SO as 10 mM stock solutions and diluted into aqueous solution, with the final solvent concentration ≤0.5%, v/v.
In reconstitution experiments, P450 membranes were replaced by purified P450 (50 pmol), NADPH-P450 reductase (100 pmol), b5 (100 pmol) (when required), and L-α-1,2-dilauroyl-sn-glycero-3-phosphocholine (50 µg) as described previously (Shimada et al., 2015; 2016a). Our previous studies have suggested that more than a 2-fold excess of NADPH-P450 reductase over P450 is required to account for full catalytic activities for drug oxidations in reconstituted systems, although the ratio of expression of the reductase and P450 is not always as high in bicistronic systems (Shimada et al., 2000; Yamazaki et al., 2001).
Incubations were carried out at 37 °C for 20 min, following a preincubation time of 1 min. Each reaction was terminated by the addition of an equal volume of ice-cold CH3CN. The mixture was vortexed vigorously and centrifuged at 10,000 × g for 10 min. After centrifugation, the mixture was kept at −20 °C for 2 h, and the upper CH3CN layer was collected; an aliquot of this layer was analyzed by HPLC.
HPLC-Flourescnece Analysis of 1-Chloropyrene Metabolites
HPLC-fluorescence analyses were performed on a liquid chromatograph (Alliance e2695, Waters) coupled to a fluorescence detector (2475 Multi λ, Waters). An injection volume of 20 µl was used for LC analyses. Chromatographic separation was performed on a C18 octadecylsilane column (CORTECS C18, 150 mm × 4.6 mm, 2.7 µm, Waters). The column temperature was maintained at 40 °C and gradient elution (70% methanol in water to 100% methanol (v/v) over 20 min) was performed at a flow rate of 1.0 ml/min. The excitation and emission wavelengths were 348 nm and 391 nm for mono-hydroxylated metabolites in the HPLC-fluorescence analyses. To determine the enzymatic kinetic parameters, we quantified each 1-chloropyrene metabolite using standard curves ranging from 0.001 to 1 µM of the compound (R2 > 0.99). The detection limit for each metabolite was 0.001 µM in HPLC-fluorescence analyses.
Other Assays
Oxidation of pyrene, 1-hydroxypyrene, naphthalene, phenanthrene, and biphenyl by P450 enzymes was determined as described previously (Shimada et a., 2016a). P450 and protein contents and coumarin 7-hydroxylation activities (CYP2A6 and 2A13) were determined by methods described previously (Omura and Sato, 1964; Brown et al., 1989).
Docking Simulations into Human P450 Enzymes
Crystal structures of CYP2A6 bound to coumarin (Protein Data Bank (PBD) 1Z10), pilocarpine (PDB 3T3R), and nicotine (PDB 4EJJ) and CYP2A13 bound to nicotine (PDB 4EJG), NNK (PDB 4EJH), indole (PDB 2P85), and pilocarpine (PDB 3T3S) have been reported and were used in this study (Yano et al., 2005; DeVore et al., 2009; 2012a; 2012b; Smith et al., 2007; Shimada et al., 2016b). A CYP2A13.3 structure was developed from that of CYP2A13.1 (PDB:4EJH) by insertion of T134 and substitution of D158E and stabilized using the Protein Builder Function in the MOE software. The crystal structures of CYP1A1 418V (Walsh et al., 2013) and CYP1B1 3PMO (Wang et al., 2011), both of which have been crystallized in complexes with α-naphthoflavone, were also used.
Simulations were carried out after removing each ligand from these P450 structures using the MMFF94x force field described in the MOE software (ver. 2015.10, Computing Group, Montreal, Canada). Ligand-interaction energies (U values) were obtained by use of the program ASEdock in MOE. Lower U values are the indication of higher interaction between a chemical and the enzyme (Shimada et al., 2010; Shimada et al., 2011; Shimada 2013).
Kinetic Analysis
Kinetic parameters were estimated by nonlinear regression analysis of hyperbolic plots using the program Kaleida-Graph (Synergy Software, Reading, PA, USA) or GraphPad Prism (GraphPad, La Jolla, CA, USA). Correlation coefficients of ligand-interaction energies (U values) were determined using the Curve Fit program of Cricket Graph verision III.
Results
Spectral Interactions of 1-Chloropyrene with Purified P450 Enzymes
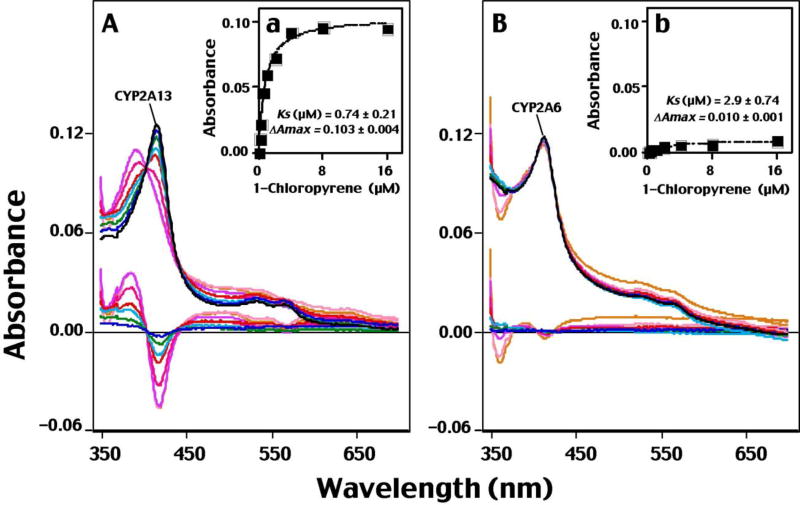
We first examined spectral interaction of 1-chloropyrene with purified CYP2A13.1 and 2A6 and found that CYP2A13 interacted more strongly with 1-chloropyrene to produce Type I binding spectra than CYP2A6 did (Figure 1). The Ks value was 0.74 µM for CYP2A13.1, lower than the value for CYP2A6 (2.9 µM), which had a much lower ΔAmax value than CYP2A13.1. The CY2A13.2, 2A13.3, 2A13.10A, and 2A13.10B allelic variants also produced Type I binding spectra with 1-chloropyrene (vide infra). It should be mentioned that all of the CYP2A13 variants, as well as CYP2A13.1 and 2A6, were shown to be almost entirely in the low-spin state in the spectral analysis. Other human P450 enzymes, including CYP1A1, 1A2, 1B1.1, 1B1.3, 2D6, 2E1, 2C9, and 3A4, did not show any detectable spectral changes in our assay conditions.
Fig. 1.
Type I binding spectra of interaction of CYP2A13 (A and a) and CYP 2A6 (B and b) with 1-chloropyrene. P450 enzymes were diluted in 100 mM potassium phosphate buffer (pH 7.4) containing 20% glycerol (v/v) to a final P450 concentration of 1 µM and, after addition of increasing concentrations of 1-chloropyrene to the sample cuvette (and vehicle to the reference cuvette), difference spectra were obtained. The substrate spectra were obtained by subtracting the blank spectra (in the absence of P450) from the P450 spectra (in the presence of P450). Spectral intensities were determined using GraphPad Prism software, as shown in Figs. 1A and 1B for CYP2A13 and CYP 2A6, respectively. Spectral dissociation constants (Fig. 1A and 1B) were estimated using GraphPad Prism software as described in Materials and Methods.
Oxidation of 1-Chloropyrene by Human P450 Enzymes
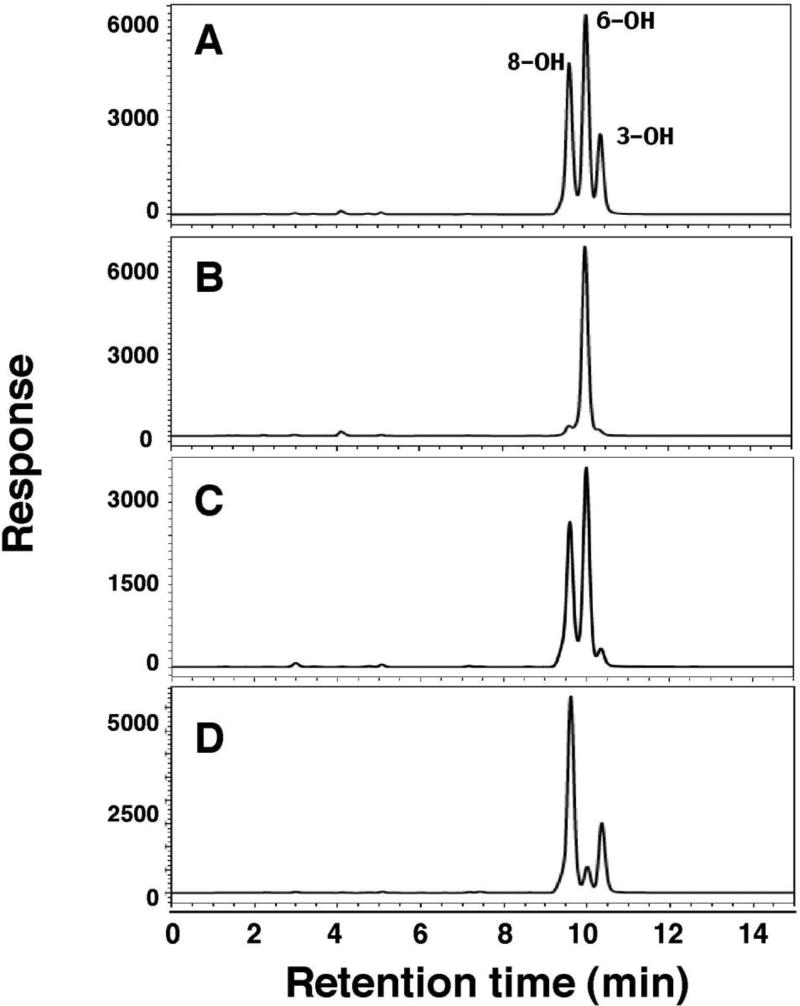
1-Chloropyrene was incubated at 37 °C for 20 min with purified CYP1A1, 1B1.1, 2A6, and 2A13.1 in reconstituted monooxygenase systems, and the products (which increased linearly for about 20 min) were determined by HPLC-fluorescence chromatography (Fig. 2). Product formation was also found to increase linearly with P450 concentrations up to 25–50 pmol in the reaction mixture. CYP1A1 produced 8-, 6-, and 3-hydroxylated products at significant levels while CYP1B1.1 only formed the 6-hydroxy product, at a rate similar to CYP1A1 (Fig. 2A and 2B). CYP2A6 oxidized 1-chloropyrene to 8- and 6-hydroxylated products, but the levels were lower than those produced by CYP1A1 (note that the scale of the y-axis was different) and CYP2A13.1 catalyzed hydroxylation at the 8- and 3-positions, although some 6-hydroxylated product was formed (Figs. 2C and 2D). Thus, form-specific product formation was different for the P450s.
Fig. 2.
HPLC-fluorescence analysis of formation of 1-chloropyrene products formed by CYP1A1 (A), 1B1.1 (B), 2A6 (C), and 2A13.1 (D) in the reconstituted monooxygenase system containing purified P450 and NADPH-P450 reductase. In the latter two P450 enzymes, b5 was also added for the reconstitution. Formation of three oxidative metabolites (i.e., 8-, 6-, and 3-hydroxylated products) is shown in the figure.
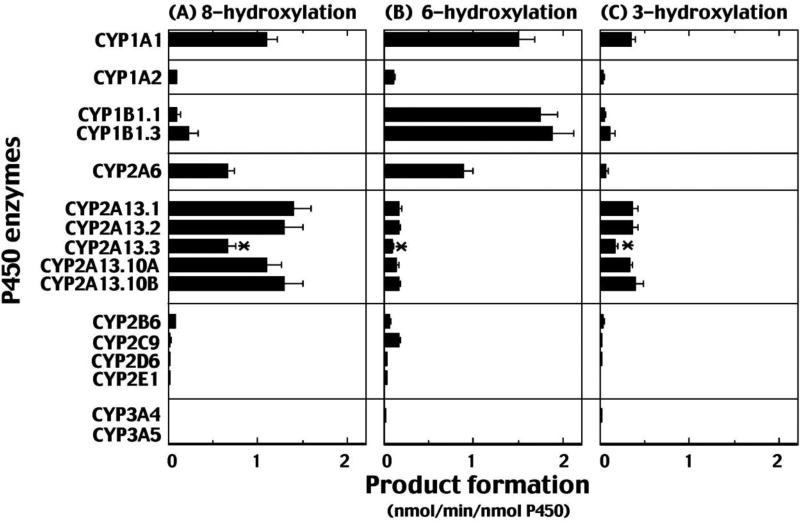
We compared 1-chloropyrene oxidation activities catalyzed by human P450 enzymes CYP1A1, 1A2, 1B1.1, 1B1.2, 2A6, 2A13.1, 2A13.2, 2A13.3, 2A13.10A, 2A13.10B, 2C9, and 3A4 using purified preparations, and CYP2B6, 2D6, 2E1, and 3A5 were recombinant enzymes in T. ni cell microsomes, as described in Materials and Methods. CYP1A1, 1B1, and 2A13 enzymes were the major enzymes that oxidized 1-chloropyrene, followed by CYP2A6 and 1A2 (Fig. 3). The pattern of formation of three hydroxylated metabolites in CYP1A2 was similar to, but slower than, CYP1A1. CYP2B6, 2C9, 2D6, 2E1, 3A4, and 3A5 were minor enzymes involved in the oxidation of 1-chloropyrene.
Fig. 3.
Oxidation of 1-chloropyrene by human P450 enzymes. 1-Chloropyrene was incubated with human P450 enzymes in a standard reaction system, as described in Materials and Methods, and the formation of 8-hydroxy (A), 6-hydroxy (B), and 3-hydroxy (C) products is shown in the figure. Results are presented as means of triplicate determinations ± S.D. An asterisk (*) indicates statistical differences from those of CYP2A13.1.
Polymorphic CYP1B1.1 and 1B1.3 catalyzed 1-chloropyrene at similar rates; however, differences were found in the catalytic activities of the five CYP2A13 allelic variants. Interestingly, formation of these three metabolites by CYP2A13.3 was significantly less than by CYP2A13.1 and the other variants (Fig. 3).
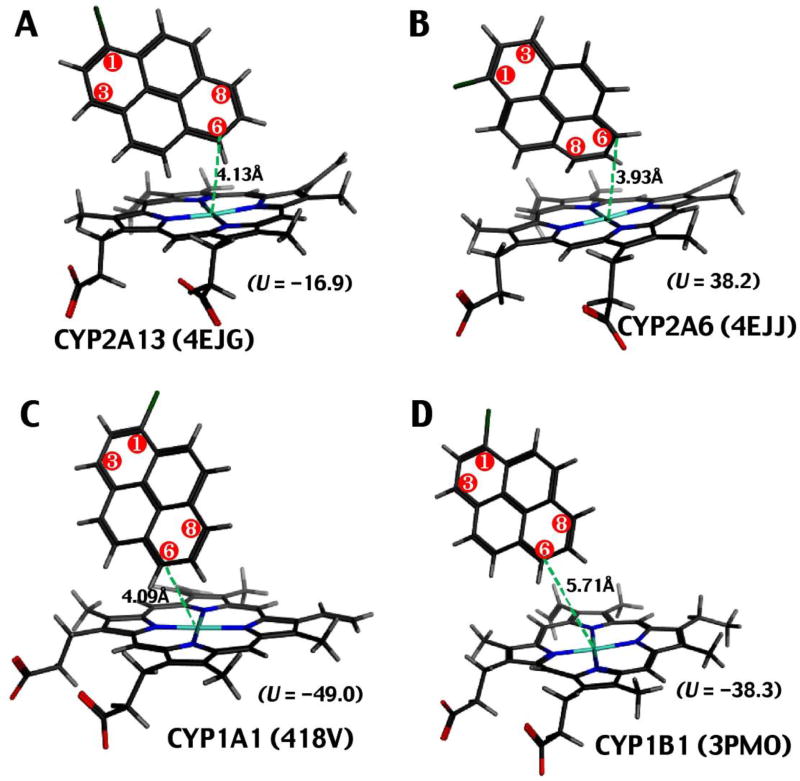
Molecular Docking of 1-Chloropyrene with CYP2A13, 2A6, 1A1, and 1B1
Since the above results suggested CYP2A13 and 2A6—as well as CYP1A1 and 1B1—were the major enzymes involved in the oxidation of 1-chloropyrene with different activities at three positions, we examined differences in the molecular interactions of 1-chloropyrene with these P450 enzymes with molecular docking simulations (Fig. 4). The distance between the C6 atom of 1-chloropyrene and the iron center of CYP2A13 4EJG (nicotine-type) was calculated to be 4.13 Å and that between C6 of the chemical and the CYP2A6 4EJJ iron (nicotine-type) was 3.93 Å (Fig. 4A and 4B). The ligand-interaction energy (U value) of interaction of 1-chloropyrene and the active site of CYP2A13 was found to be –16.9, which was low as compared with that of CYP2A6 (U = 38.2). The distances between C6 of the 1-chloropyrene molecule and the active site iron atoms of CYP1A1 and 1B1 were found to be 4.09 Å and 5.71 Å, respectively, and the U values of interaction of 1-chloropyrene and CYP1A1 and 1B1 were −49.0 and −38.3, respectively (Fig. 4C and 4D).
Fig. 4.
Molecular docking simulation of the interaction of 1-chloropyrene with CYP2A13.1 (A), CYP2A6 (B), CYP1A1 (C), and CYP1B1 (D). Model structures used for analysis were CYP2A13 4EJG (nicotine-type) (A), CYP2A6 4EJJ (nicotine-type) (B), CYP1A1 (418V) (C), and CYP1B1 (3PMO) (D). Distances between the 1-chloropyrene molecule and the active site iron (shown in blue) of the P450 heme are shown in the figure in green. The ligand interaction energies (U) were determined as described in Materials and methods.
Correlation of Ligand Interaction Energies (U values) of Binding of 1-Chloropyrene in the Active Sites of P450 Enzymes
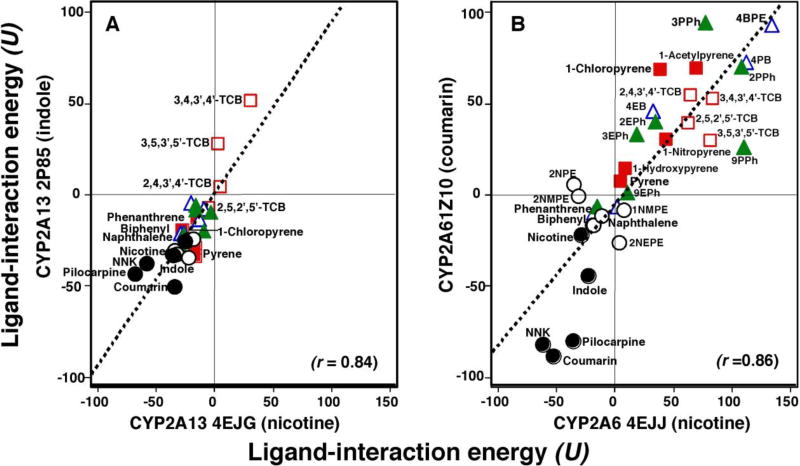
We have previously shown correlations of ligand-interaction energies (U values) obtained from molecular docking of interaction of various structures of xenobiotic chemicals with CYP2A13 and CYP2A6 (Shimada et al., 2016a; 2016b) when analyzing the crystal structures of these P450 enzymes (Yano et al., 2005; DeVore et al., 2009; 2012a; 2012b; Smith et al., 2007). In this study, we compared ligand-interaction energies using CYP2A13 4EJG (nicotine-type) and CYP2A13 2P85 (indole-type) structures for CYP2A13 analysis (Fig. 5A) and CYP2A6 4EJJ (nicotine-type) and CYP2A6 1Z10 (coumarin-type) structures for CYP2A6 analysis (Fig. 5B). The chemicals compared were five known CYP2A substrates (1-chloropyrene, pyrene, 1-hydroxypyrene, 1-nitropyrene, 1-acetylpyrene), naphthalene and its six derivatives, phenanthrene and six derivatives, biphenyl and four derivatives, and four tetrachlorobiphenyl isomers (total of 33 chemicals). Positive correlations were obtained by comparing ligand-interaction energies of interaction of these 33 chemicals with CYP2A13 (r = 0.84) and CYP2A6 structures (r = 0.86). It was also evident that the U values obtained with CYP2A13 were generally lower than those with CYP2A6, except for NNK, coumarin, pilocarpine, and indole, for which the U values were very low when CYP2A6 (1Z10) structure was used (Fig. 5B).
Fig. 5.
Correlation of ligand-interaction energies (U values) obtained with CYP2A13 4EJG (nicotine-type) and CYP2A13 2P85 (indole-type) (A) and with CYP2A6 4EJJ (nicotine-type) and CYP2A6 1Z10 (coumarin-type) (B). Chemicals shown in the figure included known CYP2A ligand (closed circles), naphthalene and derivatives (open circles), phenanthrene and the derivatives (closed triangles), pyrene and derivatives (closed squares), biphenyl and derivatives (open triangles), and TCBs (open squares). Correlation coefficients were obtained using linear analysis with the Curve Fit Program of Cricket Graph III.
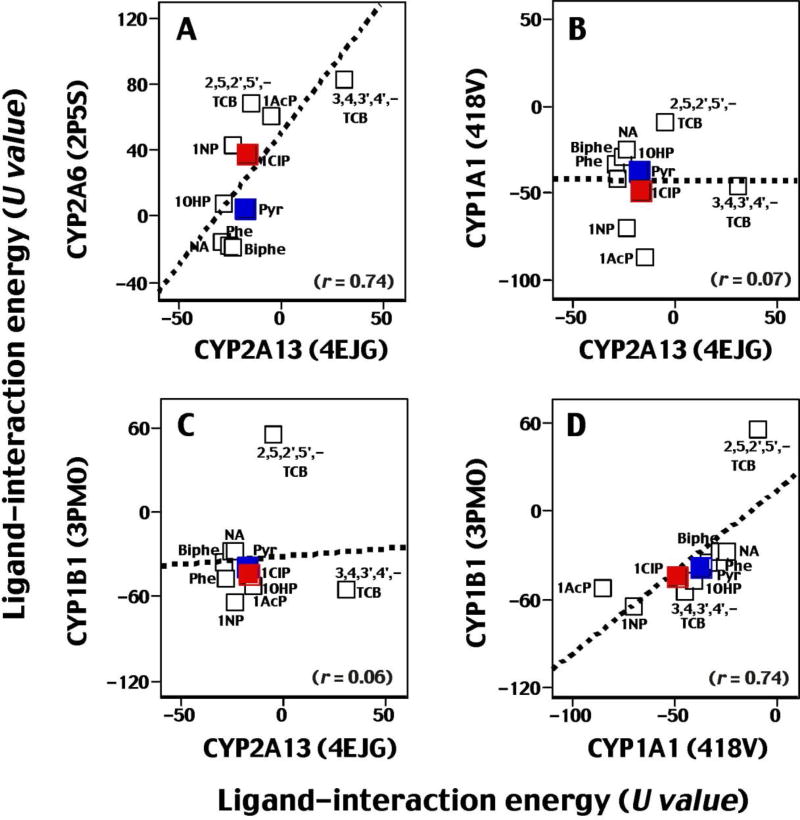
We also compared U values obtained with crystal structures of CYP2A13 4EJG, CYP2A6 2P5S, CYP1A1 (418V), and CYP1B1 (3PMO) using 10 ligands (Fig. 6). Good correlations were obtained when comparisons were made between CYP2A13 and CYP2A6 (r = 0.74) (Fig. 6A) or between CYP1A1 and CYP1B1 (r = 0.74) (Fig. 6D), but there were no correlations between U values of CYP2A13 and CYP1A1 or CYP1B1 (Fig. 6B and 6C).
Fig. 6.
Correlation of ligand-interaction energies (U values) obtained with CYP2A13 (4EJG) and CYP2A6 (2P5S) (A), CYP2A13 (4EJG) and CYP1A1 (418V) (B), CYP2A13 (4EJG) and CYP1B1 (3PMO) (C), and P450 1A1 (418V) and P450 1B1 (3PMO) (D). The chemicals used were naphthalene (NA), phenanthrene (Phe), biphenyl (Biphe), pyrene (pyr), 1-hydroxypyrene (1ONP), 1-nitropyrene (1NP), 1-acethylpyrene (1AcP), 1-chloropyrene (1ClP), 2,5,2´,5´- TCB, and 3,4,3´,4´-TCB. Other details are as in legend to Fig. 5.
Type I Binding Spectra and Oxidation of 1-Chloropyrene and Pyrene by Five CYP2A13 Variants
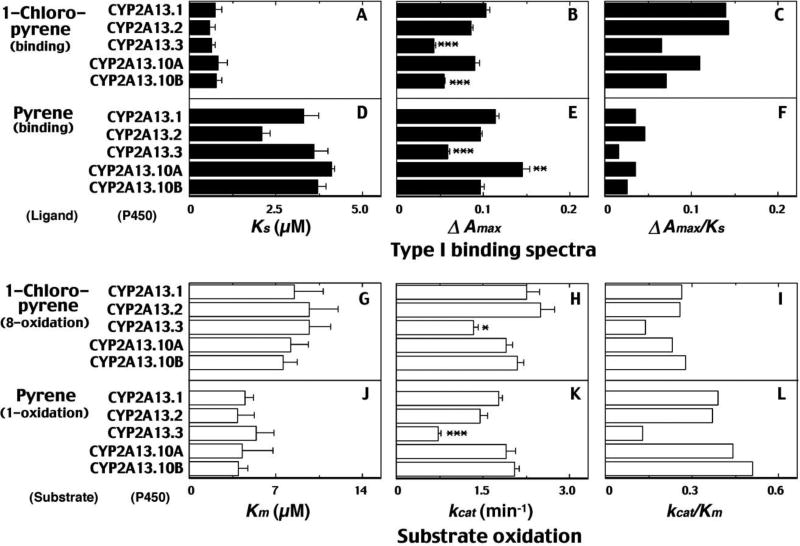
We examined spectral changes for the interaction of 1-chloropyrene and pyrene with five CYP2A13 variants and also the steady-state kinetics of 8-hydroxylation of 1-chloropyrene and 1-hydroxylation of pyrene by these P450 enzymes (Fig. 7). 1-Chloropyrene produced Type I spectral changes with all of the CYP2A13 variants, with lower ΔAmax values for CYP2A13.3 (P<0.001) and CYP2A13.10B (P<0.001) than that of CYP2A13.1. CYP2A13.3 also showed a lower ΔAmax value (P<0.001) for the interaction with pyrene than that of CYP2A13.1 (Fig. 7F). The 8-hydroxylation of 1-chloropyrene and 1-hydroxylation of pyrene were slower with CYP2A13.3, with regard to kcat values (P<0.01 in both cases) as compared with those of CYP2A13.1 (Fig. 7).
Fig. 7.
Type I binding of 1-chloropyrene and pyrene with CYP2A13 variants (parts A, B, and C for 1-chloropyrene and parts D, E, and F for pyrene) and steady-state kinetic catalytic activities of 1-chloropyrene 8-hydroxylation (parts G, H, and I) and pyrene 1-hydroxylation (parts J, K, and L) by purified preparations of CYP2A13 variants. Spectral binding constants Ks (A and D) and ΔAmax (B and E) values and kinetic parameters Km (parts G and J) (in the range of 1.25, 2.5, 5.0, 10, 20, 40, and 80 µM 1-chloropyrene) and kcat (parts H and K) values (in the range of 2.5, 5.0, 10. 20, and 30 µM 1-chloropyrene) were obtained as described in Materials and methods. Data are presented as means ± S.E. Spectral intensities were obtained by calculating ratio of ΔAmax and Ks (A and D) and catalytic efficiencies were determined by calculating the ratio of kcat to Km. Significantly different from CYP2A13.1 group. *P<0.05, **P<0.01, ***P<0.001.
Comparison of Docking Simulations of Interaction of 1-Chloropyrene with CYP2A13.1 and 2A13.3
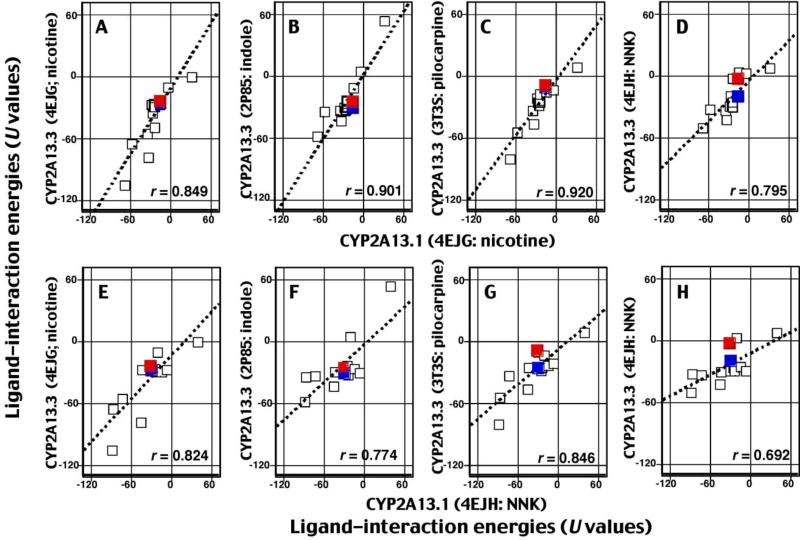
Because the above results suggested that there might be differences in the interaction of 1-chloropyrene (and also pyrene) with CYP2A13.1 and CYP2A13.3 variants, we further compared ligand-interaction energies (U values) for 17 chemicals with CYP2A13.1 and CYP2A13.3, using different CYP2A13 crystal structures (Fig. 8). The chemicals were NNK, indole, pilocarpine, nicotine, coumarin, acenaphthene, acenaphthylene, naphthalene, phenanthrene, biphenyl, pyrene, 1-hydroxypyrene, 1-nitropyrene, 1-acethylpyrene, 1-chloropyrene, 2,5,2´,5´-TCB, and 3,4,3´,4´-TCB, which we have studied in order to identify roles of CYP2A enzymes in their oxidation (Shimada et al., 2015, 2016a, 2016b, 2016c).
Fig. 8.
Effects of genetic variation of CYP2A13 on interaction of 17 chemicals with CYP2A13.1 and 2A13.3. The positions of 1-chloropyrene and pyrene are marked as red squares and blue squares, respectively. Other details are as in the legends to Figs. 5 and 6.
Good correlations of interaction (measurements of U values) were observed for 17 chemicals with CYP2A13.1 and CYP2A13.3 when the former enzyme was modeled using the CYP2A13 crystal structure (PDB) 4EJG (nicotine type) and the latter enzyme with 4EJG (nicotine-type) (Fig. 8A), 2P85 (indole-type) (Fig. 8B), 3T3S (pilocarpine-type) (Fig. 8C), or 4EJH (NNK-type) (Fig. 8D). 1-Chloropyrene (red squares) and pyrene (blue squares) were placed at similar positions among 17 chemicals in the CYP2A13 structures determined. It is interesting to note that the U values of CYP2A13.3 were increased when the CYP2A13 4EJH structure was used; the tangent at the vertical axis was increased (Fig. 8D), as compared with other cases in which the lines intersected at similar positions (Fig. 8A–8C). We also found that there were good correlations of U values using structure 4EJH (NNK-type) for CYP2A13.1 and using the four structures described above for CYP2A13.3 (Fig. 8E–8H). However, the U values of CYP2A13.3 were again increased when 4EJH (NNK-type) was used for analysis (Fig. 8H).
Comaprisons were also made by determining the U values for interaction of 17 chemicals with CYP2A13.1, 2A13.2, and 2A13.3 using different CYP2A13 crystal structures (Supplementary Figure 1). In all cases, there were good correlations between these combinations and the best one (r = 0.978) was found when CYP2A13.2 and CYP2A13.3 were compared using one CYP2A13 crystal structure (4EJG, nicotine-type) for the model (Supplementary Figure 1E). It was also evident that U values in CYP2A13.3 were higher when the crystal structure of 4EJH (NNK-type) was used (Supplementary Figure 1D, 1F, 1H, and 1J–L).
Docking Simulation of Interaction of NNK and 1-Chloropyrene with CYP2A13.1 and CYP2A13.3
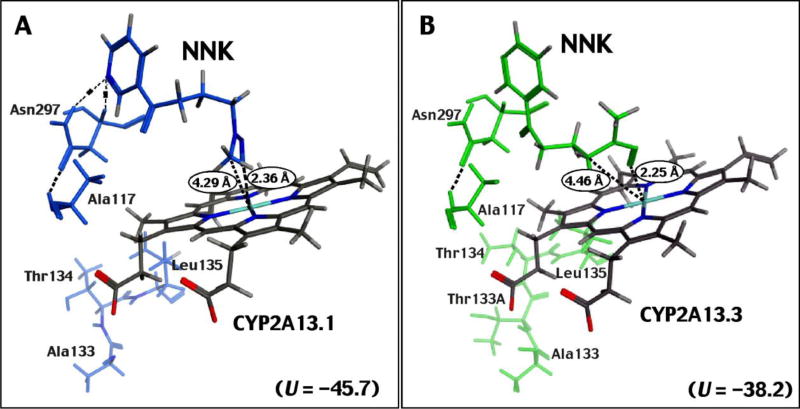
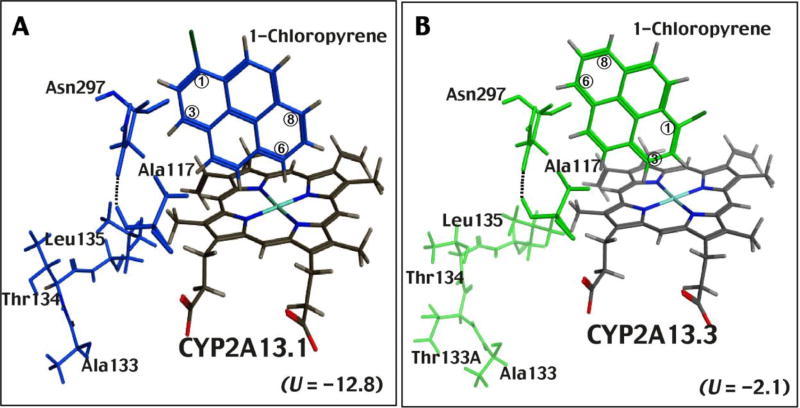
These results suggest differences in the interaction of NNK and 1-chloropyrene with CYP2A13.1 and 2A13.3 when the crystal structure CYP2A13 4EJG (NNK-type) is used for docking analysis. In fact, we found that insertion of Thr134 (133_134Thr) into CYP2A13.3 caused changes in the C-helix, compared with those in CYP2A13.1, in the absence of ligand (Supplementary Figure 2). In the presence of the ligand NNK, the Thr134 insertion in CYP2A13.3 variant significantly changed the region around the C-helix, compared with that of CYP2A13.1 (Fig. 9A and Supplementary Figure 3A) and resulted in structural changes near Asn297 in the I-helix (Figs. 9B and Supplementary Figure 3B). A hydrogen bond interaction between Asn297 and the pyridine ring nitrogen of NNK, seen in CYP2A13.1 (Fig. 9A and Supplementary Figure 3A), was lost in CYP2A13.3 (Fig. 9B and Supplementary Figure 3B) and we also found that there was no apparent interaction between the NNK methyl group and the active site of CYP2A13.3 (Fig. 9B and Supplementary Figure 3B), which has been reported to be involved in α-hydroxylation of NNK, resulting in the formaton of reactive products that bind covalently to DNA (Hecht et al., 2008). In addition, an insertion of Thr134 was also found to cause changes near the C-helix and changes at Asn297 in the I-helix, resulting in conformational changes and in the interaction of 1-chloropyrene and active site of CYP2A13.3 (Fig. 10B and Supplementary Figure 3B), as compared with those of CYP2A13.1 (Fig. 10A and Supplementary Figure 3C). The C6- and C8-positions of 1-chloropyrene were found to be far from the active site of CYP2A13.3, as compared with those of CYP2A13.1 (Fig. 10 and Supplementary Figure 3).
Fig. 9.
Effects of 133_134Thr insertion on the molecular interaction of NNK with CYP2A13.1 (A) and CYP2A13.3 (B) in the C-helix area, I-helix area, and active sites of P450 enzymes. The crystal structure of CYP2A13 4EJH (NNK-type) was used for modelling.
Fig. 10.
Effects of the 133_134Thr insertion on the molecular interaction of 1-chloropyrene with CYP2A13.1 (A) and CYP2A13.3 (B) in the C-helix area, I-helix area, and active sites of P450 enzymes. The crystal structure of CYP2A13 4EJH (NNK-type) was used for modelling.
Discussion
Our present results show that individual forms of human P450 enzymes have different but overlapping specificities in the oxidation of 1-chloropyrene at the 8-, 6-, and 3-positions. CYP1A1 preferentially oxidizes 1-chloropyrene at the 8- and 6-positions, while CYP1B1.1 and CYP1B1.3 catalyze the 6-hydroxylation reaction at similar rate to CYP1A1. CYP1A2 was slow in catalyzing 1-chloropyrene hydroxylation as compared with other CYP1A1 and CYP1B1 enzymes. 1-Chloropyrene was oxidized by CYP2A13 to form 8- and 3-hydroxy products more rapidly than CYP2A6, while the latter enzyme formed more 6-hydroxy product than CYP2A13. Molecular docking analysis supports the view that CYP1A1 and CYP1B1 enzymes interact with 1-chloropyrene in somewhat different orientations than CYP2A13 and CYP2A6, suggesting roles for these P450 enzymes in the oxidation of 1-chloropyrene at different carbon positions. Ligand-interaction energies (U values) obtained with 10 ligands indicate that there are good correlations between the U values of CYP2A13 and CYP2A6 and of CYP1A1 and CYP1B1.
1-Chloropyrene induced Type I binding spectra on interaction with CYP2A13 (Ks = 0.74 µM; Amax = 0.103) more than with CYP2A6 (Ks = 2.9 µM; Amax = 0.010), and oxidation by these P450 enzymes formed 3-, 6-, and 8-hydroxylated products at different rates. Our previous studies have shown that pyrene, 1-hydroxypyrene, 1-nitropyrene, and 1-acetylpyrene all induced Type I binding spectra with CYP2A13, with Ks values of 0.81, 5.0, 0.23, and 0.34 µM, respectively, although these chemicals do not show detectable Type I binding spectra with CYP2A6 (Shimada et al., 2016a). Although CYP2A13 has been shown to oxidize pyrene and its derivatives to several mono- and di-hydroxylated products, at different rates, CYP2A6 was also active in catalyzing these reactions, at rates equal to or more slowly than CYP2A13 in these reactions. These results indicate that the spectral intensities of Type I binding specta are not always correlated to rates of oxidation of 1-chloropyrene and related chmicals, such as pyrene and its derivatives, by these CYP2A enzymes (Shimada et al., 2016a). These results indicate that interaction of 1-chloropyrene and pyrene (and its derivatives) with active sites of CYP2A13 (and its variants) and 2A6 might be different as suggested by molecular docking analysis with 1-chloropyrene in this study and pyrene (and its derivatives) in our previous work (Shimada et al., 2017a).
In the present study, we used X-ray crystal structures of CYP2A13.1 for the comparison of interaction of various ligands (including 1-chloropyrene) with active sites of other CYP2A13 variants in molecular docking analysis, because none of the crystal structures of these CYP2A13 variants have been reported. In our previous work using various CYP2A13 and CYP2A6 crystal structures bound to several ligands, such as coumarin, nicotine, pilocarpine, indole, and NNK (Shimada et al., 2016b), we found positive correlations in ligand-interaction energies (U values) using various CYP2A ligands with several CYP2A13 and CYP2A6 crystal structures (DeVore et. al., 2009; 2012b; DeVore and Scott, 2012a; Yano et al., 2005). The usefulness of U-values for the studies of structure-function relationships of various ligands was also supported in this study, in that there were positive correlations between the U-values of CYP2A13 4EJG (nicotine-type) and CYP2A13 2P85 (indole-type) and those of CYP2A6 4EJJ (nicotine-type) and CYP2A6 1Z10 (coumarin-type) with 33 CYP2A ligands, including 1-chloropyrene (Fig. 5). The results also showed that U-values obtained with crystal structures of CYP2A13 and CYP2A6 for 10 ligands did not correlate with those obtained with the crystal structures of CYP1A1 and CYP1B1, although the values showed good correlation for CYP1A1 and CYP1B1 (Fig. 6). We also found that there were good correlations of U-values for the interaction of 17 CYP2A ligands with five CYP2A13 variants when analyzed with the reported crystal structures of CYP2A13.1 bound to ligands (Fig. 8). Interestingly, the positions of 1-chloropyrene and pyrene were similar at the centers of 17 ligands examined (Fig. 8 and Supplementary Figure 1). These results suggest the use of these X-ray crystal structures of CYP2A13.1 for the studies of structure-function relationships for other CYP2A13 variants; however, ultimately the X-ray crystal structures of CYP2A13 variants should be determined as well.
We compared catalytic activities of five CYP2A13 variants, namely CYP2A13.1 (wild-type), CYP2A13.2 (R257C), CYP2A13.3 (133_134Thr insertion), CYP2A13.10A (I331T), and CYP2A13.10B (R257C: I331T) for the oxidation of 1-chloropyrene. CYP2A13.3 showed lower activity for the oxidaiton of 1-chloropyrene and pyrene as compared with wild-type CYP2A13.1 and the other variants, i.e. CYP2A13.2, CYP2A13.10A, and CYP2A13.10B. The intensities of Type I binding spectra in the interaction of 1-chloropyrene and pyrene suggested lower activities for oxidation of these chemicals by CYP2A13.3. Molecular docking analysis suggested different orientations of 1-chloropyrene in CYP2A13.1 and 2A13.3, utilizing a reported crystal structure of CYP2A13 4EJG (NNK-bound form). The NNK pyridine ring interacts through hydrogen bonding with Asn297 of CYP2A13.1, and the α-methyl group of the N-nitrosamine is located over the heme in the active site of CYP2A13.1 (DeVore et al., 2012a); however, such interactions were lost in CYP2A13.3 in our molecular docking analysis. Furthermore, our docking studies suggest that an insertion of Thr134 in CYP2A13.3 produces conformational changes in the interaction of 1-chloropyrene with the active site of CYP2A13.3, as compared with CYP2A13.1, and that the C6- and C8-positions of 1-chloropyrene are located far from the heme in the active site of CYP2A13.3, suggesting lower activities for the oxidation of 1-chloropyrene by this variant.
It has been reported that CYP2A13*2 is associated with a substantially reduced risk for lung cancer, with an odds ratio of 0.41 (Wang et al., 2003), and that CYP2A13.2 is 20–40% less active than CYP2A13.1 with the substrates NNK, N-nitrosomethylphenylamine, and N,N-dimethylaniline (D’Agostino et al., 2008). Our present results show that the CYP2A13.2 variant had similar activities for the 8-, 6-, and 3-hydroxylation of 1-chloropyrene as wild type CYP2A13.1. It should, however, be mentioned that our CYP2A13.2 preparation does not contain the additional R25C mutation due to the N-terminal modification used for heterologous expression; the native form contains R25C as well as a R257C mutation (D’Agostino et al., 2008; Tamaki et al., 2011; Zhang et al., 2002).
Genetic variations of the human CYP1B1 gene have been reported to show individual differences in susceptibilities to toxic and carcinogenic effects caused by environmental chemicals including PAHs (Watanabe et al., 2000; Aklillu et al., 2005; Shimada, 2006; 2017). Several reports have shown that the CYP1B1.3 genetic variant L432V is associated with increased risk of prostate, colorectal, and urinary system cancers (Yang et al., 2012; Cui et al., 2012; Xie et al., 2012; Liu et al., 2014). In this study, we compared activities of oxidation of 1-chloropyrene by CYP1B1.1 and CYP1B1.3 variants and found similar activities in the oxidation of 1-chloropyrene by these enzymes, supporting our previous studies of metabolism of various procarcinogens and estradiol (Shimada et al., 1999; 2001).
In conclusion, human CYP1A1, CYP1B1, CYP2A13, and CYP2A6 catalyzed the oxidation of 1-chloropyrene at the 8-, 6-, and 3- positions. CYP1A1 preferentially oxidized 1-chloropyrene at the 8- and 6-positions and CYP1B1 catalyzed the 6-hydroxylation reaction at a much higher rate than at the 8- and 3-positions. CYP2A13, on the other hand, catalyzed 8-hydroxylation at a much higher rate than the 6- and 3-hydroxylation reactions, while CYP2A6 catalyzed 6-hydroxylation at a high rate. The CYP1B1.1 and CYP1B1.3 variants had similar catalytic rates; however, the CYP2A13.3 variant showed decreased catalytic activity in the oxidation of 1-chloropyrene, with a lower kcat value than wild-type CYP2A13.1. Molecular docking analysis suggested that a 133_134Thr insertion in CYP2A13.3 affects the orientation of interaction of 1-chloropyrene with the active site heme when a crystal structure of CYP2A13 4EJH (NNK-type) was used for the simulation. The molecular docking studies suggested that mutation of CYP2A13.3 with the 133_134Thr insertion affected both orientation at Asn297, leading to altered interaction of NNK as a result of hydrogen bonding and the interaction of NNK α-methyl and methylene groups with the P450 heme. Apparent effects of Thr134 insertion of CYP2A13.3 on NADPH-P450 reductase interactions may not be ruled out, because of decreased kcat values with the similar Km values. However, because ΔAmax values for 1-chloropyrene and pyrene were also significantly reduced as indicated in Figure 7B and 7E, chemical-P450 binding interactions should be one of main causal factors in the present results. The usefulness of molecular docking analysis is consistent with our previous work on other P450s and ligands (Shimada et al., 2016b; 2016c).
Supplementary Material
Acknowledgments
This study was supported in part by JSPS KAKENHI [Grant Number 15K07770] (to S. T.), [16K09119] (to K. K.), and [17K08425] (to H. Y.), National Research Foundation of Korea [NRF-2016R1D1A1B03932002] (to D. K.), and United States Public Health Service grant [R01 GM118122] (to F. P. G.).
ABBREVIATIONS
- P450 or CYP
cytochrome P450
- b5
cytochrome b5
- PAH
polycyclic aromatic hydrocarbon
- PDB
Protein Data Bank
- NNK
4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone
- TCB
tetrachlorobiphenyl
- 2EN
2-ethynylnaphthalene
- 1NMPE
1-naphthalene methyl propargyl ether
- 1NEPE
1-naphthalene ethyl propargyl ether
- 2NPE
2-naphthalene propargyl ether
- 2NMPE
2-naphthalene methyl propargyl ether
- 2NEPE
2-naphthalene ethyl propargyl ether
- 2EPh
2-ethynylphenanthrene
- 3EPh
3-ethynylphenanthrene
- 9EPh
9-ethynylphenanthrene
- 2PPh
2-(1-propynyl)phenanthrene
- 3PPh
3-(1-propynyl)phenanthrene
- 9PPh
9-(1-propynyl)phenanthrene
- 4EB
4-ethynylbiphenyl
- 4PB
4-propynylbiphenyl
- 2BPE
2-biphenyl propargyl ether
- 4BPE
4-biphenyl propargyl ether
- TCB
tetrachlorobiphenyl.
Footnotes
Declaration of interest
The authors are responsible for the content and writing the article and report no declarations of interest.
References
- Aklillu E, Øvrebø S, Botnen IV, Otter C, Ingelman-Sundberg M. Characterization of common CYP1B1 variants with different capacity for benzo[a]pyrene-7,8-dihydrodiol epoxide formation from benzo[a]pyrene. Cancer Res. 2005;65:5105–5111. doi: 10.1158/0008-5472.CAN-05-0113. [DOI] [PubMed] [Google Scholar]
- Bhatia AL, Tausch H, Stehlik G. Mutagenicity of chlorinated polycyclic aromatic compounds. Ecotoxicol Environ Saf. 1987;14:48–55. doi: 10.1016/0147-6513(87)90082-0. [DOI] [PubMed] [Google Scholar]
- Brown RE, Jarvis KL, Hyland KJ. Protein measurement using bicinchoninic acid: elimination of interfering substances. Anal Biochem. 1989;180:136–139. doi: 10.1016/0003-2697(89)90101-2. [DOI] [PubMed] [Google Scholar]
- Colmsjö A, Rannug A, Rannug U. Some chloro derivatives of polynuclear aromatic hydrocarbons are potent mutagens in Salmonella typhimurium. Mutat Res. 1984;135:21–29. doi: 10.1016/0165-1218(84)90144-7. [DOI] [PubMed] [Google Scholar]
- Cui L, Dillehay K, Chen W, Shen D, Dong Z, Li W. Association of the CYP1B1 Leu432Val polymorphism with the risk of prostate cancer: a meta-analysis. Mol Biol Rep. 2012;39:7465–71. doi: 10.1007/s11033-012-1579-y. [DOI] [PubMed] [Google Scholar]
- D'Agostino J, Zhang X, Wu H, Ling G, Wang S, Zhang QY, Liu F, Ding X. Characterization of CYP2A13*2, a variant cytochrome P450 allele previously found to be associated with decreased incidences of lung adenocarcinoma in smokers. Drug Metab Dispos. 2008;36:2316–2323. doi: 10.1124/dmd.108.022822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeVore NM, Smith BD, Wang JL, Lushington GH, Scott EE. Key residues controlling binding of diverse ligands to human cytochrome P450 2A enzymes. Drug Metab Dispos. 2009;37:1319–1327. doi: 10.1124/dmd.109.026765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeVore NM, Scott EE. Nicotine and 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone binding and access channel in human cytochrome P450 2A6 and 2A13 enzymes. J Biol Chem. 2012a;287:26576–26585. doi: 10.1074/jbc.M112.372813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeVore NM, Meneely KM, Bart AG, Stephens ES, Battaile KP, Scott EE. Structural comparison of cytochromes P450 2A6, 2A13, and 2E1 with pilocarpine. FEBS J. 2012b;279:1621–1631. doi: 10.1111/j.1742-4658.2011.08412.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto T, Moriuchi H, Fu X, Ikegawa T, Matsubara T, Chang G, Uno T, Morigaki K, Isshiki K, Imaishi H. The effects of single nucleotide polymorphisms in CYP2A13 on metabolism of 5-methoxypsoralen. Drug Metab Dispos. 2010;38:2110–2116. doi: 10.1124/dmd.110.034553. [DOI] [PubMed] [Google Scholar]
- Gotoh O. Substrate recognition sites in cytochrome P450 family 2 (CYP2) proteins inferred from comparative analyses of amino acid and coding nucleotide sequences. J Biol Chem. 1992;267:83–90. [PubMed] [Google Scholar]
- Guengerich FP. Human cytochrome P450 enzymes. In: Ortiz de Montellano PR, editor. Cytochrome P450: Structure, Mechanism, and Biochemistry. 4. Springer; New York: 2015. pp. 523–785. [Google Scholar]
- Han S, Choi S, Chun YJ, Yun CH, Lee CH, Shin HJ, Na HS, Chung MW, Kim D. Functional characterization of allelic variants of polymorphic human cytochrome P450 2A6 (CYP2A6*5, *7, *8, *18, *19, and *35) Biol Pharm Bull. 2012;35:394–9. doi: 10.1248/bpb.35.394. [DOI] [PubMed] [Google Scholar]
- Kakimoto K, Nagayoshi H, Konishi Y, Kajimura K, Ohura T, Hayakawa K, Toriba A. Atmospheric chlorinated polycyclic aromatic hydrocarbons in East Asia. Chemosphere. 2014;111:40–46. doi: 10.1016/j.chemosphere.2014.03.072. [DOI] [PubMed] [Google Scholar]
- Kakimoto K, Nagayoshi H, Inazumi N, Tani A, Konishi Y, Kajimura K, Ohura T, Nakano T, Tang N, Hayakawa K, Toriba A. Identification and characterization of oxidative metabolites of 1-chloropyrene. Chem Res Toxicol. 2015;28:1728–36. doi: 10.1021/acs.chemrestox.5b00173. [DOI] [PubMed] [Google Scholar]
- Kakimoto K, Nagayoshi H, Konishi Y, Kajimura K, Ohura T, Nakano T, Hata M, Furuuchi M, Tang N, Hayakawa K, Toriba A. Size distribution of chlorinated polycyclic aromatic hydrocarbons in atmospheric particles. Arch Environ Contam Toxicol. 2017;72:58–64. doi: 10.1007/s00244-016-0327-z. [DOI] [PubMed] [Google Scholar]
- Liu Y, Lin CS, Zhang AM, Song H, Fan CC. The CYP1B1 Leu432Val polymorphism and risk of urinary system cancers. Tumour Biol. 2014;35:4719–4725. doi: 10.1007/s13277-014-1617-6. [DOI] [PubMed] [Google Scholar]
- Ma J, Chen Z, Wu M, Feng J, Horii Y, Ohura T, Kannan K. Airborne PM2.5/PM10-associated chlorinated polycyclic aromatic hydrocarbons and their parent compounds in a suburban area in Shanghai, China. Environ Sci Technol. 2013;47:7615–23. doi: 10.1021/es400338h. [DOI] [PubMed] [Google Scholar]
- Ohura T, Miwa M. Photochlorination of polycyclic aromatic hydrocarbons in acidic brine solution. Bull Environ Contam Toxicol. 2016;96:524–529. doi: 10.1007/s00128-015-1723-1. [DOI] [PubMed] [Google Scholar]
- Ohura T, Kitazawa A, Amagai T. Seasonal variability of 1-chloropyrene on atmospheric particles and photostability in toluene. Chemosphere. 2004;57:831–837. doi: 10.1016/j.chemosphere.2004.08.069. [DOI] [PubMed] [Google Scholar]
- Ohura T, Kitazawa A, Amagai T, Makino M. Occurrence, profiles, and photostabilities of chlorinated polycyclic aromatic hydrocarbons associated with particulates in urban air. Environ Sci Technol. 2005;39:85–91. [PubMed] [Google Scholar]
- Ohura T, Fujima S, Amagai T, Shinomiya M. Chlorinated polycyclic aromatic hydrocarbons in the atmosphere: seasonal levels, gas-particle partitioning, and origin. Environ Sci Technol. 2008;42:3296–3302. doi: 10.1021/es703068n. [DOI] [PubMed] [Google Scholar]
- Ohura T, Morita M, Makino M, Amagai T, Shimoi K. Aryl hydrocarbon receptor-mediated effects of chlorinated polycyclic aromatic hydrocarbons. Chem Res Toxicol. 2007;20:1237–1241. doi: 10.1021/tx700148b. [DOI] [PubMed] [Google Scholar]
- Omura T, Sato R. The carbon monoxide-binding pigment of liver microsomes. I. Evidence for its hemoprotein nature. J Biol Chem. 1964;239:2370–2378. [PubMed] [Google Scholar]
- Parikh A, Gillam EMJ, Guengerich FP. Drug metabolism by Escherichia coli expressing human cytochromes P450. Nat Biotechnol. 1997;15:784–788. doi: 10.1038/nbt0897-784. [DOI] [PubMed] [Google Scholar]
- Sandhu P, Baba T, Guengerich FP. Expression of modified cytochrome P450 2C10 (2C9) in Escherichia coli, purification, and reconstitution of catalytic activity. Arch Biochem Biophys. 1993;306:443–450. doi: 10.1006/abbi.1993.1536. [DOI] [PubMed] [Google Scholar]
- Sandhu P, Guo Z, Baba T, Martin MV, Tukey RH, Guengerich FP. Expression of modified human cytochrome P450 1A2 in Escherichia coli: Stabilization, purification, spectral characterization, and catalytic activities of the enzyme. Arch Biochem Biophys. 1994;309:168–177. doi: 10.1006/abbi.1994.1099. [DOI] [PubMed] [Google Scholar]
- Shiraishi H, Pilkington NH, Otsuki A, Fuwa K. Occurrence of chlorinated polynuclear aromatic hydrocarbons in tap water. Environ Sci Technol. 1985;19:585–590. doi: 10.1021/es00137a001. [DOI] [PubMed] [Google Scholar]
- Shimada T. Xenobiotic-metabolizing enzymes involved in activation and detoxification of carcinogenic polycyclic aromatic hydrocarbons. Drug Metab Pharmacokinet. 2006;21:257–76. doi: 10.2133/dmpk.21.257. [DOI] [PubMed] [Google Scholar]
- Shimada T. Inhibition of carcinogen-activating cytochrome P450 enzymes by xenobiotic chemicals in relation to antimutagenicity and anticarcinogenicity. Toxicol Res. 2017;33:79–96. doi: 10.5487/TR.2017.33.2.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada T, Watanabe J, Kawajiri K, Sutter TR, Guengerich FP, Gillam EMJ, Inoue K. Catalytic properties of polymorphic human cytochrome P450 1B1 variants. Carcinogenesis. 1999;20:1607–1613. doi: 10.1093/carcin/20.8.1607. [DOI] [PubMed] [Google Scholar]
- Shimada T, Tsumura F, Gillam EM, Guengerich FP, Inoue K. Roles of NADPH-P450 reductase in the O-deethylation of 7-ethoxycoumarin by recombinant human cytochrome P450 1B1 variants in Escherichia coli. Protein Expr Purif. 2000;20:73–80. doi: 10.1006/prep.2000.1302. [DOI] [PubMed] [Google Scholar]
- Shimada T, Watanabe J, Guengerich FP, Inoue K, Gillam EMJ. Specificity of 17β-estradiol and benzo[a]pyrene oxidations by polymorphic human cytochrome P450 1B1 variants substituted at residues 48, 119 and 432. Xenobiotica. 2001;31:163–176. doi: 10.1080/00498250110043490. [DOI] [PubMed] [Google Scholar]
- Shimada T, Oda Y, Gillam EMJ, Guengerich FP, Inoue Metabolic activation of polycyclic aromatic hydrocarbons and other procarcinogens by cytochromes P450 1A1 and P450 1B1 allelic variants and othwe human cytochromes P450 in Salmonella typhimurium NM2009. Drug Metab Dispos. 2001;29:1176–1182. [PubMed] [Google Scholar]
- Shimada T, Tanaka K, Takenaka S, Foroozesh MK, Murayama N, Yamazaki H, Guengerich FP, Komori M. Reverse type I binding spectra of human cytochrome P450 1B1 induced by derivatives of flavonoids, stilbenes, pyrenes, naphthalenes, phenanthrenes, and biphenyls that Inhibit catalytic activity: Structure-function relationships. Chem Res Toxicol. 2009;22:1325–1333. doi: 10.1021/tx900127s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada T, Tanaka K, Takenaka S, Murayama N, Martin MV, Foroozesh MK, Yamazaki H, Guengerich FP, Komori M. Structure-function relationships of inhibition of human cytochromes P450 1A1, 1A2, 1B1, 2C9, and 3A4 by 33 flavonoid derivatives. Chem Res Toxicol. 2010;23:1921–1935. doi: 10.1021/tx100286d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada T, Murayama N, Tanaka K, Takenaka S, Guengerich FP, Yamazaki H, Komori M. Spectral modification and catalytic inhibition of human cytochromes P450 1A1, 1A2, 1B1, 2A6, and 2A13 by four chemopreventive organoselenium compounds. Chem Res Toxicol. 2011;24:1327–1337. doi: 10.1021/tx200218u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada T, Kim D, Murayama N, Tanaka K, Takenaka S, Nagy LD, Folkman LM, Foroozesh MK, Komori M, Yamazaki H, Guengerich FP. Binding of diverse environmental chemicals with human cytochromes P450 2A13, 2A6, and 1B1 and enzyme inhibition. Chem Res Toxicol. 2013;26:517–528. doi: 10.1021/tx300492j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada T, Takenaka S, Murayama N, Yamazaki H, Kim JH, Kim D, Yoshimoto FK, Guengerich FP, Komori M. Oxidation of acenaphthene and acenaphthylene by human cytochrome P450 enzymes. Chem Res Toxicol. 2015;28:268–278. doi: 10.1021/tx500505y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada T, Takenaka S, Murayama N, Kramlinger VM, Kim JH, Kim D, Liu J, Foroozesh MK, Yamazaki H, Guengerich FP, Komori M. Oxidation of pyrene, 1-hydroxypyrene, 1-nitropyrene and 1-acetylpyrene by human cytochrome P450 2A13. Xenobiotica. 2016a;46:211–224. doi: 10.3109/00498254.2015.1069419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada T, Takenaka S, Kakimoto K, Murayama N, Lim YR, Kim D, Foroozesh MK, Yamazaki H, Guengerich FP, Komori M. Structure-function studies of naphthalene, phenanthrene, biphenyl, and their derivatives in interaction with and oxidation by cytochromes P450 2A13 and 2A6. Chem. Res. Toxicol. 2016b;29:1029–1040. doi: 10.1021/acs.chemrestox.6b00083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada T, Kakimoto K, Takenaka S, Koga N, Uehara S, Murayama N, Yamazaki H, Kim D, Guengerich FP, Komori M. Roles of human CYP2A6 and monkey CYP2A24 and 2A26 cytochrome P450 enzymes in the oxidation of 2,5,2´,5´-tetrachlorobiphenyl. Drug Metab Disp. 2016c;44:1899–1909. doi: 10.1124/dmd.116.072991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith BD, Sanders JL, Porubsky PR, Lushington GH, Stout CD, Scott EE. Structure of the human lung cytochrome P450 2A13. J Biol Chem. 2007;282:17306–17313. doi: 10.1074/jbc.M702361200. [DOI] [PubMed] [Google Scholar]
- Sugiyama H, Katagiri Y, Kaneko M, Watanabe T, Hirayama T. Chlorination of pyrene in soil components with sodium chloride under xenon irradiation. Chemosphere. 1999;38:1937–1945. [Google Scholar]
- Tamaki Y, Honda M, Muroi Y, Arai T, Sugimura H, Matsubara Y, Kanno S, Ishikawa M, Hirasawa N, Hiratsuka M. Novel single nucleotide polymorphism of the CYP2A13 gene in Japanese individuals. Drug Metab Pharmacokinet. 2011;226:544–547. doi: 10.2133/dmpk.dmpk-11-sc-033. [DOI] [PubMed] [Google Scholar]
- Wang H, Tan W, Hao B, Miao X, Zhou G, He F, Lin D. Substantial reduction in risk of lung adenocarcinoma associated with genetic polymorphism in CYP2A13, the most active cytochrome P450 for the metabolic activation of tobacco-specific carcinogen NNK. Cancer Res. 2003;63:8057–61. [PubMed] [Google Scholar]
- Wang A, Savas U, Stout CD, Johnson EF. Structural characterization of the complex between α-naphthoflavone and human cytochrome P450 1B1. J Biol Chem. 2011;286:5736–43. doi: 10.1074/jbc.M110.204420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh AA, Szklarz GD, Scott EE. Human cytochrome P450 1A1 structure and utility in understanding drug and xenobiotic metabolism. J Biol Chem. 2013;288:12932–12943. doi: 10.1074/jbc.M113.452953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe J, Shimada T, Gillam EMJ, Ikuta T, Suemasu K, Higashi Y, Gotoh O, Kawajiri K. Association of CYP1B1 genetic polymorphism with incidence to breast and lung cancer. Pharmacogenetics. 2000;10:25–33. doi: 10.1097/00008571-200002000-00004. [DOI] [PubMed] [Google Scholar]
- Xie Y, Liu GQ, Miao XY, Liu Y, Zhou W, Zhong DW. CYP1B1 Leu432Val polymorphism and colorectal cancer risk among Caucasians: a meta-analysis. Tumour Biol. 2012;33:809–816. doi: 10.1007/s13277-011-0298-7. [DOI] [PubMed] [Google Scholar]
- Yamazaki H, Nakamura M, Komatsu T, Ohyama K, Hatanaka N, Asahi S, Shimada N, Guengerich FP, Shimada T, Nakajima M, Yokoi T. Roles of NADPH-P450 reductase and apo- and holo-cytochrome b5 on xenobiotic oxidations catalyzed by 12 recombinant human cytochrome P450s expressed in membranes of Escherichia coli. Protein Expr Purif. 2002;24:329–337. doi: 10.1006/prep.2001.1578. [DOI] [PubMed] [Google Scholar]
- Yang J, Xu DL, Lu Q, Han ZJ, Tao J, Lu P, Wang C, Di XK, Gu M. Prostate cancer risk and aggressiveness associated with the CYP1B1 4326C/G (Leu432Val) polymorphism: a meta-analysis of 2788 cases and 2968 controls. Asian J Androl. 2012;14:560–5. doi: 10.1038/aja.2012.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano JK, Hsu MH, Griffin KJ, Stout CD, Johnson EF. Structures of human microsomal cytochrome P450 2A6 complexed with coumarin and methoxsalen. Nat Struct Mol Biol. 2005;12:822–823. doi: 10.1038/nsmb971. [DOI] [PubMed] [Google Scholar]
- Zhang X, Su T, Zhang QY, Gu J, Caggana M, Li H, Ding X. Genetic polymorphisms of the human CYP2A13 gene: Identification of single-nucleotidepolymorphisms and functional characterization of an Arg257Cys variant. J Pharmacol Exp Ther. 2002;302:416–423. doi: 10.1124/jpet.302.2.416. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.