Abstract
Arrhythmogenic cardiomyopathy (AC) is most commonly characterized as a disease of the intercalated disc that promotes abnormal cardiac conduction. Previously, arrhythmogenic cardiomyopathy was frequently referred to as arrhythmogenic right ventricular cardiomyopathy/dysplasia (ARVC/D); however, genotype–phenotype studies have defined a broader phenotypic spectrum; with the identification of left-dominant and biventricular subtypes. Molecular insight into AC has primarily focused on mutations in desmosomal proteins and the downstream signaling pathways; however, desmosomal gene mutations can only be identified in approximately 50% of patients with AC. Animal and cellular studies have shown that in addition to abnormal biomechanical properties from changes in desmosome function, crosstalk from the desmosome to the nucleus, gap junctions, and ion channels are implicated in the pathobiology of AC. In this review, we highlight some of the newly identified genetic and epigenetic mechanisms that may lead to the development of AC including the role of the Hippo pathway and microRNAs.
Keywords: Arrhythmogenic cardiomyopathy, Hippo, MicroRNA, Desmosome, Single nucleotide polymorphism
1. Introduction
Arrhythmogenic cardiomyopathy (AC), which is commonly referred to as arrhythmogenic right ventricular cardiomyopathy/dysplasia (ARVC/D), is defined as a disease of the intercalated disc that promotes abnormal cardiac conduction, sudden cardiac death, and heart failure [1,2]. Recently, clinical definitions of AC have been refined to encompass a broader phenotypic spectrum; including left-dominant and biventricular subtypes [3,4]. AC is clinically distinguishable from idiopathic dilated cardiomyopathy (DCM) as it presents an arrhythmogenic burden that exceeds the degree of cardiac dysfunction defined by morphology, histology, and ejection fraction [5]. However, recent clinical evidence has determined that patients with atypical AC disease phenotypes are often misdiagnosed with DCM [6,7]. AC is diagnosed clinically using electrocardiogram (ECG) and morphological recordings in conjunction with genetic screening and familial disease history. Histological evidence for transmural fibrofatty replacement is considered a classical symptom for a definitive diagnosis [8–10]. However, this assessment is not possible in most patients.
AC is typically considered a hereditary cardiomyopathy as approximately half of all AC cases can be attributed to known genetic mutations. The most common mutations occur in genes encoding desmosomal proteins, which result in desmosomal dysfunction [11]. In the heart, desmosomes are critical protein complexes localized at the intercalated disc that facilitate strong myocyte–myocyte interactions. Furthermore, desmosomal proteins associate with fascia adherens proteins to provide stability and mechanical continuity throughout the myocardium. Gap junctions, which are essential for fast intermyocyte electrical propagation of the action potential, also form interactions with desmosomes at the intercalated disc [12]. Growing evidence suggests that desmosomal dysfunction directly contributes to decreased connexin expression at the intercalated disc [13–15].
Molecular mechanisms that are attributed to cardiac arrhythmias involve the dysregulation of ion channels and pumps, abnormal intracellular Ca2+ handling, and disrupted cell–cell electrical coupling. Dysfunction of the intercalated disc as well as fibrosis and adipose infiltration contribute to slowed or blocked cell-cell electrical coupling within the ventricular myocardium. This gives rise to conditions favorable to re-entrant circuits and arrhythmogenesis [16]. The majority of known genetic mutations in humans that directly contribute to dysfunction of the intercalated disc and the AC phenotype include: Plakophilin-2, Desmoplakin, Desmoglein-2, Desmocollin-2, Plakoglobin, and αT-catenin. Other genetic mutations that promote AC have also been described in the extra-desmosomal proteins TMEM43 and TGFβ3, which have been shown to indirectly affect intercalated disc function (Table 1) [17]. As these genetic mechanisms have been reviewed extensively [1,4,18,19], these will not be included in this review as to focus on recent findings.
Table 1.
Genes implicated in arrhythmogenic cardiomyopathy.
| Gene | Protein | Function | Phenotype |
|---|---|---|---|
| JUP | Plakoglobin | Desmosomal protein | ARVD-cutaneous syndrome |
| DSP | Desmoplakin | Desmosomal protein | ARVD-cutaneous syndrome |
| PKP2 | Plakophilin-2 | Desmosomal protein | ARVD |
| DSG2 | Desmoglein-2 | Desmosomal protein | ARVD |
| DSC2 | Desmocollin-2 | Desmosomal protein | ARVD |
| RYR2 | Ryanodine receptor 2 | SR calcium channel | CPVT |
| DES | Desmin | Intermediate filament | DCM/HCM, conduction anomaly |
| TTN | Titin | Sacromeric protein | DCM, conduction anomaly |
| TMEM43 | Transmembrane protein 43 (LUMA) | Nuclear membrane protein | ARVD |
| TGFβ3 | Transforming growth factor β3 | Cytokine | ARVD |
To date, research has focused primarily on the genetic factors underlying AC pathogenesis. Since a number of the defined mutations that yield an AC phenotype are frame-shift or non-sense mutations, haploinsufficiency of desmosomal proteins is suggested as an underlying mechanism of AC [20,21]. Moreover, instances of AC patients with and without known genetic mutations have shown evidence for downregulation of desmosomal proteins [22,23]. This suggests that other mechanisms, potentially epigenetic factors, may also contribute to desmosomal dysfunction and an AC phenotype.
2. Hippo pathway in arrhythmogenic cardiomyopathy
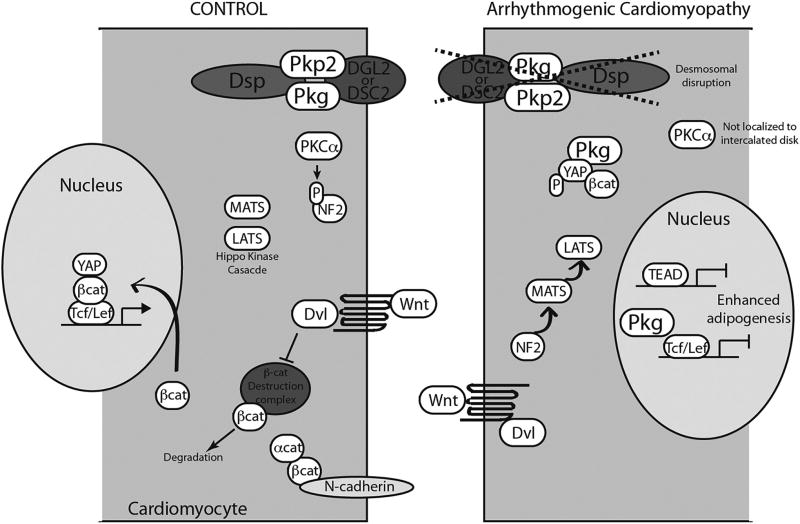
The histopathological characteristics of AC include adipose infiltration, fibrosis, and increased cell death [24,25]. These features, which are likely a secondary effect of intercalated disc dysfunction, also contribute to impaired cell-cell conduction and arrhythmogenesis. Several studies have sought to discover how changes to the desmosome could lead to fibro-fatty replacement of myocytes and investigated changes in cell signaling. In mutations resulting in AC, it has been noted that there is reduced plakoglobin localization to IDs [22]. Garcia-Gras et al. showed suppression of desmoplakin led to nuclear localization of plakoglobin and a 2-fold reduction in canonical Wnt/β-catenin signaling through Tcf/Lef1 transcription factors. Nuclear plakoglobin was shown to suppress canonical Wnt signaling and thereby enhance adipogenesis driven by PPARγ and C/EBPα. Recently, Chen et al. implicated the Hippo/YAP and its link to the canonical Wnt/β-catenin pathways to the pathogenesis of AC [26]. The Hippo pathway plays a key role in the regulation of organ growth and cellular proliferation, and regulates cardiomyocyte proliferation and heart size. Both pathways play critical roles in tissue development and homeostasis by regulating cell growth, differentiation, and proliferation [27–30]. Recent studies have described cross-regulation between these two pathways, which suggests that an important negative feedback role exists for Hippo/YAP in the Wnt/β-catenin pathway. YAP (Yes-associated protein) is known to interact with β-catenin to prevent its nuclear localization, thereby inhibiting Wnt signaling [31]. Two of the major upstream stimuli regulating the activity of the Hippo pathway are cell adhesion and polarity [27]. In the cardiomyocyte, not only is cell adhesion at the intercalated disc essential for mechanical continuity and electrical propagation throughout the myocardium, but it also serves as a critical focus for cell signaling. Important cell signaling molecules that scaffold here are β-catenin and neurofibromin 2 (NF2), which are important upstream molecules in the Wnt and Hippo pathways, respectively [26,32]. Work by Chen et al. described a mechanism by which disruption of the desmosome in human AC patients or mouse and cell models of AC causes activation of NF2 and subsequent activation of downstream Hippo pathway kinases, MATS and LATS (Fig. 1). Elevated levels of phosphorylated YAP, as a result of this Hippo kinase activation, led to sequestration of β-catenin by YAP and inhibition of Wnt signaling resulting in adipogenesis. Furthermore, by knocking down Hippo kinase LATS1/2 in the HL-1 cell line model of AC, adipogenesis was ameliorated. These findings suggest that canonical Wnt inhibition and Hippo activation play an important role in the pathogenesis of AC [26]. It remains unclear whether changes in the Hippo and Wnt/β-catenin pathways are present in all mutations causing AC or if these changes are restricted to specific to any particular genotype. Additionally, this certainly does not clarify mechanisms for without identified desmosome gene mutation.
Fig. 1.
Schematic of Wnt/β-catenin and Hippo pathways in arrhythmogenic cardriomyopathy.
At the intercalated disc, PKCα is localized near the membrane allowing inactivation of NF2 via phosphorylation. YAP phosphorylation state is low and plakoglobin is localized at the ID. Wnt/β-catenin signaling can proceed normally. Disruption of the intercalated disc in AC by a desmosomal mutation effects aberrant signaling via loss of PKCα localization to the submembrane and allowing NF2 activation of the Hippo kinase cascade (including MATS and LATS). Nuclear plakoglobin, perhaps in the form of a complex with β-catenin and YAP, interferes with nuclear β-catenin.
β-cat, β-catenin; Dvl, disheveled; LEF, lymphoid enhancer- binding factor-1; NF2, neurofibromin 2; PKCα, protein kinase C-α isoform; TCF, T-cell factor; and TEAD, TEA domain family member 1; YAP, Yes-associated protein.
3. MicroRNAs
Epigenetic mechanisms, in particular microRNAs, are gaining more attention as potential regulators of the aforementioned molecular mechanisms of arrhythmogenesis [33]. MicroRNAs are small noncoding RNAs whose physiological role is the regulation of gene expression at the level of mRNA translation. Selective binding of microRNAs to complementary target sites of mRNAs functions as repressor of protein translation and, possible destabilization and degradation of the bound mRNA. The complementary nucleotide sequences bound by miRNAs are found within the 3′ untranslated region (3′UTR) or protein-coding regions of mRNA transcripts [33,34]. A majority of miRNA “target sites” are conserved throughout mammalian species, which gives credence to their importance in normal physiological processes. The epigenetic mechanism by which microRNAs control gene expression is imperative in maintaining control of complex genetic pathways. Aberrant microRNA levels are implicated in a number of pathophysiological conditions, including cardiovascular disease.
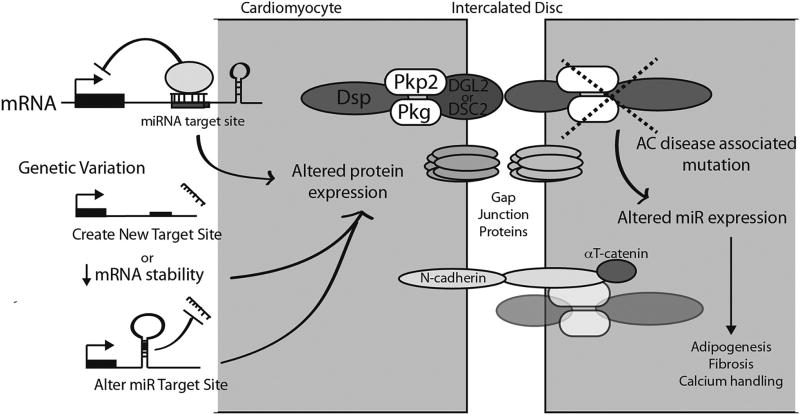
New research now suggests that epigenetic factors contribute to adipogenesis in AC (Fig. 2). Recent work by Gurha et al. investigated epigenetic changes in mouse and cell models of AC. Out of the 750 miRNAs screened for differential expression, 59 were significantly changed in plakophilin 2 deficient HL-1 cells. Of these, the most robust change was the downregulation of mir-184, which was also associated with DNA hypermethylation upstream of miR-184. Data from qPCR results also showed significant reduction in miR-184 levels in hearts of two independent mouse models of AC, suggesting that desmosomal dysfunction was the underlying cause of miR-184 reduction. Analysis of Hippo and Wnt pathways determined that changes in miR-184 expression had no significant effect on transcriptional activities of either pathway or expression levels of their targets. However, overexpressing of miR-184 in plakophilin 2 deficient HL-1 cells caused a significant reduction in adipogenesis. Further investigation identified miR-184 associated changes in the transcript levels of Agpat1 and Agpat3, which are important enzymes involved lipid biosynthesis. These results suggest an important role for miR-184 in the pathogenesis of AC that is likely independent of Hippo and Wnt pathways [35].
Fig. 2.
Potential mechanisms of microRNA mediated development of arrhythmogenic cardriomyopathy.
SNP-associated changes in miRNA target sites located in AC-associated genes 3’UTRs may alter the expression of known proteins implicated in the development of arrhythmogenic cardiomyopathy. Alternatively, changes or mutations in desmosomal proteins may also alter expression of microRNAs important in the development of the myocardial changes seen in arrhythmogenic cardiomyopathy.
PKP2, Plakophillin-2; Pkg, Plakoglobin; DSG2, Desmoglein-2; DSC2, Desmocollin-2; miR, microRNA.
Similarly, a study by Zhang et al. investigated epigenetic changes in AC by profiling miRNA expression level from 24 unrelated end-stage AC patients who underwent cardiac transplant. They hypothesized that miRNAs are involved in the pathophysiology of ARVD and contribute to the fibro-fatty formation. Out of 1078 miRNAs screened, a total of 24 significantly varied microRNAs were identified, with 12 up-regulated microRNAs (miR-21-3p, miR-21-5p, miR-34a-5p, miR-212-3p, miR-216a, miR-584-3p, miR-1251, miR-3621, miR-3674, miR-3692-3p, miR-4286, miR-4301) and 12 down-regulated microRNAs (miR-135b, miR-138-5p, miR-193b-3p, miR-302b-3p, miR-302c-3p, miR-338-3p, miR-451a, miR-491-3p, miR-575, miR-3529-5P, miR-4254, miR-4643). Of these, miR-21 and miR-135b were determined by gene ontology analysis to target multiple genes within the Wnt and Hippo pathways. In contrast to the in vitro HL-1 cell and mouse studies from Gurha et al., the expression level of miR-184 was not significantly different in this selected ARVC patient group. Indeed, the expression profiles found in both studies did not demonstrate any overlap in any single miRNA. Though these results suggest a potential role in miRNA expression and Wnt/HIPPO signaling [36], further study is necessary to determine the exact functional consequences caused by upregulation of miR-21 and downregulation of miR-135b in the pathogenesis of AC and to determine if there is indeed a differential expression signature consistent among the variable phenotypes of AC. It is possible that similar to the highly variable differential microRNA expression profile in cardiomyopathy, AC profiles will be unique.
3.1. Myocarditis and microRNA-21
In addition to the inherited ultrastructural-defect concept underlying AC, cardiomyocyte necrosis seen in AC may also be due to myocarditis. Because the appearance of inflammatory cells are a common finding in pathology studies of hearts with AC, this concept of myocardial inflammation remains under consideration [8]. Viral myocarditis (VM), an underlying cause of DCM and sudden cardiac death, is an infection of the cardiac muscle by cardiotropic viruses that include: adenovirus, enterovirus, and cytomegalovirus [37–39]. Ventricular arrhythmias are a common occurrence with acute VM [40]. Additionally, viral infection is commonly present as a comorbidity with AC [41]. VM alone often leads to cardiomyopathy with clinical symptoms that mimic the electrophysiological and functional abnormalities of AC [42]. Given these commonalities, it has been proposed that VM may provoke or unmask disease symptoms in individuals that are genetically susceptible to AC [43]. Recent studies examining the pathogenesis of VM have described mechanisms responsible for disrupting the integrity of the intercalated disc. These mechanisms are evidenced by dysregulated desmosomes, fascia adherens, and gap junctions as the result of impaired expression of desmin, vinculin, and connexin43, respectively [44,45].
Several miRNAs have been identified as differentially expressed in VM myocardium. While the function of miR-21 has yet to be characterized in AC, recent work has identified an important pathophysiological and potentially arrhythmogenic role for miR-21 in viral myocarditis. In particular, miR-21 has been shown to upregulated in a variety of viral infections including CVB3, an enterovirus known to cause VM [46–48]. Recent work by Ye et al. suggests an underlying role for miR-21 in the pathogenesis of VM. Here, they define an indirect mechanism by which miR-21 mediates degradation of desmin resulting in disrupted desmosome structure. In CVB3 infected myocardium, the deubiquitination enzyme YOD1 was targeted by miR-21 resulting in polyubiquitination of desmin and degradation by the proteasome. Furthermore, the authors provide evidence for miR-21 mediated downregulation of vinculin and a concomitant disruption of fascia adherens. Interstingly, miR-21 has been implicated in the inhibition of apoptosis of cardiac fibroblasts, leading to cardiac hypertrophy and myocardial fibrosis. It has also been implicated in the pathogenesis of aortic aneurysm formation [49]. Altogether, these new findings on miR-21 in myocardidtis define a critical role in the integrity of intercalated disc and the pathogenesis of VM [45].
3.2. MicroRNA-130a
Recently, work from our group demonstrated that overexpression of miR-130a in adult mouse myocardium could induce a highly arrhythmogenic substrate manifested as both inducible and spontaneous ventricular arrhythmias, along with a dilated cardiomyopathy. We demonstrated that miR-130a was a direct target for the myocardial gap junction protein, connexin43 [50]. In addition to Cx43, miR-130a was also found to target the desmosomal protein DSC2. In transgenic hearts, many of the characteristic features of AC were demonstrated with histologic findings of fibrosis and lipid accumulation within both ventricles. Taken together, overexpression of miR-130a in myocardium resulted in a disease phenotype resembling AC and therefore, may serve as potential model for microRNA-induced AC [51]. It remains to be seen whether or not miR-130a expression is altered in human AC.
4. Other relevant epigenetic mechanisms
DNA methylation is the most common epigenetic modification in the mammalian genome and involves the addition of a methyl group to the 5′ carbon of a cytosine by DNMT enzymes and mostly occurs at the CpG (cytosine preceding guanosine) dinucleotide sequences, also known as CpG islands. It is possible that in those patients with no known pathogenic gene mutation, DNA methylation of CpG islands may represent another mechanism for gene regulation in the pathogenesis of AC.
However, to date, this has not been reported in AC. Similarly, modifications to histones and the subsequent remodeling of chromatin have not been studied in the pathogenesis of arrhythmogenic cardiomyopathy. The eukaryotic DNA is tightly compact and organized in chromatin. The nucleosome is the central unit of chromatin and is composed of histone proteins (H2A, H2B, H3, and H4) around which a DNA segment is looped. Amino-terminal tails from each histone protein can undergo a variety of posttranscriptional modifications such as methylation, phosphorylation, acetylation, sumoylation, ubiquitination, ADP-ribosylation, proline isomerization, and deimination. These modifications effect conformational changes in the chromatin allowing for regions of DNA to be accessible or inaccessible for trasnscription and thereby altering gene expression.
Recent studies have found that a large proportion of the genome is transcribed as long noncoding RNAs (lncRNAs), a heterogeneous group of noncoding RNA transcripts greater than 200 nucleotides in length, residing within or between coding genes. Previously thought to be simply degradation products of messenger RNA transcripts, many lncRNAs are functional RNAs and have been found to be involved in regulation of gene expression through transcriptional or posttranscriptional regulatory mechanisms. Unlike microRNAs, lncRNAs are able to confer a complex 3-dimensional structure enabling many of them to bind specific sets of proteins. Additionally, segments of single-strand RNA sequences within lncRNA structures allow them to bind to complementary sequences in the genome. Functionally, lncRNAs are predominately known as regulators of transcription, including epigenetic modification of chromatin structure. However, recent data has implicated lncRNAs as regulators of gene expression using a wide array of mechanisms including pre-mRNA splicing, mRNA degradation, and mRNA translation. lncRNAs differ from miRNAs as their function is not dependent exclusively on sequence complementarity with a target mRNA. lncRNAs can function using sequence complementarity with other nucleic acids and can also function using structure, forming molecular scaffolds for the assembly of macromolecular complexes. Given their recent appearance on our understanding of the transcriptional landscape, there is yet no data on the role of lncRNAs in arrhythmogenic cardiomyopathy.
Although these important epigenetic pathways have not been studied in arrhythmogenic cardiomyopathy, their prevalence in regulation of gene expression in disease states make it a likely contributor and deserves further exploration.
5. The role of variants in arrhythmogenic cardiomyopathy
Genetic variations, such as single nucleotide polymorphisms (SNPs), have the potential to contribute to an individual's susceptibility to developing disease. Disease associated SNPs can affect either coding and non-coding regions of genes, which can ultimately change protein function or gene expression levels. Investigators have determined that SNPs lying within the mRNA transcript could yield a gain or loss of miRNA target sites [52]. In fact, 52% of the known SNPs in the dbSNP database have been defined as creating novel miRNA binding sites [53]. Over the past decade, several pathologies have now described SNP-associated changes in miRNA target sites [54]. It is conceivable that SNPs in regions of miRNA target sites in AC-associated genes may also contribute to the development of the AC phenotype without directly affecting the genetic sequence [53]. Studies are still required to validate this mechanism of disease-associated SNPs in the development of AC. To date, no SNPs identified from human GWAS have been characterized in UTRs of AC-associated gene transcripts.
However, instances of UTR mutations have been identified that associate with the pathogenesis of AC. Work done by Beffagna et al. identified mutations in both the 5′ and 3′ UTR of the TGFβ3 transcript in patients with AC. Using luciferase reporter assays, they determined that either one of these mutations was sufficient to cause a significantly higher translation efficiency. While the 5′ UTR mutation was determined to cause an amino acid substitution in the coding region of TGFβ3, the mechanism by which the 3′ UTR mutation enhanced translation efficiency of TGFβ3 and ultimately an AC phenotype has not yet been characterized [55].
Recent work by Meurs et al. investigated genetic changes in adult boxer dogs with heritable AC. By performing GWAS, they identified an eight base pair deletion that resided in the 3′UTR of the extra-desmosomal gene striatin The presence of the mutation significantly downregulated total striatin expression and its localization at the intercalated disc. With the prediction of change in secondary structure of the striatin transcript, the authors suggest reduced expression of striatin is likely associated with destabilization of the mRNA [[56].].
As clinical genetic testing becomes more routine in clinical practice, the extent of this genetic variation is increasingly being appreciated. It is widely recognized a significant proportion of genetic variation is not relevant for determining health or risk for disease and collectively has been referred to as “genetic noise.” Kapplinger et al. reported background genetic variation in ARVC. They performed a genetic analysis of PKP2, DSP, DSG2, DSC2, and TMEM43 for 92 affected individuals diagnosed with ARVC and 427 control subjects. Not surprisingly, radical mutations resulting in stop codons were identified in 43% of affected individuals versus 0.5% in control subjects. However, the frequency of missense mutations was similar between probands and controls (21% of affected individuals and 16% of controls). Further analysis of ARVC related genes and locations of missense mutations that were highly detected in controls demonstrated that missense mutations in DSP and DSG2, especially at the C-terminals of both genes, were highly detected in controls [57]. In recent years, the expansion of technology, bioinformatics, and scalability of next generation platforms have provided a more in-depth and expansive coverage of the human genetic landscape. Through the development of next-generation sequencing, study of the global molecular changes in the failing myocardium has begun to be explored. As this analysis will eventually be extended to arrhythmogenic cardiomyopathy, separation of the “signal” from the “noise” and distinguishing a diagnostic genetic change from background genetic variation will remain a difficult task for researchers and clinicians.
6. Conclusions
Altered expression of demosomal proteins and dysfunction of the intercalated disc has been the hallmark of AC. However, the characterization of extra-desmosomal proteins, including those involved in downstream signaling pathways, has given new insight into the pathogenesis of AC. The broad phenotypic variability suggests that AC is highly complex and likely influenced by both genetic and epigenetic factors. In some cases, desmosomal genetic mutations have been identified in patients diagnosed with DCM that never met the histopathological criteria for AC [7]. Moreover, clinical diagnosis of AC with evidence of desmosomal dysfunction has occurred in patients not carrying a known genetic mutation [22,23]. Given the intricate nature of cardiomyopathies and arrhythmia, future studies that define common molecular mechanisms responsible for compromising the integrity of the intercalated disc are imperative for development of more comprehensive diagnostic tools and potential novel therapeutics.
Footnotes
Transparency document
The http://dx.doi.org/10.1016/j.bbadis.2017.04.020 associated with this article can be found, in online version.
References
- 1.Calore M, Lorenzon A, De Bortoli M, Poloni G, Rampazzo A. Arrhythmogenic cardiomyopathy: a disease of intercalated discs. Cell Tissue Res. 2015;360(3):491–500. doi: 10.1007/s00441-014-2015-5. [DOI] [PubMed] [Google Scholar]
- 2.Rigato I, Bauce B, Rampazzo A, Zorzi A, Pilichou K, Mazzotti E, et al. Compound and digenic heterozygosity predicts lifetime arrhythmic outcome and sudden cardiac death in desmosomal gene-related arrhythmogenic right ventricular cardiomyopathy. Circ. Cardiovasc. Genet. 2013;6(6):533–542. doi: 10.1161/CIRCGENETICS.113.000288. [DOI] [PubMed] [Google Scholar]
- 3.Saguner AM, Brunckhorst C, Duru F. Arrhythmogenic ventricular cardiomyopathy: a paradigm shift from right to biventricular disease. World J. Cardiol. 2014;6(4):154–174. doi: 10.4330/wjc.v6.i4.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Basso C, Bauce B, Corrado D, Thiene G. Pathophysiology of arrhythmogenic cardiomyopathy. Nat. Rev. Cardiol. 2011;9(4):223–233. doi: 10.1038/nrcardio.2011.173. [DOI] [PubMed] [Google Scholar]
- 5.Sen-Chowdhry S, McKenna WJ. Sudden death from genetic and acquired cardiomyopathies. Circulation. 2012;125(12):1563–1576. doi: 10.1161/CIRCULATIONAHA.111.025528. [DOI] [PubMed] [Google Scholar]
- 6.Sen-Chowdhry S, Syrris P, Prasad SK, Hughes SE, Merrifield R, Ward D, et al. Left-dominant arrhythmogenic cardiomyopathy: an under-recognized clinical entity. J. Am. Coll. Cardiol. 2008;52(25):2175–2187. doi: 10.1016/j.jacc.2008.09.019. [DOI] [PubMed] [Google Scholar]
- 7.Garcia-Pavia P, Syrris P, Salas C, Evans A, Mirelis JG, Cobo-Marcos M, et al. Desmosomal protein gene mutations in patients with idiopathic dilated cardiomyopathy undergoing cardiac transplantation: a clinicopathological study. Heart. 2011;97(21):1744–1752. doi: 10.1136/hrt.2011.227967. [DOI] [PubMed] [Google Scholar]
- 8.Basso C, Thiene G, Corrado D, Angelini A, Nava A, Valente M. Arrhythmogenic right ventricular cardiomyopathy. Dysplasia, dystrophy, or myocarditis? Circulation. 1996;94(5):983–991. doi: 10.1161/01.cir.94.5.983. [DOI] [PubMed] [Google Scholar]
- 9.Basso C, Corrado D, Marcus FI, Nava A, Thiene G. Arrhythmogenic right ventricular cardiomyopathy. Lancet. 2009;373(9671):1289–1300. doi: 10.1016/S0140-6736(09)60256-7. [DOI] [PubMed] [Google Scholar]
- 10.Marcus FI, McKenna WJ, Sherrill D, Basso C, Bauce B, Bluemke DA, et al. Diagnosis of arrhythmogenic right ventricular cardiomyopathy/dysplasia: proposed modification of the task force criteria. Circulation. 2010;121(13):1533–1541. doi: 10.1161/CIRCULATIONAHA.108.840827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Murray B. Arrhythmogenic right ventricular dysplasia/cardiomyopathy (ARVD/C): a review of molecular and clinical literature. J. Genet. Couns. 2012;21(4):494–504. doi: 10.1007/s10897-012-9497-7. [DOI] [PubMed] [Google Scholar]
- 12.Sheikh F, Ross RS, Chen J. Cell-cell connection to cardiac disease. Trends Cardiovasc. Med. 2009;19(6):182–190. doi: 10.1016/j.tcm.2009.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lyon RC, Mezzano V, Wright AT, Pfeiffer E, Chuang J, Banares K, et al. Connexin defects underlie arrhythmogenic right ventricular cardiomyopathy in a novel mouse model. Hum. Mol. Genet. 2014;23(5):1134–1150. doi: 10.1093/hmg/ddt508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kaplan SR, Gard JJ, Protonotarios N, Tsatsopoulou A, Spiliopoulou C, Anastasakis A, et al. Remodeling of myocyte gap junctions in arrhythmogenic right ventricular cardiomyopathy due to a deletion in plakoglobin (Naxos disease) Heart Rhythm. 2004;1(1):3–11. doi: 10.1016/j.hrthm.2004.01.001. [DOI] [PubMed] [Google Scholar]
- 15.Fidler LM, Wilson GJ, Liu F, Cui X, Scherer SW, Taylor GP, et al. Abnormal connexin43 in arrhythmogenic right ventricular cardiomyopathy caused by plakophilin-2 mutations. J. Cell. Mol. Med. 2009;13(10):4219–4228. doi: 10.1111/j.1582-4934.2008.00438.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kleber AG, Saffitz JE. Role of the intercalated disc in cardiac propagation and arrhythmogenesis. Front. Physiol. 2014;5:404. doi: 10.3389/fphys.2014.00404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mizusawa Y. Recent advances in genetic testing and counseling for inherited arrhythmias. J. Arrhythm. 2016;32(5):389–397. doi: 10.1016/j.joa.2015.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Awad MM, Calkins H, Judge DP. Mechanisms of disease: molecular genetics of arrhythmogenic right ventricular dysplasia/cardiomyopathy. Nat. Clin. Pract. Cardiovasc. Med. 2008;5(5):258–267. doi: 10.1038/ncpcardio1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Corrado D, Link MS, Calkins H. Arrhythmogenic right ventricular cardiomyopathy. N. Engl. J. Med. 2017;376(1):61–72. doi: 10.1056/NEJMra1509267. [DOI] [PubMed] [Google Scholar]
- 20.Delmar M, McKenna WJ. The cardiac desmosome and arrhythmogenic cardiomyopathies: from gene to disease. Circ. Res. 2010;107(6):700–714. doi: 10.1161/CIRCRESAHA.110.223412. [DOI] [PubMed] [Google Scholar]
- 21.Rasmussen TB, Nissen PH, Palmfeldt J, Gehmlich K, Dalager S, Jensen UB, et al. Truncating plakophilin-2 mutations in arrhythmogenic cardiomyopathy are associated with protein haploinsufficiency in both myocardium and epidermis. Circ. Cardiovasc. Genet. 2014;7(3):230–240. doi: 10.1161/CIRCGENETICS.113.000338. [DOI] [PubMed] [Google Scholar]
- 22.Asimaki A, Tandri H, Huang H, Halushka MK, Gautam S, Basso C, et al. A new diagnostic test for arrhythmogenic right ventricular cardiomyopathy. N. Engl. J. Med. 2009;360(11):1075–1084. doi: 10.1056/NEJMoa0808138. [DOI] [PubMed] [Google Scholar]
- 23.Vite A, Gandjbakhch E, Prost C, Fressart V, Fouret P, Neyroud N, et al. Desmosomal cadherins are decreased in explanted arrhythmogenic right ventricular dysplasia/cardiomyopathy patient hearts. PLoS One. 2013;8(9):e75082. doi: 10.1371/journal.pone.0075082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Burke AP, Farb A, Tashko G, Virmani R. Arrhythmogenic right ventricular cardiomyopathy and fatty replacement of the right ventricular myocardium: are they different diseases? Circulation. 1998;97(16):1571–1580. doi: 10.1161/01.cir.97.16.1571. [DOI] [PubMed] [Google Scholar]
- 25.Yang Z, Bowles NE, Scherer SE, Taylor MD, Kearney DL, Ge S, et al. Desmosomal dysfunction due to mutations in desmoplakin causes arrhythmogenic right ventricular dysplasia/cardiomyopathy. Circ. Res. 2006;99(6):646–655. doi: 10.1161/01.RES.0000241482.19382.c6. [DOI] [PubMed] [Google Scholar]
- 26.Chen SN, Gurha P, Lombardi R, Ruggiero A, Willerson JT, Marian AJ. The hippo pathway is activated and is a causal mechanism for adipogenesis in arrhythmogenic cardiomyopathy. Circ. Res. 2014;114(3):454–468. doi: 10.1161/CIRCRESAHA.114.302810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yu FX, Guan KL. The Hippo pathway: regulators and regulations. Genes Dev. 2013;27(4):355–371. doi: 10.1101/gad.210773.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Clevers H, Nusse R. Wnt/beta-catenin signaling and disease. Cell. 2012;149(6):1192–1205. doi: 10.1016/j.cell.2012.05.012. [DOI] [PubMed] [Google Scholar]
- 29.Kim W, Kim M, Jho EH. Wnt/beta-catenin signalling: from plasma membrane to nucleus. Biochem. J. 2013;450(1):9–21. doi: 10.1042/BJ20121284. [DOI] [PubMed] [Google Scholar]
- 30.Meng Z, Moroishi T, Guan KL. Mechanisms of Hippo pathway regulation. Genes Dev. 2016;30(1):1–17. doi: 10.1101/gad.274027.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Imajo M, Miyatake K, Iimura A, Miyamoto A, Nishida E. A molecular mechanism that links Hippo signalling to the inhibition of Wnt/beta-catenin signalling. EMBO J. 2012;31(5):1109–1122. doi: 10.1038/emboj.2011.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wheeler MA, Warley A, Roberts RG, Ehler E, Ellis JA. Identification of an emerin-beta-catenin complex in the heart important for intercalated disc architecture and beta-catenin localisation. Cell. Mol. Life Sci. 2010;67(5):781–796. doi: 10.1007/s00018-009-0219-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim GH. MicroRNA regulation of cardiac conduction and arrhythmias. Transl. Res. 2013;161(5):381–392. doi: 10.1016/j.trsl.2012.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ambros V. The functions of animal microRNAs. Nature. 2004;431(7006):350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 35.Gurha P, Chen X, Lombardi R, Willerson JT, Marian AJ. Knockdown of plakophilin 2 downregulates miR-184 through CpG hypermethylation and suppression of the E2F1 pathway and leads to enhanced adipogenesis in vitro. Circ. Res. 2016;119(6):731–750. doi: 10.1161/CIRCRESAHA.116.308422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang H, Liu S, Dong T, Yang J, Xie Y, Wu Y, et al. Profiling of differentially expressed microRNAs in arrhythmogenic right ventricular cardiomyopathy. Sci. Rep. 2016;6:28101. doi: 10.1038/srep28101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bowles NE, Ni J, Kearney DL, Pauschinger M, Schultheiss HP, McCarthy R, et al. Detection of viruses in myocardial tissues by polymerase chain reaction. evidence of adenovirus as a common cause of myocarditis in children and adults. J. Am. Coll. Cardiol. 2003;42(3):466–472. doi: 10.1016/s0735-1097(03)00648-x. [DOI] [PubMed] [Google Scholar]
- 38.Kuhl U, Pauschinger M, Noutsias M, Seeberg B, Bock T, Lassner D, et al. High prevalence of viral genomes and multiple viral infections in the myocardium of adults with “idiopathic” left ventricular dysfunction. Circulation. 2005;111(7):887–893. doi: 10.1161/01.CIR.0000155616.07901.35. [DOI] [PubMed] [Google Scholar]
- 39.Cioc AM, Nuovo GJ. Histologic and in situ viral findings in the myocardium in cases of sudden, unexpected death. Mod. Pathol. 2002;15(9):914–922. doi: 10.1097/01.MP.0000024291.37651.CD. [DOI] [PubMed] [Google Scholar]
- 40.Baksi AJ, Kanaganayagam GS, Prasad SK. Arrhythmias in viral myocarditis and pericarditis. Card. Electrophysiol. Clin. 2015;7(2):269–281. doi: 10.1016/j.ccep.2015.03.009. [DOI] [PubMed] [Google Scholar]
- 41.Bowles NE, Ni J, Marcus F, Towbin JA. The detection of cardiotropic viruses in the myocardium of patients with arrhythmogenic right ventricular dysplasia/cardiomyopathy. J. Am. Coll. Cardiol. 2002;39(5):892–895. doi: 10.1016/s0735-1097(02)01688-1. [DOI] [PubMed] [Google Scholar]
- 42.Pieroni M, Dello Russo A, Marzo F, Pelargonio G, Casella M, Bellocci F, et al. High prevalence of myocarditis mimicking arrhythmogenic right ventricular cardiomyopathy differential diagnosis by electroanatomic mapping-guided endomyocardial biopsy. J. Am. Coll. Cardiol. 2009;53(8):681–689. doi: 10.1016/j.jacc.2008.11.017. [DOI] [PubMed] [Google Scholar]
- 43.Lopez-Ayala JM, Pastor-Quirante F, Gonzalez-Carrillo J, Lopez-Cuenca D, Sanchez-Munoz JJ, Oliva-Sandoval MJ, et al. Genetics of myocarditis in arrhythmogenic right ventricular dysplasia. Heart Rhythm. 2015;12(4):766–773. doi: 10.1016/j.hrthm.2015.01.001. [DOI] [PubMed] [Google Scholar]
- 44.Xu HF, Ding YJ, Shen YW, Xue AM, Xu HM, Luo CL, et al. MicroRNA-1 represses Cx43 expression in viral myocarditis. Mol. Cell. Biochem. 2012;362(1–2):141–148. doi: 10.1007/s11010-011-1136-3. [DOI] [PubMed] [Google Scholar]
- 45.Ye X, Zhang HM, Qiu Y, Hanson PJ, Hemida MG, Wei W, et al. Coxsackievirus-induced miR-21 disrupts cardiomyocyte interactions via the downregulation of intercalated disk components. PLoS Pathog. 2014;10(4):e1004070. doi: 10.1371/journal.ppat.1004070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Anastasiadou E, Garg N, Bigi R, Yadav S, Campese AF, Lapenta C, et al. Epstein-Barr virus infection induces miR-21 in terminally differentiated malignant B cells. Int. J. Cancer. 2015;137(6):1491–1497. doi: 10.1002/ijc.29489. [DOI] [PubMed] [Google Scholar]
- 47.Zhang Q, Xiao Z, He F, Zou J, Wu S, Liu Z. MicroRNAs regulate the pathogenesis of CVB3-induced viral myocarditis. Intervirology. 2013;56(2):104–113. doi: 10.1159/000343750. [DOI] [PubMed] [Google Scholar]
- 48.Marquez RT, Bandyopadhyay S, Wendlandt EB, Keck K, Hoffer BA, Icardi MS, et al. Correlation between microRNA expression levels and clinical parameters associated with chronic hepatitis C viral infection in humans. Lab. Investig. 2010;90(12):1727–1736. doi: 10.1038/labinvest.2010.126. [DOI] [PubMed] [Google Scholar]
- 49.Calway T, Kim GH. Harnessing the therapeutic potential of microRNAs for cardiovascular disease. J. Cardiovasc. Pharmacol. Ther. 2015;20(2):131–143. doi: 10.1177/1074248414552902. [DOI] [PubMed] [Google Scholar]
- 50.Osbourne A, Calway T, Broman M, McSharry S, Earley J, Kim GH. Downregulation of connexin43 by microRNA-130a in cardiomyocytes results in cardiac arrhythmias. J. Mol. Cell. Cardiol. 2014;74:53–63. doi: 10.1016/j.yjmcc.2014.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mazurek SR, Calway T, Harmon C, Farrell P, Kim GH. MicroRNA-130a regulation of desmocollin 2 in a novel model of arrhythmogenic cardiomyopathy. Microrna. 2016 doi: 10.2174/2211536605666161109111031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Haas U, Sczakiel G, Laufer SD. MicroRNA-mediated regulation of gene expression is affected by disease-associated SNPs within the 3′-UTR via altered RNA structure. RNA Biol. 2012;9(6):924–937. doi: 10.4161/rna.20497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gong J, Tong Y, Zhang HM, Wang K, Hu T, Shan G, et al. Genome-wide identification of SNPs in microRNA genes and the SNP effects on microRNA target binding and biogenesis. Hum. Mutat. 2012;33(1):254–263. doi: 10.1002/humu.21641. [DOI] [PubMed] [Google Scholar]
- 54.Bruno AE, Li L, Kalabus JL, Pan Y, Yu A, Hu Z. miRdSNP: a database of disease-associated SNPs and microRNA target sites on 3′UTRs of human genes. BMC Genomics. 2012;13:44. doi: 10.1186/1471-2164-13-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Beffagna G, Occhi G, Nava A, Vitiello L, Ditadi A, Basso C, et al. Regulatory mutations in transforming growth factor-beta3 gene cause arrhythmogenic right ventricular cardiomyopathy type 1. Cardiovasc. Res. 2005;65(2):366–373. doi: 10.1016/j.cardiores.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 56.Meurs KM, Mauceli E, Lahmers S, Acland GM, White SN, Lindblad-Toh K. Genome-wide association identifies a deletion in the 3′ untranslated region of striatin in a canine model of arrhythmogenic right ventricular cardiomyopathy. Hum. Genet. 2010;128(3):315–324. doi: 10.1007/s00439-010-0855-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kapplinger JD, Landstrom AP, Salisbury BA, Callis TE, Pollevick GD, Tester DJ, et al. Distinguishing arrhythmogenic right ventricular cardiomyopathy/ dysplasia-associated mutations from background genetic noise. J. Am. Coll. Cardiol. 2011;57(23):2317–2327. doi: 10.1016/j.jacc.2010.12.036. [DOI] [PMC free article] [PubMed] [Google Scholar]