Abstract
In a survey of undiscovered taxa in Korea, three zygomycete fungal strains–EML-W31, EML-HGD1-1, and EML-RUS1-1–were isolated from freshwater, grasshopper fecal, and soil samples in Korea. On the basis of the morphological characteristics and phylogenetic analysis of internal transcribed spacer and 28S rDNA, the isolates of EML-W31, EML-HGD1-1, and EML-RUS1-1 were confirmed to be Cunninghamella bertholletiae, Cunninghamella echinulata, and Cunninghamella elegans, respectively. These species have not been previously described in Korea.
Keywords: Cunninghamella bertholletiae, Cunninghamella echinulata, Cunninghamella elegans, Undiscovered taxa, Zygomycota
Mucorales is the largest order of fungi, classified into the subphylum Mucoromycotina [1], which consist of at least 57 genera within 15 families comprising 334 species [2]. Members of Mucorales can be easily isolated from soil, dung, water, stored grains, plants, and fungal masses [3,4]. Many members in the order are characterized by rapid growth and the ability to colonize and sporulate on diverse, carbohydrate-rich, and terrestrial substrates.
The genus Cunninghamella (Cunninghamellaceae, Mucorales) was established in 1903 by Matruchot, and is comprised of species characterized by the formation of the pedicellate and unispored sporangia on the surface of the entire vesicle [5]. There are 14 accepted species in this genus [6,7,8]. Cunninghamella species are often found in soil, stored grains, and other organic substrates [3,7,8,9,10]. Some are reported as human mucormycosis [11,12], and some species are known to produce compounds that can destroy tumors [13]. The taxonomy of Cunninghamella has been determined based on morphological characteristics such as colony color, pattern of sporophore branching, vesicle shape and size, sporangiola shape, and the length of the spines in the sporangiola [5,14]. In the study by Zheng and Chen [5], the genus Cunninghamella was monographed according to the morphological characteristics and maximum growth temperatures. Liu et al. [15], Walther et al. [16], and Yu et al. [6] sequenced the internal transcribed spacer (ITS), 28S, and elongation factor 1α gene regions of the accepted taxa and showed the phylogeny to be compatible with Zeng and Chen's taxonomical conclusions [5].
Knowledge of the taxonomy of some important species of mucoralean fungi in Korea has been limited. Only 60 taxa of zygomycetes, belonging to 21 genera and 9 families, have been found in Korea [17,18,19]. Within the Cunninghamellaceae, only 8 species have been roughly described [4,17,18,19]. However, there is no published record of species belonging to this genus in Korea.
The objective of the present study was to perform morphological and molecular analyses to characterize three unrecorded zygomycete species–Cunninghamella bertholletiae, Cunninghamella echinulata, and Cunninghamella elegans–in Korea.
MATERIALS AND METHODS
Isolation of fungal strains from grasshopper feces, freshwater, and soil samples
In July, grasshoppers were collected at the CNU Arboretum located in Chonnam National University, Gwangju, Korea and put into 50 mL conical tubes (SPL Life Sciences Co., Pocheon, Korea). Grasshoppers were transferred to the laboratory and after 6 hr, feces were obtained from the bottom of tube using sterile forceps, placed onto water agar (20 g/L agar in deionized water), and incubated at 25℃ for 3–7 days. Under a stereomicroscope, hyphal tips were transferred to potato dextrose agar (PDA) plates, using tips of heat-stretched capillary tubes.
Soil samples were collected from the garden of Chonnam National University located in Gwangju. Freshwater samples were collected from a branch stream of the Nakdong River located in Gyeongsangbuk-do, Korea. These samples were transferred to sterile 50-mL conical tubes, and stored at 4℃ until examination. Fungi were isolated by a serial dilution plating method. In this technique, 1 g of soil or 1 mL water was mixed with 9 mL of sterile distilled water and the solution was shaken for 15 min at room temperature, and serial dilutions were made ranging from 10−1 to 10−4. A 0.1 mL aliquot from each dilution was transferred onto PDA and incubated at 25℃ for 3–7 days.
To isolate pure cultures, individual colonies of varied morphologies were transferred to PDA plates. Pure isolates were maintained in PDA slant tubes and stored in 20% glycerol at −80℃ at the Environmental Microbiology Laboratory Fungarium, Chonnam National University, Gwangju, Korea.
Morphological studies
For detailed morphological studies, EML-W31, EML-HGD1-1, and EML-RUS1-1 strains were cultured on synthetic mucor agar (SMA; 40 g dextrose, 2 g asparagine, 0.5 g KH2PO4, 0.25 g MgSO4 · 7H2O, 0.5 g thiamine chloride, and 15 g agar in 1 L of deionized water). The plates were incubated at 25℃ in the dark for 5 days. Samples were mounted in a lactophenol solution (Junsei Chemical Co. Ltd., Tokyo, Japan) and observed under an Olympus BX51 microscope with DIC optics (Olympus, Tokyo, Japan). Fine fungal structures were observed by scanning electron microscopy (SEM) (Hitachi S4700; Hitachi, Tokyo, Japan). The isolates were fixed in 2.5% paraformaldehyde-glutaraldehyde in 0.05 M phosphate buffer (pH 7.2) for 2 hr, and then washed with cacodylate buffer (Junsei Chemical Co. Ltd.). Cellular membranes were preserved by fixing the samples in 1% osmium tetroxide (Electron Microscopy Sciences, Hatfield, PA, USA) diluted in cacodylate buffer for 1 hr. Samples were then washed again in cacodylate buffer, dehydrated in graded ethanol (Emsure, Darmstadt, Germany) and isoamyl acetate (Junsei Chemical Co. Ltd.), and dried in a fume hood. Finally, samples were sputter-coated with gold and observed under a Hitachi S4700 field emission scanning electron microscope at the Korea Basic Science Institute, Gwangju, Korea.
To investigate growth rates, EML-W31, EML-HGD1-1, and EML-RUS1-1 strains were incubated on PDA medium at 10℃, 20℃, 25℃, 30℃, 35℃, 37℃, 40℃, and 42℃ in the dark for 7 days.
DNA extraction, PCR, and sequencing
Genomic DNA was extracted directly from the mycelia of fungal isolates, using a genomic DNA prep kit (Solgent, Daejeon, Korea). The ITS region and large subunit of 28S rDNA were amplified with the primer pairs ITS4 and ITS5 [20]; LROR and LR5F [21,22], respectively. The PCR amplification mixture (total volume, 20 µL) contained the fungal DNA template, 5 pmol/µL of each primer, and the Accupower PCR Premix (Bioneer, Daejeon, Korea). PCR products were purified using the Accuprep PCR Purification Kit (Bioneer) according to the manufacturer's instructions. DNA sequencing was performed on an ABI 3700 Automated DNA sequencer (Applied Biosystems Inc., Foster City, CA, USA). The EML-W31, EML-HGD1-1, EML-HGD1-2, EML-RUS1-1, and EML-RUS1-2 sequences were deposited in the NCBI database under accession numbers as shown in Table 1.
Table 1. Sequences used in this study and GenBank accession numbers.
Bold letters indicate isolates and accession numbers determined in our study.
ITS, internal transcribed spacer; CBS, Centraalbureau voor Schimmelcultures, Utrecht, The Netherlands; NRRL, ARS Culture Collection, Peoria, Illinois, USA; NT, ex-neotype strain; ATCC, American Type Culture Collection, Manassas, VA, USA; EML, Environmental Microbiology Laboratory Fungarium, Chonnam National University, Gwangju, South Korea; T, ex-type strain; TUFC, Tottori University Fungal Culture Collection; URM, Universidade Federal de Pernambuco, Recife, Brazil.
Phylogenetic analysis
Sequence data obtained from the GenBank database (Table 1) were aligned using Clustal_X v.1.83 [23] and Bioedit v. 5.0.9.1 software [24]. Phylogenetic analyses were performed using MEGA 6 software [25], and maximum likelihood was constructed by Kimura's two-parameter correction method. Mortierella parvispora was used as the outgroup. Reliability of the internal branches was assessed using the p-distance substitution model, with 1,000 bootstrap replications.
RESULTS
Phylogenetic analysis
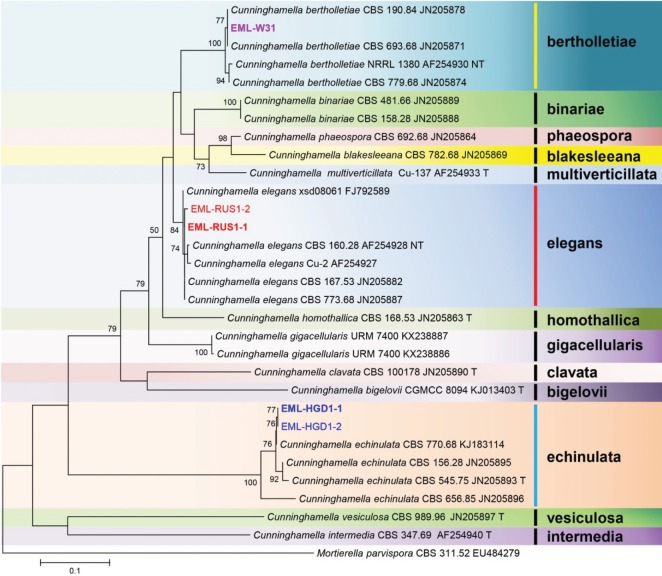
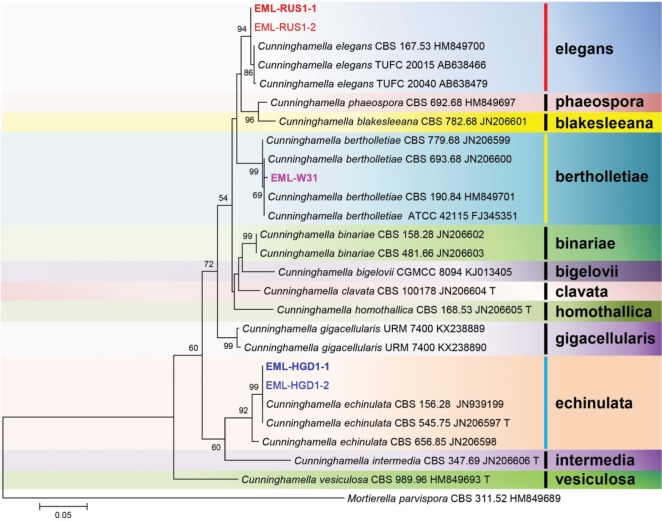
BLASTN searches were performed against the ITS sequences of EML-W31, EML-HGD1-1, and EML-RUS1-1. Sequence similarity comparison revealed 100% (700/700 bp), 99.5% (657/660 bp), and 99.6% (674/677 bp) homologies with the three most closely related species, Cunninghamella polymorpha CBS 693.68 (current name: C. bertholletiae) (GenBank accession No. JN205871), C. echinulata CBS 770.68 (GenBank accession No. KJ183114), and C. elegans xsd08061 (GenBank accession No. FJ792589), respectively. On the basis of the 28S rDNA sequence analysis, EML-W31, EML-HGD1-1, and EML-RUS1-1 strains showed 99.8% (905/907 bp), 100% (628/628 bp), and 99.8% (659/660 bp) homologies with C. bertholletiae ATCC 42115 (GenBank accession FJ345351), C. echinulata CBS 156.28 (GenBank accession No. JN939199), and C. elegans CBS 167.53 (GenBank accession No. HM849700), respectively. On the basis of the ITS and 28S sequence analysis, the three isolates EML-W31, EML-HGD1-1, and EML-RUS1-1 were identical to C. bertholletiae, C. echinulata, and C. elegans (Figs. 1 and 2), respectively.
Fig. 1. Phylogenetic tree based on the maximum likelihood analysis of internal transcribed rDNA sequences for Cunninghamella bertholletiae EML-W31, C. echinulata EML-HGD1-1, C. echinulata EML-HGD1-2, C. elegans EML-RUS1-1, and C. elegans EML-RUS1-2. Mortierella parvispora was used as the outgroup. Bootstrap support values of ≥ 50% are indicated at the nodes. The bar indicates the number of substitutions per position.
Fig. 2. Phylogenetic tree based on the maximum likelihood analysis of 28S rDNA sequences for Cunninghamella bertholletiae EML-W31, C. echinulata EML-HGD1-1, C. echinulata EML-HGD1-2, C. elegans EML-RUS1-1, and C. elegans EML-RUS1-2. Mortierella parvispora was used as the outgroup. Bootstrap support values of ≥ 50% are indicated at the nodes. The bar indicates the number of substitutions per position.
Taxonomy of EML-W31
Cunninghamella bertholletiae Stadel, Über einen neuen Pilz, Cunninghamella bertholletiae: 1 (1911) (Table 2, Fig. 3).
Table 2. Morphological characteristics of EML-W31 and the reference species Cunninghamella bertholletiae on synthetic mucor agar medium at 25℃.
aFrom the description by Weitzman and Crist [14].
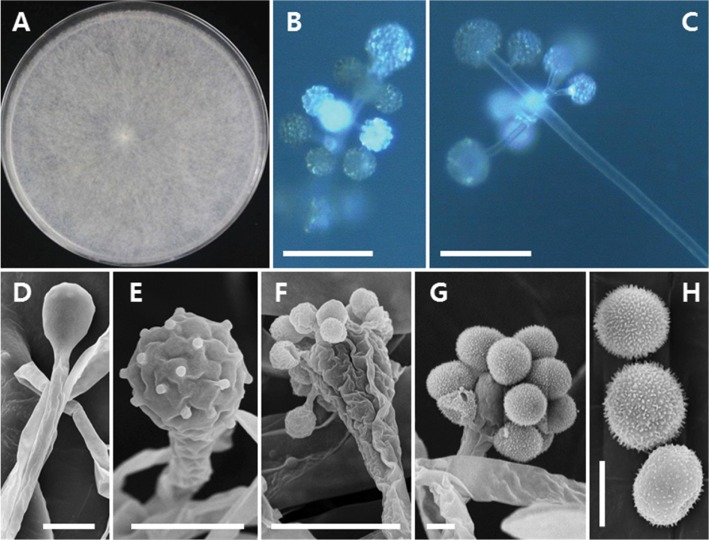
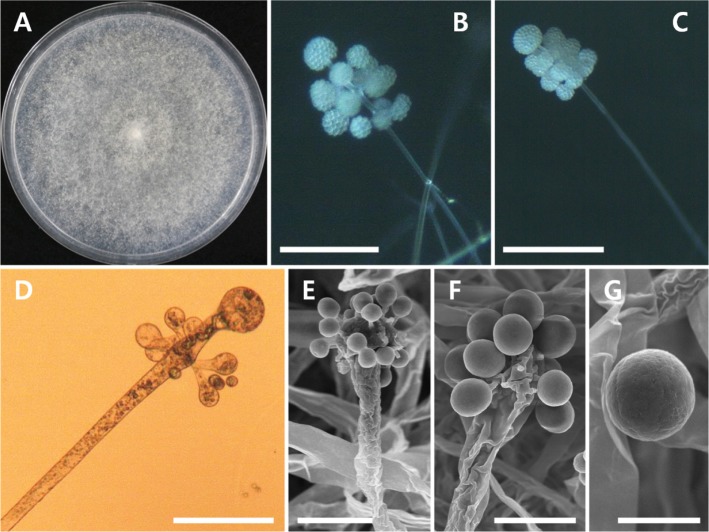
Fig. 3. Morphology of Cunninghamella bertholletiae EML-W31. A, Colonies on synthetic mucor agar; B, C, Sporangiola on branched sporangiophores (observed under light microscope); D, Sporangiophore with vesicle (observed under scanning electron microscopy [SEM]); E, Development of sporangiola on vesicles (observed under SEM); F, G, Vesicles bearing sporangiola (observed under SEM); H, Sporangiola with spines (observed under SEM) (scale bars: B, C = 100 µm, D–G = 10 µm, H = 5 µm).
Description: Colonies exhibited rapid growth on SMA, attaining a diameter of 76–78 mm after 4 days at 25℃. The colony color was initially white, later turning to gray. The colony reverse was pale gray and regularly zonate. Sporangiophores were 6.3–12.5 µm wide, erect, diversely branched, irregular, and verticillate. Sporangiola were subglobose to ovoidal, and measured 7.1–13.3 × 5.5–10.0 µm. Terminal vesicles were globose to subglobose, and measured 16.5–45.5 µm in diameter. Lateral vesicles were the same shape as terminal vesicles, and measured 11.6–30.5 µm in diameter. Chlamydospores and zygospores were not observed.
Taxonomy of EML-HGD1-1
Cunninghamella echinulata (Thaxt.) Thaxt., Rhodora 5: 98 (1903) (Table 3, Fig. 4).
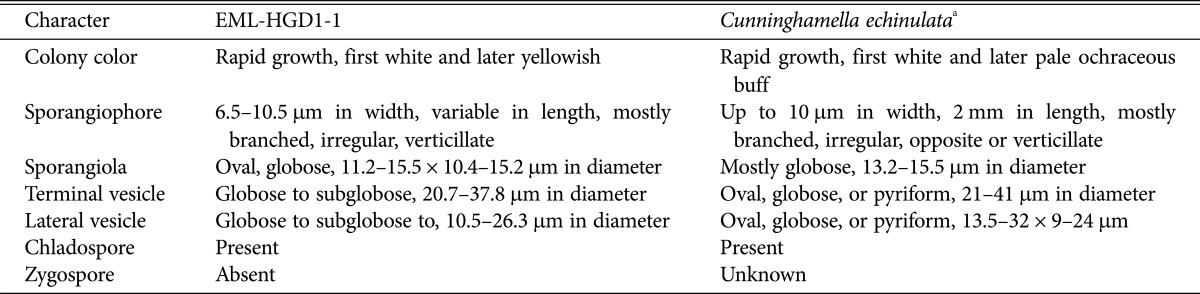
Table 3. Morphological characteristics of EML-HGD1-1 and the reference species Cunninghamella echinulata on synthetic mucor agar medium at 25℃.
aFrom the description by Baijal and Mehrotra [10].
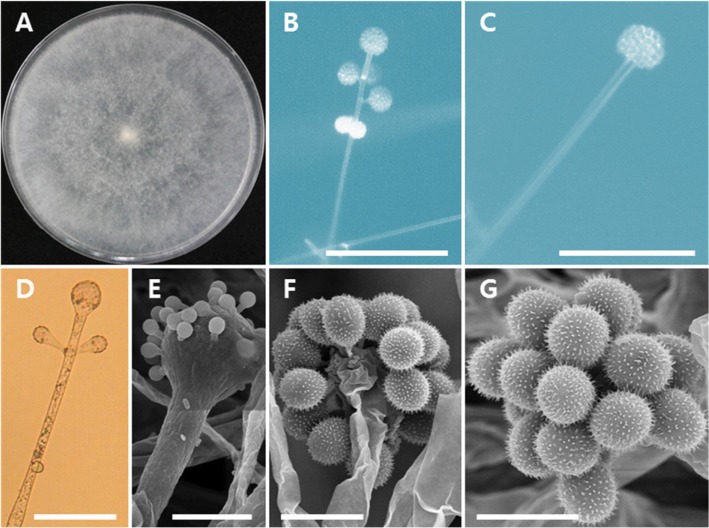
Fig. 4. Morphology of Cunninghamella echinulata EML-HGD1-1. A, Colonies on synthetic mucor agar; B, C, Sporangiophores with sporangiola (observed under light microscope); D, Sporangiophores with vesicles; E, F, Vesicles bearing sporangiola (observed under scanningelectron microscopy [SEM]); G, Smooth sporangiolum (observed under SEM) (scale bars: B, C = 100 µm, D = 50 µm, E, F = 20 µm, G = 10 µm).
Description: Colonies exhibited rapid growth on SMA, attaining a diameter of 82–84 mm after 4 days at 25℃. The colony color was initially white, later becoming yellowish. Sporangiophores were 6.5–10.5 µm wide, erect, mostly branched, irregular, and verticillate. Sporangiola were oval, globose, and measured 11.2–15.5 × 10.4–15.2 µm. Terminal vesicles were globose to subglobose, and measured 20.7–37.8 µm in diameter. Lateral vesicles were a similar shape to the terminal vesicles, but smaller in size, and measured 10.5–26.3 µm in diameter. Chlamydospores were oval. Zygospores were not observed.
Taxonomy of EML-RUS1-1
Cunninghamella elegans Lendn., Bull. Herb. Boissier 7: 250 (1907) (Table 4, Fig. 5).
Table 4. Morphological characteristics of EML-RUS1-1 and the reference species Cunninghamella elegans on synthetic mucor agar medium at 25℃.
aFrom the description by Baijal and Mehrotra [10].
Fig. 5. Morphology of Cunninghamella elegans EML-RUS1-1. A, Colonies on synthetic mucor agar; B, C, Sporangiophores with sporangiola (observed under the light microscope); D, Sporangiophores with vesicles; E, Young sporangiola formed on a sporangiophore vesicle (observed under scanning electron microscopy [SEM]); F, G, Sporangiola with spines (observed under SEM) (scale bars: B = 200 µm, C = 100 µm, D = 50 µm, E–G = 10 µm).
Description: Colonies exhibited rapid growth on SMA, covering the petri dish after 4 days at 25℃. The colony color was initially white, later turning to a stormy gray. Mycelial growth on SMA was sparse. Sporangiophores were 5.5–9.3 µm wide and erect and had solitary, opposite, or verticillate branches. Sporangiola were globose to ellipsoidal, and measured 7.3–11.6 × 6.1–10.5 µm. Terminal vesicles were subglobose to oval and measured 18.3–35.5 µm in diameter. Lateral vesicles were oval to pyriform and measured 13.1–19.8 µm in diameter. Chlamydospores and zygospores were not observed.
Mycelial growth
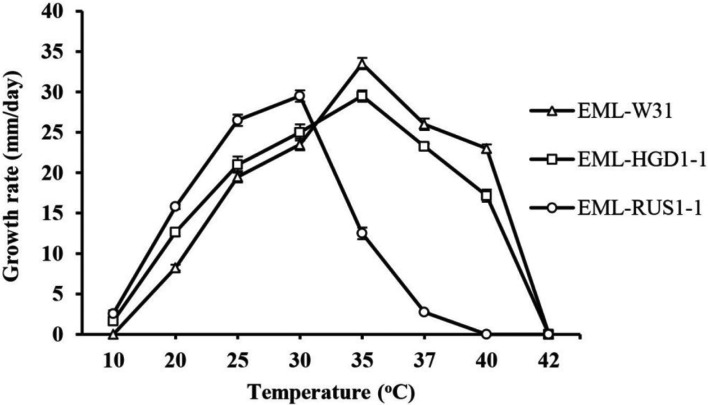
The results regarding mycelial growth at different temperatures are shown in Fig. 6. The average growth rates of EML-W31, EML-HGD1-1, and EML-RUS1-1 on PDA were 19.5 mm, 21 mm, and 26.5 mm per day, respectively, at 25℃. The optimal temperature for growth was 30℃ for EML-RUS1-1 and 35℃ for EML-HGD1-1 and EML-RUS1-1. Slow growth was observed at 10℃ and no growth was observed at 40℃ for EML-RUS1-1 and 42℃ for EML-W31 and EML-HGD1-1.
Fig. 6. Effect of temperature on mycelial growth of Cunninghamella bertholletiae EML-W31, C. echinulata EML-HGD1-1, and C. elegans EML-RUS1-1. Mycelia were grown on potato dextrose agar at different temperatures of 10℃, 20℃, 25℃, 30℃, 35℃, 37℃, 40℃, and 42℃.
DISCUSSION
In this paper, the detailed morphologies of Cunninghamella species were described based on ultrastructures revealed by SEM and light microscopy. Observing the detailed morphologies of sporangiola and sporangiophores under SEM are valuable tools in classifying a diverse group within Cunninghamellaceae.
In the studies by Samson [26], Baijal and Mehrotra [10], and Lunn and Shipton [27], C. bertholletiae and C. elegans were considered to be the same species because there were no significant differences in the morphological characteristics. Generally, temperature plays an important role in fungal growth. In this study, the effect of temperature on the growth of different Cunninghamella species varied with the species. According to Weitzman and Crist [14], growth capacity at 40℃, or at temperatures above this was a key parameter used to distinguish the species of C. elegans and C. bertholletiae. These authors showed that the species of C. elegans has a maximum growth temperature of 37℃; whereas the maximum growth temperature of C. bertholletiae is between 40℃ and 45℃.
Of the three species studied in this work, only C. bertholletiae and C. echinulata had a maximum growth temperature at 40℃, and the maximum growth temperature for C. elegans was 37℃. The observed growth temperature of C. bertholletiae and C. echinulata suggested that these species of Cunninghamella have thermotolerance. Our results are in agreement with those reported by Weitzman and Crist [14] in that temperature tolerance is a criterion for distinguishing the difference between C. bertholletiae and C. elegans species.
Although C. bertholletiae and C. echinulata have a similar temperature tolerance, C. bertholletiae is morphologically different from C. echinulata because it forms long branches on the sporangiophore.
The morphological characteristics of the Cunninghamella echinulata isolate studied were similar to those previously described by Baijal and Mehrotra [10]. However, our Cunninghamella echinulata isolate had sporangiophores with swellings, which was not described by Baijal and Mehrotra [10].
Despite the wide intraspecific variation found among some taxa, the ITS region is the barcode marker for mucoralean species identification. The identification of the isolates was confirmed by DNA sequencing of ITS and 28S rDNA. In the phylogenetic trees based on the ITS and 28S sequences, EML-W31 strain was placed into the bertholletiae clade, EML-HGD1-1 and EML-HGD1-2 strains into the echinulata clade, and EML-RUS1-1 and EML-RUS1-2 strains into the elegans clade, along with representative species of C. bertholletiae, C. echinulata, and C. elegans (Figs. 1 and 2). The results of our molecular data analysis were consistent with the phylogeny presented by Walther et al. [16] and Yu et al. [6].
Pispek [28] described a new species, C. polymorpha, on the basis of the morphological characteristics of variously branched sporangiophores and globose or ovoid to ellipsoidal conidia. Weitzman and Crist [14,29] later demonstrated that C. polymorpha was a synonym of C. bertholletiae on the basis of mating and morphology studies. Recently, Walther et al. [16] and Yu et al. [6], who performed phylogenetic analysis of sequences of C. polymorpha, indicated that this species was a synonym of C. bertholletiae.
Morphologically, the species of C. bertholletiae reported here had a close similarity with the description by Weitzman and Crist [14], although there were differences in the size of the terminal vesicles. The sizes of terminal vesicles were potentially larger (15–52 µm according to Weitzman and Crist [14]) than those (16.5–45.5 µm) observed in our isolate.
In recent studies on zygomycetous fungi belonging to undiscovered taxa in Korea, eight new species have been registered in Index Fungorum [4,18,30,31].
Our results suggest that our knowledge on the diversity of zygomycetous fungi is small and more undescribed and novel taxa in zygomycota remain to be discovered.
ACKNOWLEDGEMENTS
This work was supported by the Graduate Program for the Undiscovered Taxa of Korea, and by the Project on Survey and Discovery of Indigenous Fungal Species of Korea funded by NIBR and by the Project on Discovery of Fungi from Freshwater and Collection of Fungarium funded by NNIBR of the Ministry of Environment (MOE), and in part carried out with the support of Cooperative Research Program for Agriculture Science and Technology Development (PJ012957), Rural Development Administration, Republic of Korea.
References
- 1.Hibbett DS, Binder M, Bischoff JF, Blackwell M, Cannon PF, Eriksson OE, Huhndorf S, James T, Kirk PM, Lücking R, et al. A higher-level phylogenetic classification of the Fungi. Mycol Res. 2007;111(Pt 5):509–547. doi: 10.1016/j.mycres.2007.03.004. [DOI] [PubMed] [Google Scholar]
- 2.Benny GL, Humber RA, Voigt K. The zygomycetous fungi: the Phylum Entomophthoromycota and subphyla Kickxellomycotina, Mortierellomycotina, Mucoromycotina, and Zoopagomycotina. In: McLaughlin DJ, Blackwell M, Spatafora JW, editors. The Mycota. Vol. VII, part A. Systematics of fungi. 2nd ed. New York: Springer-Verlag; 2014. pp. 209–250. [Google Scholar]
- 3.Benny GL. Methods used by Dr. R. K. Benjamin, and other mycologists, to isolate zygomycetes. Aliso. 2008;26:37–61. [Google Scholar]
- 4.Ariyawansa HA, Hyde KD, Jayasiri SC, Buyck B, Chethana KW, Dai DQ, Dai YC, Daranagama DA, Jayawardena RS, Lücking R, et al. Fungal diversity notes 111-252: taxonomic and phylogenetic contributions to fungal taxa. Fungal Divers. 2015;75:27–274. [Google Scholar]
- 5.Zheng RY, Chen GQ. A monograph of Cunninghamella. Mycotaxon. 2001;80:1–75. [Google Scholar]
- 6.Yu J, Walther G, Van Diepeningen AD, Gerrits Van Den Ende AH, Li RY, Moussa TA, Almaghrabi OA, De Hoog GS. DNA barcoding of clinically relevant Cunninghamella species. Med Mycol. 2015;53:99–106. doi: 10.1093/mmy/myu079. [DOI] [PubMed] [Google Scholar]
- 7.Guo J, Wang H, Liu D, Zhang JN, Zhao YH, Liu TX, Xin ZH. Isolation of Cunninghamella bigelovii sp. nov. CGMCC 8094 as a new endophytic oleaginous fungus from Salicornia bigelovii. Mycol Prog. 2015;14:11. [Google Scholar]
- 8.Alves AL, de Souza CA, de Oliveira RJ, Cordeiro TR, de Santiago AL. Cunninghamella clavata from Brazil: a new record for the western hemisphere. Mycotaxon. 2017;132:381–389. [Google Scholar]
- 9.Liu CW, Liou GY, Chien CY. New records of the genus Cunninghamella (Mucorales) in Taiwan. Fungal Sci. 2005;20:1–9. [Google Scholar]
- 10.Baijal U, Mehrotra BS. The genus Cunninghamella: a reassessment. Sydowia. 1980;33:1–13. [Google Scholar]
- 11.Gomes MZ, Lewis RE, Kontoyiannis DP. Mucormycosis caused by unusual mucormycetes, non-Rhizopus, -Mucor, and -Lichtheimia species. Clin Microbiol Rev. 2011;24:411–445. doi: 10.1128/CMR.00056-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kwon-Chung KJ. Taxonomy of fungi causing mucormycosis and entomophthoramycosis (zygomycosis) and nomenclature of the disease: molecular mycologic perspectives. Clin Infect Dis. 2012;54(Suppl 1):S8–S15. doi: 10.1093/cid/cir864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Betts RE, Walters DE, Rosazza JP. Microbial transformations of antitumor compounds. 1. Conversion of acronycine to 9-hydroxyacronycine by Cunninghamella echinulata. J Med Chem. 1974;17:599–602. doi: 10.1021/jm00252a006. [DOI] [PubMed] [Google Scholar]
- 14.Weitzman I, Crist MY. Studies with clinical isolates of Cunninghamella. II. Physiological and morphological studies. Mycologia. 1980;72:661–669. [PubMed] [Google Scholar]
- 15.Liu XY, Huang H, Zheng RY. Relationships within Cunninghamella based on sequence analysis of ITS rDNA. Mycotaxon. 2001;80:77–95. [Google Scholar]
- 16.Walther G, Pawłowska J, Alastruey-Izquierdo A, Wrzosek M, Rodriguez-Tudela JL, Dolatabadi S, Chakrabarti A, de Hoog GS. DNA barcoding in Mucorales: an inventory of biodiversity. Persoonia. 2013;30:11–47. doi: 10.3767/003158513X665070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee YS, Jung HY, Lee HB, Kim SH, Shin KS, Eom AH, Kim C, Lee SY Korean Society of Mycology. National list of species of Korea. Ascomycota, Glomeromycota, Zygomycota, Myxomycota, Oomycota. Incheon: National Institute of Biological Resources; 2015. [Google Scholar]
- 18.Li GJ, Hyde KD, Zhao RL, Hongsanan S, Abdel-Aziz FA, Abdel-Wahab MA, Alvarado P, Alves-Silva G, Ammirati JF, Ariyawansa HA, et al. Fungal diversity notes 253-366: taxonomic and phylogenetic contributions to fungal taxa. Fungal Divers. 2016;78:1–237. [Google Scholar]
- 19.Nguyen TT, Lee SH, Bae S, Jeong SJ, Mun HY, Lee HB. Characterization of two new records of zygomycete species belonging to undiscovered taxa in Korea. Mycobiology. 2016;44:29–37. doi: 10.5941/MYCO.2016.44.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.White TJ, Bruns T, Lee S, Taylor J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ, editors. PCR protocols: a guide to methods and applications. New York: Academic Press; 1990. pp. 315–322. [Google Scholar]
- 21.Vilgalys R, Hester M. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Crytococcus species. J Bacteriol. 1990;172:4238–4246. doi: 10.1128/jb.172.8.4238-4246.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee HB. Molecular phylogenetic status of Korean strain of Podosphaera xanthii, a causal pathogen of powdery mildew on Japanese thistle (Cirsium japonicum) in Korea. J Microbiol. 2012;50:1075–1080. doi: 10.1007/s12275-012-2618-z. [DOI] [PubMed] [Google Scholar]
- 23.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- 25.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Samson RA. Revision of the genus Cunninghamella (Fungi, Mucorales) Proc K Ned Akad Wet C. 1969;72:322–335. [Google Scholar]
- 27.Lunn JA, Shipton WA. Re-evaluation of taxonomic criteria in Cunninghamella. Trans Br Mycol Soc. 1983;81:303–312. [Google Scholar]
- 28.Pispek PA. Edafske mukorineje Jugoslavije. Acta Bot Inst Bot Regalis Univ Zagrebensis. 1929;4:77–112. [Google Scholar]
- 29.Weitzman I, Crist MY. Studies with clinical isolates of Cunninghamella. I. Mating behavior. Mycologia. 1979;71:1024–1033. [PubMed] [Google Scholar]
- 30.Hyde KD, Hongsanan S, Jeewon R, Bhat DJ, McKenzie EH, Gareth Jones EB, Phookamsak R, Ariyawansa HA, Boonmee S, Zhao Q, et al. Fungal diversity notes 367–490: taxonomic and phylogenetic contributions to fungal taxa. Fungal Divers. 2016;80:1–270. [Google Scholar]
- 31.Tibpromma S, Hyde KD, Jeewon R, Maharachchikumbura SS, Liu JK, Bhat DJ, Gareth Jones EB, McKenzie EH, Camporesi E, Bulgakov TS, et al. Fungal diversity notes 491–602: taxonomic and phylogenetic contributions to fungal taxa. Fungal Divers. 2017;83:1–261. [Google Scholar]