Abstract
Peanut yield and quality are seriously affected by pod rot pathogens worldwide, especially in China in recent years. The goals of this study are to analyze the structure of fungal communities of peanut pod rot in soil in three peanut cultivars and the correlation of pod rot with environmental variables using 454 pyrosequencing. A total of 46,723 internal transcribed spacer high-quality sequences were obtained and grouped into 1,706 operational taxonomic units at the 97% similarity cut-off level. The coverage, rank abundance, and the Chao 1 and Shannon diversity indices of the operational taxonomic units were analyzed. Members of the phylum Ascomycota were dominant, such as Fusarium, Chaetomium, Alternaria, and Sordariomycetes, followed by Basidiomycota. The results of the heatmap and redundancy analysis revealed significant variation in the composition of the fungal community among the three cultivar samples. The environmental conditions in different peanut cultivars may also influence on the structure of the fungal community. The results of this study suggest that the causal agent of peanut pod rot may be more complex, and cultivars and environmental conditions are both important contributors to the community structure of peanut pod rot fungi.
Keywords: Cultivar, Environmental parameters, Fungal community, Peanut, Pod rot pathogens
Plant growth and-crop quality are significantly affected by microorganisms in the soil [1]. A wide range of soil microorganisms are known to provide many functions in an ecosystem [2], such as mineral nutrient cycling, organic matter turnover, soil structure formation, and toxin removal or accumulation [3]. Fungi are an important and diverse group of these microorganisms. Soil fungi are critical to the ecosystem functions of soil quality and plant health. Because of their close relationships with plants, soil fungal communities are shaped by a number of biotic and abiotic factors [4]. Of particular note is that plant genotypes (different genotypes, i.e., different plant species or even different crop cultivars) also have been reported to have an impact on the genetic diversity of fungi based on some preliminary studies [5].
Peanut (Arachis hypogaea L.), an important oil and food crop, widely grows in many different environments between the 40th parallels [6]. China is the world's largest peanut producer, contributing one-third of overall production [7]. Unfortunately, the production of peanut is seriously threatened by pod rot pathogens worldwide and especially in China in recent years. Furthermore, it appears that all commercial cultivars of peanut are susceptible to pod rot [8]. Management of pod rot has been difficult in part because of the wide host range of the pathogens [9] and limited cultivar resistance [10]. To date, there are few studies on the diversity of fungi associated with peanut pod rot. The complete fungal community structure and possible impact factors, such as habitat environment and peanut genotypes or cultivars on peanut pod rot, remain unexplored.
Recently, high-throughput DNA sequencing technologies, particularly 454 deep amplicon sequencing, have been widely used in determining the fungal community associated with plants. These studies have shown that different environmental factors including climate, soil properties, plant genotypes, and plant habitats, may have significant effects on their soil fungi community structures [5]. Hence, the impact factors of fungal communities in peanut pod rot need to be further studied using high-throughput DNA sequencing technologies.
In this study, we first assessed the genetic diversity of the fungal community by 454 pyrosequencing of the internal transcribed spacer (ITS) region amplified from soil samples of three peanut cultivars, which was infected by pod rot pathogens. Then, to evaluate the possible effects of peanut cultivars and environmental factors on the fungal community in the soil samples, we performed redundancy analysis (RDA) using the operational taxonomic units (OTU) abundance of each of the samples. This study aims to provide baseline data about differences among fungal community structures for different peanut cultivars affected by pod rot in soil in China and the effect of physicochemical parameters.
MATERIALS AND METHODS
Site description and soil sampling analyses
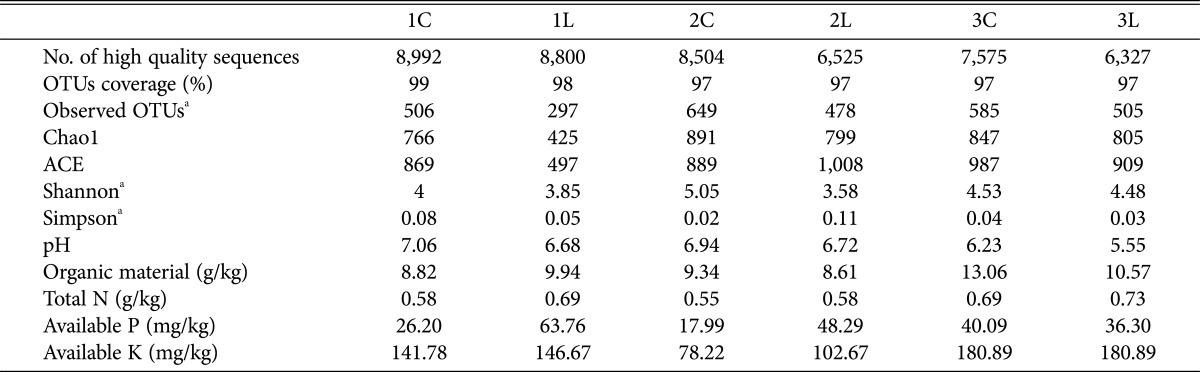
Peanuts are planted in May and harvested in October. Six soil samples were collected from a peanut field at the Breeding and Experimental Station of the Shandong Peanut Research Institute in Qingdao, China (36.10°N, 120.41°E) during the 2013 cropping season. Three peanut cultivars with different known levels of resistance to pod rot pathogens were employed in the experimental field. HuaYu912 is the most susceptible to peanut pod rot pathogens, followed by HuaYu33 and HuaYu20 is the least susceptible peanut cultivar. The soil samples (1L, 2L, and 3L) being contaminated with pod rot were collected from root soils of the peanut cultivars HuaYu33, HuaYu20, and HuaYu912, respectively. The soil samples (1C, 2C, and 3C) are planted in the same peanut field but not infected by pod rot pathogens, served as the control soil samples respectively. The peanut cultivars HuaYu33 and HuaYu912 are a large-pod cultivar, while HuaYu20 is a small-pod cultivar. The soils were collected by a soil probe (1.5 cm diameter) at depth of 5–10 cm. Three replicates of each sample were mixed and filtered together. Then, soil samples were frozen at −80℃ in the laboratory and were sent for fungal community analysis. A total of five physicochemical soil parameters were analyzed, including pH, organic material, total nitrogen (N), phosphorus (P), and available potassium (K), by the College of Resources and Environment Testing Group of Qingdao Agricultural University.
Pyrosequencing of fungal ITS regions
Total soil DNA was extracted from subsamples of soil was used to amplify the ITS regions using the fusion primer pair ITS5 (5′-A [6-bp MID] ACCCGCTGAACTTAAGC-3′) and ITS4 (5′-BTCCTGAGGGAAACTTCG-3′). After being purified, the target PCR products were separated by electrophoresis through a 0.8% agarose gel and then were quantified. The product pool was sequenced in one quarter of a sequencing plate on a Roche 454 GS-FLX sequencer (454-Life Sciences, Basel, Switzerland) at Personal Biotechnology Co., Ltd. (Shanghai, China).
OTU-based sequence analysis
Quality filtering and analysis of the 454 ITS sequences were performed mainly using mothur open-source software. In brief, raw sequences containing ambiguous (N) bases, homo-polymers more than eight bases, and those with a quality score lower than 20 and less than 150-bp nucleotides were removed. Then, sequences were checked for chimeric sequences using the Uchime algorithm implemented in mothur. The trimmed, high-quality sequences were clustered into OTUs using the QIIME (Quantitative Insights into Microbial Ecology, ver. 1.50; http://qiime.sourceforge.net/) implementation at a threshold of 97% identity. Taxonomic assignment of the representative sequences for the OTUs was done with the mother command “classify.seq” applied to the Unite fungal ITS reference database, version 6. To improve the taxonomical resolution, the OTUs with abundance less than 0.001% of the total raw sequences were removed. Then, the SILVA database (v108, http://www.arb-silva.de/) was used to identify representative sequences.
Sequence statistical analysis
A rarefaction analysis was performed by the QIIME implementation. The mothur software was utilized to calculate the pod rot fungal diversity, using the Shannon and Simpson indices, and richness, using the ACE and Chao 1 indices, of different samples. R software (ver. 2.15.3) was used to plot the Venn diagrams based on the number of OTUs. In order to analyze the influence of cultivar on the pod rot fungal community structure, Heatmap analysis was performed based on the OTU abundance of each sample, by using the R software data of all samples. The relevance of environmental parameters in explaining the distribution patterns of fungal communities between the pod rot and control soil samples was analyzed by RDA using R software.
RESULTS
Soil environmental parameters of the samples
Six soil samples of three peanut cultivars were collected from the city of Laixi in Shandong province in China. We are sampling peanut pod rot soil, general using agronomic traits—the incidence of peanut pods as fundamental basis. We took the soil within more than 70% of the incidence of pod as soil sample contamination of the pathogens. We took non incidence of peanut pod attaching soil as the sample free from pathogens. Unfortunately, we do not have image data. Table 1 summarizes the physiochemical parameters measured. Compared with the control samples, most of the physiochemical parameters showed differences after peanut was infected by pod rot pathogens. The susceptible soil samples showed that the pH value decreased a little in both three cultivars, whereas the content of total N increased. The content of organic material in cultivar 1 susceptible soil showed a difference compared with the two others. The content of available P and K increased in cultivars 1 and 2, but decreased in cultivar 3. These results showed that different peanut cultivars have different environmental parameters, especially when peanut is infected by pod rot pathogens.
Table 1. Physicochemical property and information of sequences obtained and OTUs richness and diversity index for 454 FLX+ pyrosequencing in six soil samples.
OTU, operational taxonomic units.
aData were calculated at 3% genetic level based on normalized sequences with mothur.
Sequence data
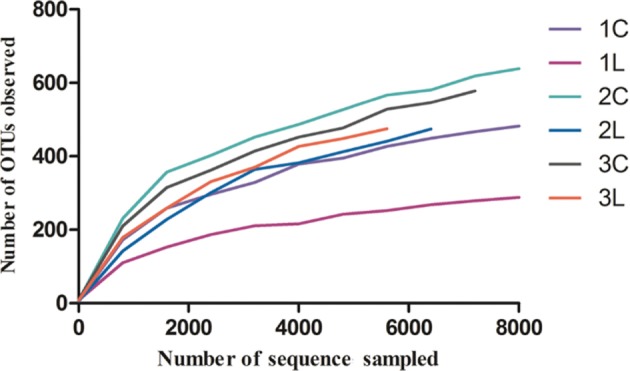
Data the sequences obtained in these experiments are available in the NCBI Sequence Read Archive under bioproject (SRP066284) with sample accession numbers SRS1165854, SRS1165881, SRS1165883, SRS1165884, SRS1165885, and SRS1165886 for the six soil samples 1C, 1L, 2C, 2L, 3C, and 3L, respectively. After filtering and quality control processes were applied to the sequences, 46,723 ITS high-quality full-length sequences were obtained from six soil samples (Table 1), of which 98.36% had a hit in the NCBI. Of these hits, 98.03% were assigned to fungi. Among these sequences, about 86.3% had a length of > 400 bp, with most ranging between 450 and 600 bp. All the sequences were grouped into 1,706 OTUs at the 97% similarity cut-off level. Rarefaction analyses showed that the number of recorded OTUs tended to plateau at 6,000 sequences reads (Fig. 1). Further rank abundance analyses showed that these soil samples had high fungal community richness (data not shown).
Fig. 1. Rarefaction curves showing the relationship between sampling intensity and the number of recovered operational taxonomic units (OTUs) from soil of a peanut pod rot field at harvest time.
Fungal community diversity
The OTUs coverage as well as the Chao 1 and Shannon indices were used to evaluate and compare the diversity of the fungal communities among the six soil samples (Table 1). The OTUs coverage ranged from 97% to 99%, which showed that 454 pyrosequencing captured the dominant phenotypes. The Chao 1 (59.82–72.84%) and ACE (47.42–73%) indices, shown in Table 1, indicated that the level of diversity varied among the six soil samples. The number of OTUs as well as the Shannon and Simpson indices of the six samples showed high variation within each sample. At a genetic distance of 3%, the Shannon index showed lower fungal community diversity of pod rot soil samples, ranging from 3.58 to 4.48, than that of the control samples, which ranged from 4.00 to 5.05 (Table 1). The same result was shown by the Simpson index. The highest fungal community diversity was found in control soil 2C, while the lowest fungal community diversity was found in the pod rot soil sample 2L (Table 1). Taken together, the Chao 1 index, the Shannon index, and the number of OTUs in both the three control soils were higher than in the susceptible cultivar soils.
Fungal community composition
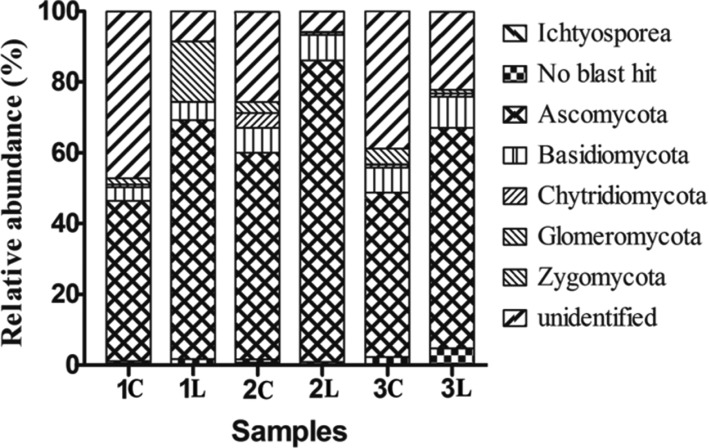
The fungal OTUs were affiliated to six phyla: Ascomycota, 770 OTUs (45.13% of all sequences); Basidiomycota, 189 OTUs (11.08% of all sequences); Chytridiomycota, 49 OTUs (2.87% of all sequences); Glomeromycota, 5 OTUs (0.29% of all sequences); Zygomycota, 57 OTUs (3.34% of all sequences); unidentified, 525 OTUs (30.77% of all sequences), 24 classes, and 272 genera. The remaining 16 OTUs (0.94% of all sequences) were affiliated to Ichtyosporea, and 95 OTUs (5.57% of all sequences) had no BLASTN hit against the GenBank database. The relative abundance and structure of the fungi observed among the three cultivars control and susceptible samples showed high variation in each one. The structures of the fungal communities in the control soil samples and the susceptible soil samples at harvest time were compared at the level of fungal classes (98.03% of all sequences) (Fig. 2).
Fig. 2. Relative abundances of the main fungal classes in the six soil samples (1C, 1L, 2C, 2L, 3C, and 3L) collected from Laixi, Shandong, China at harvest time. The relative abundances of those fungal DNA sequences could be ascribed to a class of fungal. Besides, many sequences were “unidentified.” The “no blast hit and other” means operational taxonomic units could not BLASTN hit against the GenBank database. Many DNA sequences could be ascribed to a class of Ichtyosporea but not fungi.
Compared to the control samples, the relative abundances of Ascomycota and Basidiomycota significantly increased, while the relative abundance of Fungal-unidentified significantly decreased in the susceptible samples. The relative abundance of Chytridiomycota detected from 2L was sharply lower compared to 2C. The relative abundances of Zygomycota detected in 1L increased, but decreased in 2L and 3L. The no blast hit OTUs in the pod rot samples increased its abundance in 1L and 3L, but decreased in 2L. Interestingly, the relative abundance of Ichtyosporea was detected in 1C and 2C, but none were detected in 1L and 2L (Fig. 2). These results mean that fungi community structures differ among the soil samples.
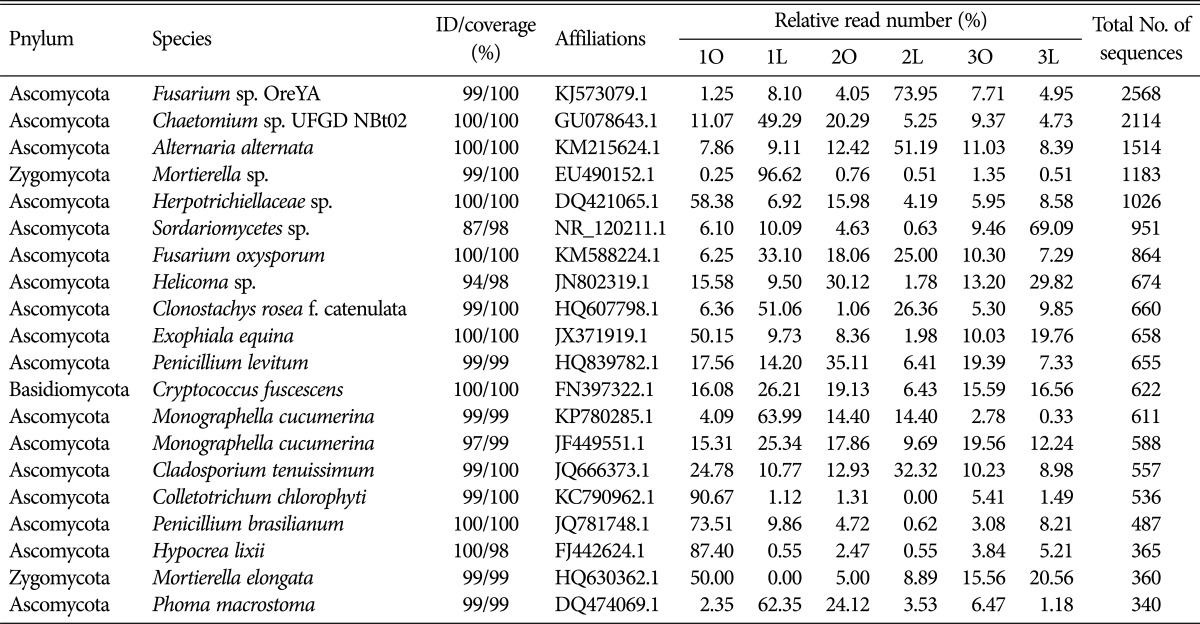
At the species level, according to the hits to the GenBank database, the 20 most abundant OTUs accounted for 37% of the total number of sequences and are listed in Table 2 in which the most abundance (> 2%) was Fusarium sp. OreYA, followed by Chaetomium sp. UFGD NBt02, Alternaria alternata, Mortierella, and Herpotrichiellaceae. In addition, blasted in the GenBank database, the other abundant OTUs (fungi-unidentified) accounted for 21% of the total number of sequences that are not listed in Table 2.
Table 2. Taxonomic affiliations of the 20 most abundant OTUs in six soil samples of three cultivars collected from Laixi, Shandong, China.
OTU, operational taxonomic units.
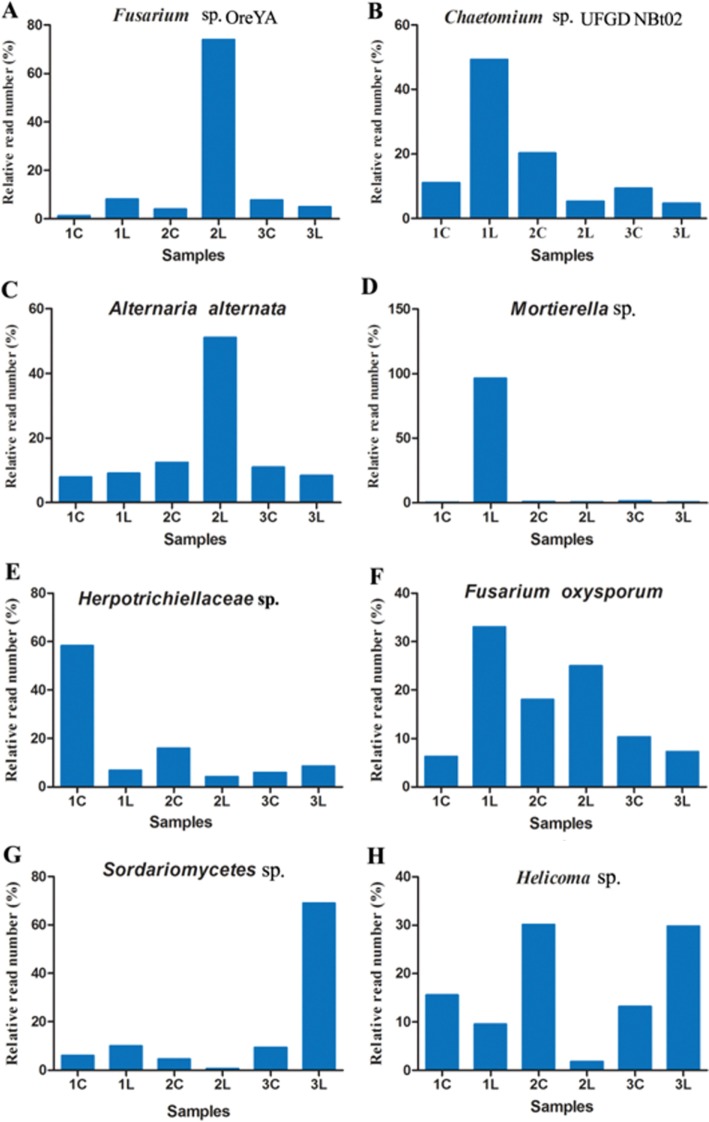
The most abundant OTU belonged to the fungal species Fusarium sp. OreYA, which had higher relative abundance in 1L than 1C, as well as Chaetomium sp. UFGD NBt02, Mortierella, and Fusarium oxysporum (Fig. 3A, 3B, 3D, and 3F). In contrast, lower relative abundance of Herpotrichiellaceae and Helicoma was observed in 1L (Fig. 3D and 3E). The relative abundance of Sordariomycetes and Alternaria alternata had no difference between 1C and 1L (Fig. 3). The relative abundance of species mentioned above, except for Chaetomium, Mortierella, and Sordariomycetes, had similar abundance in 2C and 2L (Fig. 3B, 3D, and 3G). Except for the species of Sordariomycetes and Helicoma, the relative abundance of other species had no significant difference between 3L and 3C as shown in Fig. 3G and 3H. Interestingly, the relative abundance of Mortierella increased significantly only in 1L. In general, there were many differences of species abundance among the six soil samples. Additionally, 30.77% of all sequences could not be classified into any species, which indicated that a large number of unknown fungi existed during the peanut pod rot pathogens infection.
Fig. 3. A–H, The eight species with the most abundant operational taxonomic units coverage in the six soil samples.
Fungal community similarity
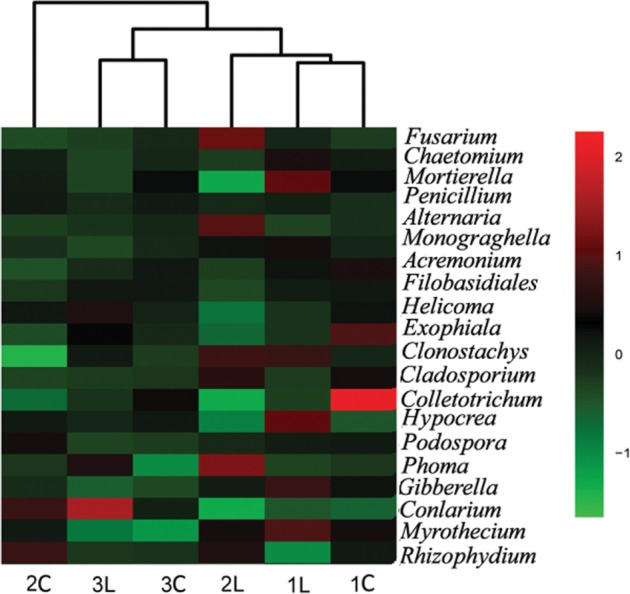
Based on the Bray-Curtis dissimilarity, hierarchical clustering was employed to analyze the community similarities of fungi (Fig. 4). The results showed that 1C and 1L clustered together, also together with 2L, indicating that the fungi communities from 1C and 1L were similar to 2L. In addition to the above group, 3L and 3C were grouped together, meaning that the fungal communities of 3C and 3L were similar, but were different from 1C and 1L. Particularly, 2C formed an independent branch, which indicated that the fungi community from 2C was clearly different from the others. The possible cause of this result is that peanut cultivar 2 may be a disease resistant peanut cultivar, which is different with other cultivars.
Fig. 4. Heatmap displaying the relative abundances of the greatest 20 genera (> 0.1%), using the soft R (pheatmap).
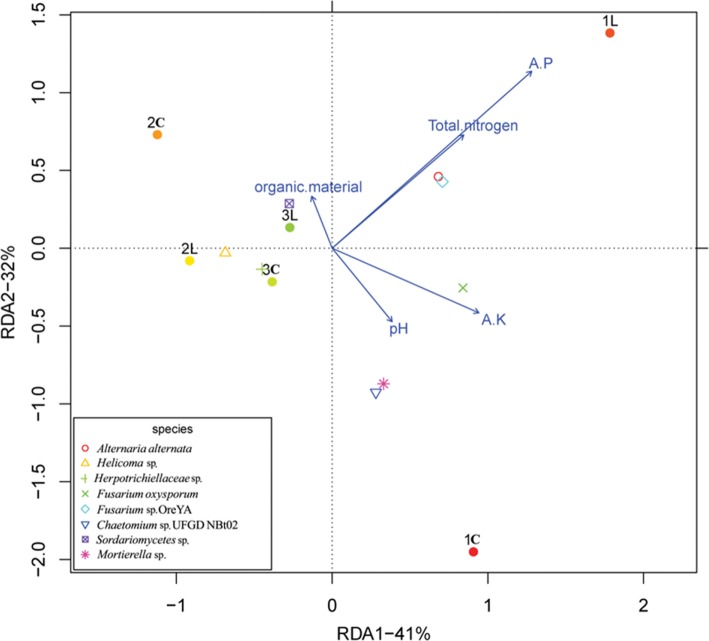
Fungal community correlation with peanut cultivars and environmental parameters
To further understand the correlation among fungi, samples, and environmental parameters, RDA analysis was performed (Fig. 5). The results showed that the fungal communities in cultivar 3 (3C and 3L) and cultivar 2 (2L) were clustered, whereas the fungal communities in cultivar 1 (1C and 1L) and cultivar 2 (2C) were separated from each other and from those in cultivars 2 and 3. It was also shown that soil environmental parameters (available P, total N, organic materials) in cultivars 2 and 3 were more similar to each other than to the ones in cultivar 1. At the same time, the RDA analysis showed that the fungi Fusarium sp. OreYA and Alternaria alternata were clustered, and significantly related to available P and total N. The fungi Chaetomium sp. UFGD NBt02 and Mortierella were clustered, having a relationship with pH. The fungi Herpotrichiellaceae, Helicoma, and Sordariomycetes were also clustered, while Fusarium oxysporum and Mortierella were separated from each other and from others. Furthermore, Fusarium oxysporum was significantly related to available K. Sordariomycetes was related to organic material. All the results demonstrated that environmental parameters, samples, and fungi have a complicated relationship with each other.
Fig. 5. Redundancy analysis of sample-fungi-physicochemical factors correlations on abundance data of operational taxonomic units. Arrows represent physicochemical factors. The different colors/symbols represent the most eight fungi species. Three samples of cultivar 1, 2, and 3 are indicated by red, orange, and green dots, respectively.
DISCUSSION
Fungal OTU characterization and diversity
The significance of fungal communities in the susceptible soils of the peanut cultivars is unclear, mainly because data on fungal species in this habitat are limited. The goals of this study were to analyze fungal communities in the soils of the three peanut cultivars which experience pod rot and find its correlation with environmental variables using high-throughput sequencing within the city of Laixi in Shandong province. The results of this study have provided much more information on fungal diversity and distribution patterns in soil in which peanut pod rot exists than previous studies [11,12]. By using the 454 pyrosequencing methods, we obtained over 46,000 effective reads falling into 1,706 OTUs. As known before, the fungal ITS region varies roughly between approximately 450 and 750 bp in length and consists of three subregions: the variable spacers ITS1 and ITS2 and the intercalary 5.8S gene. The length of sequences also showed that about 86.3% had a length of > 400 bp, with most ranging between 450 and 600 bp, which covered the ITS1 and most ITS2 regions. The results of this study imply that this sequence depth and scale should enable identification almost all possible fungi, and can be sufficiently informative in the context of ecological and microbiological studies [13].
In early studies, in the fungi associated with peanut shells and seeds, 70 genera and 146 species of fungi were identified [14], which was fewer than in the present results of 272 genera. In recent years, researchers were mostly focused on the separation of some known pathogenic fungi [15,16] due to peanut pod rot, while little study of the fungi community and diversity and its impact factors occurred. In this study, high levels of fungal diversity were found, and the taxonomic classification revealed that there were a lot of unknown taxa (8.628% unclassified sequences at species level), implying that many novel fungi existed in pod rot soils.
In analysis of the OTUs at the phylum level, in all of the soil samplings, sequences of Ascomycotas were dominant, followed by Basidiomycota in all six soil samples. This result was the same as Xu et al.'s findings [17,18], who also used deep amplicon sequencing to detect soil fungal communities. The low abundances of Zygomycota (3.34% of all sequences), Chytridiomycota (2.87% of all sequences), and Glomeromycota (0.29% of all sequences) were in accordance with the results reported by Li et al. [19] in peanut monocropping soils and also with the results of Sugiyama et al. [20] in potato fields. These results mean that fungal community structures at the phylum level appear to be similar across soil types.
Among fungal genera detected in the present study, the most abundant OTU belongs to the fungi Fusarium, including Fusarium sp. OreYA and Fusarium oxysporum, which cause wilt or root or pod rot. In particular, peanut pod rot caused by Fusarium lead to a great loss in pod yield [21]. Its relative abundance significantly increased in susceptible peanut soils, suggesting that this pathogen might be the main causal agent of peanut pod rot in our study fields.
The relative abundance of Chaetomium, Alternaria, Mortierella, and Sordariomycetes also significantly increased in peanut susceptible soils, in contrast to Herpotrichiellaceae and Helicoma. Among them, Chaetomium, a soft-rot fungi, was found to be an endophyte fungi which could colonize cotton [22]. Alternaria and Mortierella were separated from the soils [23]. Sordariomycetes were important fungi associated with plants [24]. Taken together, these results suggest that the causal agent of peanut pod rot might be more complex than recent reports have suggested.
In addition, previous studies have reported that the fungi of Cylindrocladium, Rhizoctonia solani, and Pythium could result in peanut pod rot [25]. But in this study, the fungi of Cylindrocladium was not found, which was the same result reported by Chen [26] in the same field for peanut continuous cropping, and the fungi of Rhizoctonia solani and Pythium were found in very low abundance. A possible reason may be that these fungi are obligate parasites and thus are predominately located in roots but not necessarily in our study field.
Community correlation with peanut cultivars and environmental variables. Studies on the soil microbial communities of some crops have reported that microbial communities differed between different pathogen-resistant cultivars [27]. However, in this study, the three peanut cultivars were all susceptible to pod rot pathogens. However, the present results revealed remarkable differences among the three peanut cultivars after pod rot pathogens infection and conducted a comprehensive investigation of the genetic diversity of peanut pod rot-related fungi with a high range of Shannon's diversity indices (H′ = 3.58–5.05). The Chao 1 index and the number of OTUs in both the three control soils were higher than in the susceptible cultivar soils (Table 1). Therefore, the cultivation-based evidence gave partial support to the result of the 454 pyrosequencing analysis.
Furthermore, a distinct spatial distribution of fungal communities in different susceptible cultivar samples was observed in the present study. In the phyla level, the abundances of Ascomycotas and Basidiomycotas were higher in the control samples than in the susceptible cultivars (Fig. 2). The relative read number of the eight most prevalent fungi detected in each sample indicated that they had a distinct distribution in the fungal communities in different cultivar samples (Fig. 3). The results of the heatmap and RDA analysis revealed significant variation in the composition of the fungal community among the three cultivar samples (Fig. 4 and 5). All of the above results suggest that cultivars did affect the soil fungal community diversity and abundance which may affect the resistance to fungal soil-borne pathogens [27]. In addition, the presence of Fusarium fungal pathogen “Fusarium sp. OreYA” and “Fusarium oxysporum” in all soil samples. The data can be explained in different way as following: pathogen potential (density) is low enough not to cause symptom or damage to peanut in 1L and 3L, not just by cultivar sensitivity.
What led to this distinct spatial distribution of the fungal community? It is conceivable that soil fungi may be shaped not only by host cultivars or genotypes but also by the habitat environment. Indeed, previous studies reported that the structure of fungal communities can be driven by many environmental factors, such as pH, organic matter content [5], nutrient concentration, total N, phosphorus, salinity, and temperature [23]. We thus collected basic information on the soil during the peanut pod rot experiment for all six samples. Interestingly, the lower pH value and higher content of total N in the susceptible samples could additionally explain the distinctness of the fungal community as compared to those in the control samples. The results on the effect of the soil pH are in line with a number of reports, and had been mentioned by Alceklett and Hart [28] in shaping fungal communities in soil ecosystems. Soil from susceptible cultivar 1 had lower level of organic matter content, but a higher level in susceptible cultivar 2 and 3. Higher organic matter content had also been detected in citrus AMF soil [5]. The environment conditions in different peanut cultivars may also influence the structure of fungal communities. In contrast to the cultivar 3 of the control sample (e.g., higher content of P and K), cultivars 1 and 2 have similar conditions regarding the content of P and K. These results suggest that environmental conditions may significantly influence community structure of pod rot fungi among different cultivars.
In summary, our data demonstrated the high diversity of fungal communities on peanut pod rot soil and indicated that the peanut cultivar is an important factors that harbors many fungal taxa, most of which are possibly influenced by environmental conditions. However, the study of ecological roles of these fungi and their mechanism remain poorly investigated. The mechanism of peanut resistance to pod rot pathogens is unclear. Therefore, in future studies, more information concerning the biogeography and the mechanism of peanut resistance to pod rot may be generated at both the taxonomic and the functional levels.
ACKNOWLEDGEMENTS
This study was supported by grants from the National Ten Thousand Youth alents Plan of 2014 (W02070268 to XC), China Agriculture Research System (CARS13), the National Natural Science Foundation of China (31000728 and 31200211), the Natural Science Fund of Shandong Province (ZR2014YL011 and ZR2014YL012), The Youth Scientific Research Foundation of Shandong Academy of Agricultural Sciences (2016YQN14), Agricultural scientific and technological innovation project of Shandong Academy of Agricultural Sciences (CXGC2016B02). We thank LetPub (www.letpub.com) for its linguistic assistance during the preparation of this manuscript.
References
- 1.Nayyar A, Hamel C, Lafond G, Gossen BD, Hanson K, Germida J. Soil microbial quality associated with yield reduction in continuous-pea. Appl Soil Ecol. 2009;43:115–121. [Google Scholar]
- 2.Deacon JW. Fungal biology. New York: John Wiley & Sons; 2009. [Google Scholar]
- 3.Brussaard L, De Ruiter PC, Brown GG. Soil biodiversity for agricultural sustainability. Agric Ecosyst Environ. 2007;121:233–244. [Google Scholar]
- 4.Nehls U. Mastering ectomycorrhizal symbiosis: the impact of carbohydrates. J Exp Bot. 2008;59:1097–1108. doi: 10.1093/jxb/erm334. [DOI] [PubMed] [Google Scholar]
- 5.Song F, Pan Z, Bai F, An J, Liu J, Guo W, Bisseling T, Deng X, Xiao S. The scion/rootstock genotypes and habitats affect arbuscular mycorrhizal fungal community in citrus. Front Microbiol. 2015;6:1372. doi: 10.3389/fmicb.2015.01372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sharma KK, Bhatnagar-Mathur P. Peanut (Arachis hypogaea L.) Methods Mol Biol. 2006;343:347–358. doi: 10.1385/1-59745-130-4:347. [DOI] [PubMed] [Google Scholar]
- 7.Fabra A, Castro S, Tauriani T, Angelini J, Ibañez F, Dardanelli M, Tonelli M, Bianucci E, Valetti L. Interaction among Arachis hypogaea L. (peanut) and beneficial soil microorganisms: how much is it known? Crit Rev Microbiol. 2010;36:179–194. doi: 10.3109/10408410903584863. [DOI] [PubMed] [Google Scholar]
- 8.Saleh OI. Wilt, root rot and seed diseases of groundnut in El-Minia Governorate, Egypt. Egypt J Phytopathol. 1997;25:1–18. [Google Scholar]
- 9.Melouk HA, Backman PA. Management of soilborne fungal pathogens. In: Melouk HA, Shokes FM, editors. Peanut health management. St. Paul (MN): APS Press; 1995. pp. 75–82. [Google Scholar]
- 10.Woodward JE, Baughman TA. Evaluation of Rhizoctonia diseases on peanut cultivars and advanced breeding lines in west Texas. Phytopathology. 2007;97:S183. [Google Scholar]
- 11.Sanogo S, Puppala N. Microorganisms associated with Valencia peanut affected by pod rot in New Mexico. Peanut Sci. 2012;39:95–104. [Google Scholar]
- 12.Wheeler TA, Howell CR, Cotton J, Porter D. Pythium species associated with pod rot on west Texas peanuts and in vitro sensitivity of isolates to mefenoxam and azoxystrobin. Peanut Sci. 2005;32:9–13. [Google Scholar]
- 13.Schoch CL, Seifert KA, Huhndorf S, Robert V, Spouge JL, Levesque CA, Chen W Fungal Barcoding Consortium; Fungal Barcoding Consortium Author List. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for fungi. Proc Natl Acad Sci U S A. 2012;109:6241–6246. doi: 10.1073/pnas.1117018109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hanlin RT. The distribution of peanut fungi in the southeastern United States. Mycopathol Mycol Appl. 1973;49:227–241. doi: 10.1007/BF02050716. [DOI] [PubMed] [Google Scholar]
- 15.Crous PW, Groenewald JZ, Risède JM, Simoneau P, Hyde KD. Calonectria species and their Cylindrocladium anamorphs: species with clavate vesicles. Stud Mycol. 2006;55:213–226. doi: 10.3114/sim.55.1.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lombard L, Crous PW, Wingfield BD, Wingfield MJ. Species concepts in Calonectria (Cylindrocladium) Stud Mycol. 2010;66:1–13. doi: 10.3114/sim.2010.66.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu L, Ravnskov S, Larsen J, Nilsson RH, Nicolaisen M. Soil fungal community structure along a soil health gradient in pea fields examined using deep amplicon sequencing. Soil Biol Biochem. 2012;46:26–32. [Google Scholar]
- 18.Xu L, Ravnskov S, Larsen J, Nicolaisen M. Linking fungal communities in roots, rhizosphere, and soil to the health status of Pisum sativum. FEMS Microbiol Ecol. 2012;82:736–745. doi: 10.1111/j.1574-6941.2012.01445.x. [DOI] [PubMed] [Google Scholar]
- 19.Li XG, Ding CF, Zhang TL, Wang XX. Fungal pathogen accumulation at the expense of plant-beneficial fungi as a consequence of consecutive peanut monoculturing. Soil Biol Biochem. 2014;72:11–18. [Google Scholar]
- 20.Sugiyama A, Vivanco JM, Jayanty SS, Manter DK. Pyrosequencing assessment of soil microbial communities in organic and conventional potato farms. Plant Dis. 2010;94:1329–1335. doi: 10.1094/PDIS-02-10-0090. [DOI] [PubMed] [Google Scholar]
- 21.Thiessen LD, Woodward JE. Diseases of peanut caused by soilborne pathogens in the southwestern United States. ISRN Agron. 2012;2012:517905 [Google Scholar]
- 22.Zhou W, Starr JL, Krumm JL, Sword GA. The fungal endophyte Chaetomium globosum negatively affects both above- and belowground herbivores in cotton. FEMS Microbiol Ecol. 2016;92:fiw158. doi: 10.1093/femsec/fiw158. [DOI] [PubMed] [Google Scholar]
- 23.Zhang T, Wei XL, Zhang YQ, Liu HY, Yu LY. Diversity and distribution of lichen-associated fungi in the Ny-Alesund Region (Svalbard, High Artic) as revealed by 454 pyrosequencing. Sci Rep. 2015;5:14850. doi: 10.1038/srep14850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Řehulka J, Kubátová A, Hubka V. Cephalotheca sulfurea (Ascomycota, Sordariomycetes), a new fungal pathogen of the farmes raibow trout Oncorhychus mykiss. J Fish Dis. 2016;39:1413–1419. doi: 10.1111/jfd.12477. [DOI] [PubMed] [Google Scholar]
- 25.Lechat C, Crous PW, Groenewald JZ. The enigma of Calonectria species occurring on leaves of llex aquifolium in Europe. IMA Fungus. 2010;1:101–108. doi: 10.5598/imafungus.2010.01.02.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen M, Li X, Yang Q, Chi X, Pan L, Chen N, Yang Z, Wang T, Wang M, Yu S. Soil eukaryotic microorganism succession as affected by continuous of peanut-pathogenic and beneficial fungi were selected. PLoS One. 2012;7:e40659. doi: 10.1371/journal.pone.0040659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yu C, Hu X, Deng W, Li Y, Han G, Ye C. Soil fungal community comparison of different mulberry genotypes and the relationship with mulberry fruit sclerotiniosis. Sci Rep. 2016;6:28365. doi: 10.1038/srep28365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aleklett K, Hart M. The root microbiota: a fingerprint in the soil? Plant Soil. 2013;370:671–686. [Google Scholar]