Abstract
The colonisation of new suitable habitats is crucial for species survival at evolutionary scale under changing environmental conditions. However, colonisation potential may be limited by philopatry that facilitates exploiting successful habitats across generations. We examine the mechanisms of long distance dispersal of the philopatric loggerhead sea turtle (Caretta caretta) by analysing 40 sporadic nesting events in the western Mediterranean. The analysis of a fragment of the mitochondrial DNA and 7 microsatellites of 121 samples from 18 of these nesting events revealed that these nests were colonising events associated with juveniles from distant populations feeding in nearby foraging grounds. Considering the temperature-dependent sex determination of the species, we simulated the effect of the incubation temperature and propagule pressure on a potential colonisation scenario. Our results indicated that colonisation will succeed if warm temperature conditions, already existing in some of the beaches in the area, extend to the whole western Mediterranean. We hypothesize that the sporadic nesting events in developmental foraging grounds may be a mechanism to overcome philopatry limitations thus increasing the dispersal capabilities of the species and the adaptability to changing environments. Sporadic nesting in the western Mediterranean can be viewed as potential new populations in a scenario of rising temperatures.
Introduction
Philopatry, or natal homing, has been defined as the return of the individuals to the natal location, usually to reproduce1,2 and thus exploit areas successfully used in past generations3. This strategy would proliferate in a species due to the ‘multiplier effect’, in which individuals with the ‘philopatric’ genotype increase in numbers in very successful reproductive areas in comparison to other behavioural genotypes4. Many advantages have been proposed as evolutionary drivers for this behaviour, including a higher probability of finding multiple mates for reproduction5, the use of optimal areas for raising the offspring3, an increase of the local adaptability6, or greater global genetic diversity7. However in some situations this strategy has also limitations that would favour an opposed ‘dispersal’ strategy, in which the individuals search for new areas8. Philopatry would limit the recovery of areas on the verge of extinction, increase kin competition9, favour habitat-dependent mortality10, or prevent the dispersal of the species8.
Philopatry has been found in very different taxa11–14 but marine turtles are one of the best examples of this strategy15. Early tag studies demonstrated that female turtles return to the same beaches to lay their eggs, sometimes with differences of only a few metres among nesting seasons16,17. Posterior genetic studies18,19 demonstrated that this site fidelity was in fact a true philopatry, as the reused nesting beaches corresponded to the beaches where the nesting females hatched, thus generating a strong female-mediated genetic structuring20. Recent studies have shown that males would also show high degree of philopatry21,22. The combination of natal imprinting and accurate navigation mechanism has been proposed as the key elements maintaining the philopatry in marine turtles and in other taxa2. Newborn hatchlings would memorize different chemical and magnetic cues of the nesting beaches where they hatch and use this information in adulthood to find the natal nesting beaches to reproduce15,23.
The potential limitations of philopatry are especially accentuated in marine turtle species, most of them of conservation concern24. Furthermore, many researchers have suggested the possible impact of the predicted climate change on marine turtles25 due to their Temperature Sex Determination (TSD)26. Global warming could increase feminisation of the populations and philopatry might prevent dispersal to colder nesting beaches to counteract this effect27–29. Nonetheless, the circumtropical distribution of most marine turtle species suggests the existence of mechanisms for long distance dispersal30. Consequently it has been proposed for marine turtles that ‘non-philopatric exploratory behaviours are needed to colonize new nesting environments on evolutionary time scales’31, ‘strays and wandering must occur, and are no doubt adaptively advantageous aberrations, necessary for colony proliferation’16 as ‘absolute natal homing, over the 100-million-year history of this group, would be a strategy for extinction’ 20.
Tagging has revealed that the typical distance between successive nesting sites of loggerhead turtle (Caretta caretta) individual females is 5 km or less32, although interchange of nesting females among more distant localities has also been reported33–35. In fact, the females nesting in the north-western Atlantic have a remigration rate close to 70%35, meaning that a significant portion of the nesting females are not strictly philopatric and lay their clutches in other nesting beaches. These deviations are of tens to hundreds of kilometres from the original nesting beach and could easily explain the lateral spread of the nesting areas along continuous or semi-continuous nesting habitats and thus the existence of Regional Management Units (RMU)36. However, these strays are not enough to explain the presence of very distant nesting beaches separated by vast marine areas. Thus, colonisation through long distance dispersal across oceans or seas is the only likely explanation for the current circumtropical distribution of most marine turtle species but these transoceanic dispersal strays have not yet been described in detail.
The Mediterranean Sea offers a unique scenario to ascertain how this long distance dispersal might operate, since the major nesting aggregations of the loggerhead sea turtle in the central and eastern part of this sea are the result of at least two independent colonization events during the late Pleistocene and the Holocene37. Furthermore, some sporadic nesting events of this species have been recently detected in the western Mediterranean38–42, defined as rare nesting events in an area where low or no nesting activity has been recorded to date. These clutches have always been found in the vicinity of developmental habitats for juveniles of Atlantic and Mediterranean nesting populations43–46 and most of them thousands of kilometres away from any known regular nesting area. One possible explanation for these sporadic nesting events is that they are relicts of ancient nesting populations in these locations, as some sporadic nesting had been reported in the past47,48. However, another possibility could be that these nests are examples of contemporary long distance dispersal events from distant nesting populations38.
The aim of the present study is to contextualize these sporadic nesting events in Caretta caretta under a philopatric scenario and examine the role of sporadic nesting in the long distance dispersal of this species. We thus aim to reconcile a philopatric strategy with a circumtropical distribution and evaluate the possible adaptability of this species under different global warming scenarios through long distance colonisation.
Results
Genetic analysis
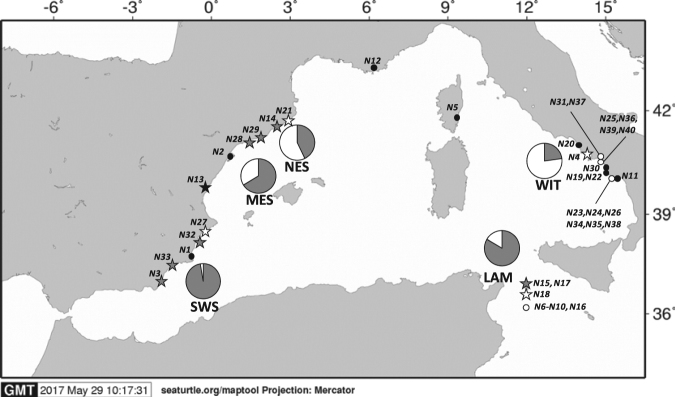
Evidence of a total of 40 sporadic nesting events were reported in the western and central Mediterranean in 1870, 1990 and from 2001 to 2015 (Table 1, Fig. 1). Almost all nesting events with biometric data (86%, Table 1) produced viable hatchlings, although almost half of the nests with information about the incubation duration (54%, Table 1) suggested a null or low production of females. Furthermore, it was not possible to obtain samples from all the clutches, considering that some were laid before starting the present study, while in other cases no samples for genetic studies were collected by the discoverers or were preserved in formalin. For the genetic analyses we obtained 121 samples (120 hatchlings plus one nesting female) from 18 different clutches in the western Mediterranean (Table 2, Supplementary Data S1). A total of six mitochondrial DNA haplotypes were found among the samples, all of them described in previous studies and found in the GenBank (Table 2). From these six haplotypes, two of them are reported as exclusive from the Atlantic nesting beaches (CC-A1.1 and CC-A9.1) and three of them are common in both Atlantic and Mediterranean nesting beaches (CC-A2.1, CC-A3.1 and CC-A20.1)49. The remaining haplotype (CC-A10.4) has only been reported from the nesting population of Melbourne beach (Florida, USA)50 and from an adult individual foraging in Drini bay (Albania)51. However, the short (~380 bp) sequence of this haplotype (CC-A10) had been also found in the nesting population of Zakynthos island (Greece)52 where no long version of this haplotype has yet been described. Hence, this haplotype may be present as well in the eastern Mediterranean nesting populations. Individual assignments using microsatellites revealed different origins for the samples from different clutches (Table 2, Figs 1 and 2). Hatchlings from the nests with Atlantic mtDNA haplotypes were assigned to the Atlantic while those from the nest with the CC-A10.4 haplotype were assigned to the Mediterranean. Hatchlings from nests with common mtDNA haplotypes were assigned either to the Atlantic or Mediterranean nesting beaches (Table 2) or could not be assigned, perhaps because they have an admixed ancestry resulting from the reproduction of individuals from different origin. However, we cannot discard that the mixed probabilities found are due to the lack of resolution of the markers as observed in previous studies46,53.
Table 1.
Evidence of sporadic nests recorded in the western and central Mediterranean.
| Code | Site | Description | Date | Hs | Eg | Tª | Id | Of | Habitat | Predicted Hs | Reference | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1950–2000 | 2020 | 2050 | 2080 | |||||||||||
| N1 | Mar Menor (Spain) | Possible nest | 1870 | — | — | — | — | Good | 35.9 | +10 | +5 | +5 | 48 | |
| N2 | Ebro delta (Spain) | Dead embryo | 09/1990 | — | — | — | — | — | Moderate | 21.7 | +10 | +10 | +10 | 47 |
| N3 | Vera (Spain) | Full nest | 27/07/2001 | 43.3 | 97 | — | 58 | 10–72 | Good | 35.9 | +10 | +10 | +5 | Present study41 |
| N4 | Baia Domizia (Italy) | Full nest# | 11/07/2002 | 49.4 | 92 | 27.5 | 64 | 0–22 | Good | 35.9 | +10 | +10 | +10 | Present study39 |
| N5 | Palombaggia (France) | 4 shells, 2 eggs | 20/11/2002 | — | — | — | — | — | Marginal¢ | 14.5 | +10 | +10 | +10 | 40 |
| N6 | Conigli (Italy) | Full nest# | 04/06/2002 | 69.5 | 128 | — | 77 | 0–0 | Good | 79.6 γ | 0 | −5 | −15 | Present study67 |
| N7 | Conigli (Italy) | Full nest# | 20/06/2002 | 71.3 | 94 | — | 69 | 0–5 | Good | 79.6 γ | 0 | −5 | −15 | Present study67 |
| N8 | Conigli (Italy) | Full nest | 20/06/2002 | 90.8 | 130 | — | 73 | 0–2 | Good | 79.6 γ | 0 | −5 | −15 | Present study67 |
| N9 | Conigli (Italy) | Full nest# | 04/07/2002 | 64.5 | 138 | — | 65 | 0–16 | Good | 79.6 γ | 0 | −5 | −15 | Present study67 |
| N10 | Conigli (Italy) | Full nest# | 11/07/2002 | 99.1 | 106 | — | 64 | 0–21 | Good | 79.6 γ | 0 | −5 | −15 | Present study67 |
| N11 | Marina di Camerota (Italy) | Egg remains | 17/10/2004 | — | — | — | — | — | Good | 35.9 | +10 | +10 | +10 | 64 |
| N12 | Sain Tropez (France) | Full nest | 18/07/2006 | 0 | 141 | 22–28 | — | 0§ | Marginal | 14.5 | +10 | +10 | +10 | 42 |
| N13 | Puzol (Spain) | Full nest# | 11/08/2006 | 36.8 | >78 | — | >50 | — | Moderate | 21.7 | +10 | +10 | +10 | Present study38 |
| N14 | Premià de Mar (Spain) | Full nest | 27/10/2006 | 68.3 | 82 | — | — | — | Marginal | 14.5 | +10 | +10 | +10 | Present study38 |
| N15 | Conigli (Italy) | Full nest# | 19/07/2006 | 67.3 | 110 | 28.8 | 57 | 10–85 | Good | 79.6 γ | 0 | −5 | −15 | Present study67 |
| N16 | Conigli (Italy) | Full nest# | 23/07/2006 | 85.1 | 94 | 28.8 | 56 | 10–93 | Good | 79.6 γ | 0 | −5 | −15 | Present study67 |
| N17 | Conigli (Italy) | Full nest | 07/08/2006 | 92.6 | 94 | 27.9 | 67 | 0–10 | Good | 79.6 γ | 0 | −5 | −15 | Present study67 |
| N18 | Conigli (Italy) | Full nest# | 26/08/2006 | 0 | 85 | 26.7 | — | 0§ | Good | 79.6 γ | 0 | −5 | −15 | Present study67 |
| N19 | Ogliastro (Italy) | Full nest# | 26/07/2006 | 33.3 | 93 | 27.1 | 73 | 0–16 | Good | 35.9 | +10 | +10 | +10 | 64 |
| N20 | Lucrino (Italy) | Full nest# | 15/07/2008 | 92.2 | 115 | 34.5 | 46 | 100–100 | Good | 35.9 | +10 | +10 | +10 | 64 |
| N21 | Malgrat de Mar (Spain) | Full nest | 01/10/ 2011 | 70.8 | 120 | — | — | — | Marginal | 14.5 | +10 | +10 | +10 | Present study |
| N22 | Ogliastro (Italy) | Full nest | 28/08/2012 | 75.3 | 73 | — | — | — | Good | 35.9 | +10 | +10 | +10 | 64 |
| N23 | Palinuro (Italy) | Full nest | 18/08/2013 | 62.1 | 132 | — | — | — | Good | 35.9 | +10 | +10 | +10 | 64 |
| N24 | Palinuro (Italy) | Full nest | 15/07/2013 | 99.0 | 96 | 28.9 | 56 | 15–93 | Good | 35.9 | +10 | +10 | +10 | 64 |
| N25 | Battipaglia (Italy) | Full nest | 12/10/2013 | — | 110 | — | — | — | Good | 35.9 | +10 | +10 | +10 | 64 |
| N26 | Palinuro (Italy) | Full nest | 06/12/2013 | Predated | 48 | — | — | — | Good | 35.9 | +10 | +10 | +10 | 64 |
| N27 | Alicante (Spain) ω | Full nest# | 30/6/2014 | 79 | 131 | 29.1 | 59 | 3–57 | Moderate | 21.7 | +10 | +10 | +10 | Present study |
| N28 | Tarragona (Spain) | Full nest# | 31/10/2014 | 0.0 | 89 | — | — | 0§ | Marginal | 14.5 | +10 | +10 | +10 | Present study |
| N29 | Tarragona (Spain) | Full nest | 30/10/2014 | 62.1 | 58 | — | — | — | Marginal | 14.5 | +10 | +10 | +10 | Present study |
| N30 | Acciaroli (Italy) | Full nest | 30/07/2014 | 95.8 | 118 | 28.4 | 60 | 7–50 | Good | 35.9 | +10 | +10 | +10 | 64 |
| N31 | Capaccio (Italy) | Full nest | 25/08/2014 | 91.5 | 117 | — | — | — | Good | 35.9 | +10 | +10 | +10 | 64 |
| N32 | Torrevieja (Spain) | Full nest# | 31/07/2015 | 52.9 | 85 | — | 69 | 0–5 | Moderate | 21.7 | +10 | +10 | +10 | Present study |
| N33 | Pulpi (Spain) | Full nest# | 17/07/2015 | 32.5 | 80 | — | 53.5 | 87–100 | Good | 35.9 | +10 | +10 | +10 | Present study |
| N34 | Marina di Camerota (Italy) | Full nest | 19/06/2015 | 83.0 | 99 | 31.1 | 50 | 90–100 | Good | 35.9 | +10 | +10 | +10 | 64 |
| N35 | Marina di Camerota (Italy) | Full nest | 07/07/2015 | 80.0 | 60 | — | 58 | 22–72 | Good | 35.9 | +10 | +10 | +10 | 64 |
| N36 | Ascea Marina (Italy) | Full nest# | 18/07/2015 | 80.5 | 87 | 29.0 | 56 | 20–93 | Good | 35.9 | +10 | +10 | +10 | 64 |
| N37 | Eboli (Italy) | Full nest | 28/08/2015 | Predated | — | — | — | — | Good | 35.9 | +10 | +10 | +10 | 64 |
| N38 | Marina di Camerota (Italy) | Full nest | 29/07/2015 | 89.3 | 56 | 29.9 | 58 | 10–95 | Good | 35.9 | +10 | +10 | +10 | 64 |
| N39 | Ascea Marina (Italy) | Full nest | 29/07/2015 | 78.2 | 55 | 29.1 | 57 | 30–85 | Good | 35.9 | +10 | +10 | +10 | 64 |
| N40 | Ascea Marina (Italy) | Full nest# | 30/07/2015 | 25.9 | 58 | 29.2 | 62 | 0–59 | Good | 35.9 | +10 | +10 | +10 | 64 |
Review of the evidence of sporadic nesting of the loggerhead turtle in the western and central Mediterranean and summary of the data collected in each report. Hs: hatchling success (%); Eg: number of eggs reported; Tª: mean temperature reported, Id: incubation duration (days); Of: minimum and maximum estimated percentage of female offspring using Id and four different models73,100–102 with the corresponding corrections when needed103; Habitat: habitat suitability as predicted in the literature63 and classified in quartiles as Excellent, Good, Moderate and Marginal; Predicted Hs: present and future Hatchling succes as predicted in the literature77, future values are expressed as the expected percentage of variation related to current values. (#) Nest relocated or incubated; (ω) Nest laid at San Juan, (Alicante) and relocated to a protected beach ~200 km north (Valencia), in the same place as nest (N13) (Fig. 1); (§) All the eggs died; (¢) No habitat modelling available for this area so the value of the closest modelled area is reported; (γ) No modelling data on hatchling success specifically reported for Lampedusa Island, so predictions for this site may not be accurate.
Figure 1.
Western Mediterranean sporadic nests and foraging areas. Location of the sporadic nesting events coded as in Table 1. Stars indicate nesting events with genetic data and reliable assignation to the Atlantic (grey), to the Mediterranean (white) or mixed (black). White dots indicate nesting events with genetic data but with no reliable assignment due to resulting low (<0.80) probabilities or due to not having microsatellite data when published64. Black dots indicate nesting events without genetic data. Pie graphs show the percentage of Atlantic (grey) and Mediterranean (white) turtles visiting the developmental foraging grounds located in the vicinity of the sporadic nesting events. SWS: south Western Spain; MES: mid Eastern Spain; NES; north Eastern Spain; WIT: Western Italy; LAM: Lampedusa43–46,52,71. Map created using the free software MAPTOOL (SEATURTLE.ORG Maptool. 2002. SEATURTLE.ORG, Inc. http://www.seaturtle.org/maptool/ 29 May 2017), that uses GMT (The Generic Mapping Tool)99.
Table 2.
Summary of the genetic results of the sporadic nest in the western Mediterranean.
| Code | Nest | Hatchlings assayed | nDNA | mtDNA | |||
|---|---|---|---|---|---|---|---|
| Origin | Probability of Atlantic assignment | Minimum n° fathers | Haplotypec | Haplotype origin | |||
| N3 | Vera | 15 | Atlantic | Fig. 2 | 2 | CC-A3.1 | Shared |
| N4 | Dominizia | 1 | Mediterranean | 0.009 | na | CC-A10.4 | Atlanticd |
| N6 | Conigli | 1 | Unknown | 0.668 | na | CC-A2.1 | Shared |
| N8 | Conigli | 1 | Unknown | 0.686 | na | CC-A2.1 | Shared |
| N9 | Conigli | 1 | Unknown | 0.781 | na | CC-A2.1 | Shared |
| N10 | Conigli | 1 | Unknown | 0.720 | na | CC-A2.1 | Shared |
| N13 | Puzol | 6 | Mixed | Fig. 2 | 2 | CC-A2.1 | Shared |
| N14 | Premià de Mar | 23 | Atlantic | Fig. 2 | 2 | CC-A1.1 | Atlantic |
| N15 | Conigli | 1 | Atlantic | 0.926 | na | CC-A9.1 | Atlantic |
| N16 | Conigli | 2 | Unknown | 0.576b | na | CC-A2.1 | Shared |
| N17 | Conigli | 2 | Atlantic | 0.8144b | na | CC-A2.1 | Shared |
| N18 | Conigli | 14 | Mediterranean | Fig. 2 | 1 | CC-A2.1 | Shared |
| N21 | Malgrat de Mar | 2 | Mediterranean | 0.118 | na | CC-A2.1 | Shared |
| N27 | Alicante | 14 | Mediterranean | Fig. 2 | 2 | CC-A20.1 | Shared |
| N28 | Tarragona | 1 | Atlantic | 0.888 | na | CC-A3.1 | Shared |
| N29 | Tarragona | 18 | Atlantic | Fig. 2 | 2 | CCA2.1 | Shared |
| N32 | Torrevieja | 10 | Atlantic | Fig. 2 | 2 | CCA2.1 | Shared |
| N33 | Pulpi | 8a | Atlantic | Fig. 2 | 1 | CCA2.1 | Shared |
| TOTAL | 121 | ||||||
Nests coded as in Table 1. nDNA: results of the individual assignments of the hatchlings including the probabilities to be associated to the Atlantic nesting beaches and the minimum number of fathers detected. Bold values indicate high probability (>0.8) of individual assignments. mtDNA: haplotypes found and nesting area from where they have been reported (shared refers to both Atlantic and Mediterranean). na: not applicable as no more than two hatchlings were sampled for these nests as needed by GERUD v 2.0. aThis includes one sample of the nesting female. bMean value of the two samples. cGenBank accession numbers for the identified haplotypes (CC-A1.1 = EU179436, CC-A2.1 = EU179445, CC-A3.1 = EU179455, CC-A9.1 = EU179463, CC-A10.4 = JQ340912, CC-A20.1 = EU179452) dA 380 bp short version of this haplotype has been found in the Mediterranean nesting beach of Zakynthos (Greece)52.
Figure 2.
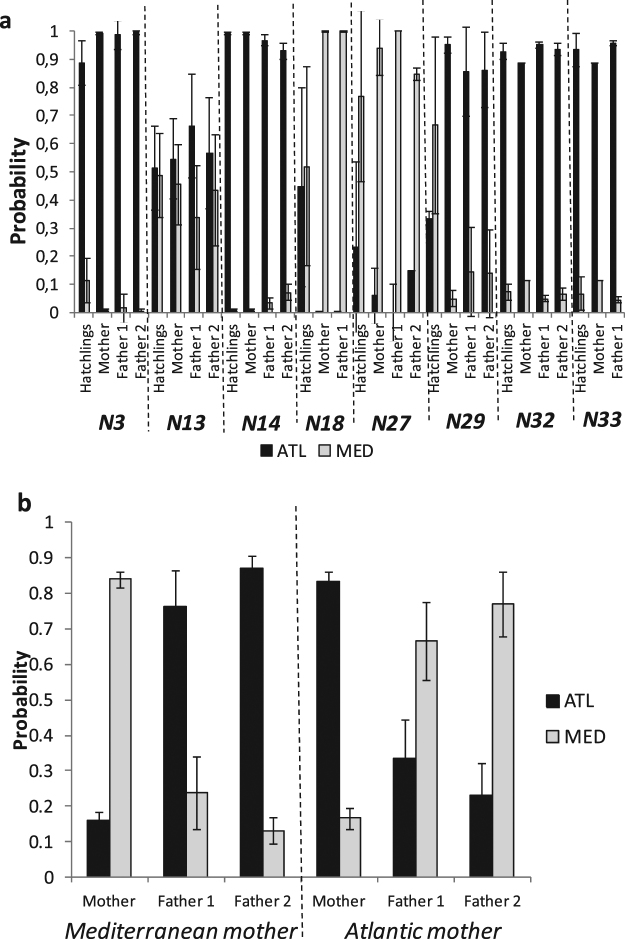
Individual Assignments of the sporadic nests with multiple samples assayed. (a) mean individual assignments of the sampled hatchlings and the inferred possible mothers and fathers of each one of the nests with more than two hatchling samples (b) mean individual assignments from Puzol nest (N13) for the two different possible fathers after assigning the putative the mother (Mediterranean or Atlantic). The Y axis represents the probability of belonging to either the Mediterranean (MED: grey) or the Atlantic (ATL: black) groups created by STRUCTURE46. Standard deviations are indicated by error bars. Nests coded as in Table 1.
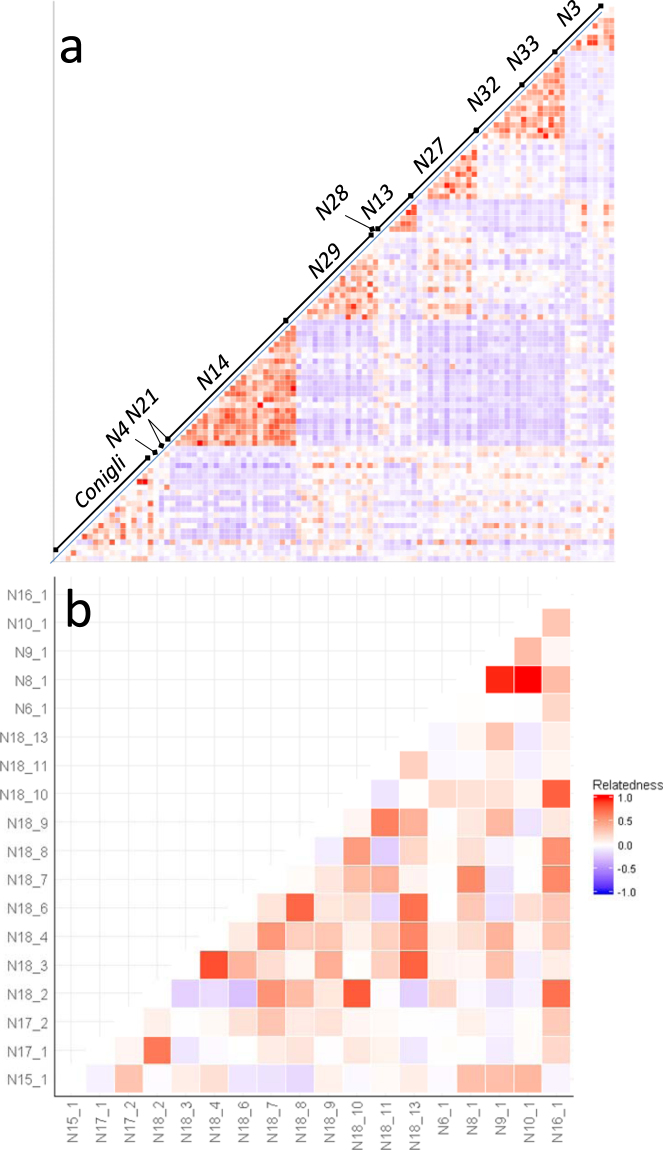
We collected samples from more than two hatchlings for 8 of the 18 clutches. Multiple paternities were found by GERUD in all but two of these clutches, resulting in a minimum number of 2 fathers per nest (Table 2). The inferred genotypes of all possible mothers and fathers presented high assignment probabilities to the same nesting area as the offspring. The assignation of the parents were generally better than the offspring (e.g. nest N18, Fig. 2a) probably because all the possible combinations of parents integrated the allele information of all the sampled hatchlings. For instance, rare alleles with high discriminating power may appear only in some hatchlings, and thus only the hatchlings with these alleles will have good assignations, but they will always be present in any parent combination. The nest N13 (Fig. 2a) was an exception as both the hatchlings and the possible parents yielded mixed assignment values resulting in inconclusive mean values. In this case, we separated the putative mothers that were assigned to the Atlantic from those assigned to the Mediterranean and we then reassigned the corresponding pair of fathers inferred by GERUD. If the mother was assigned to the Mediterranean with a good probability, the fathers were assigned to the Atlantic and vice versa (Fig. 2b), thus indicating that the parents had different origin. The relatedness analysis showed that the values obtained among samples within the same clutch were higher than the values obtained among samples of different clutches (Fig. 3a). However, the values obtained within samples of some specific pairs of sporadic nesting events were of the same magnitude than the values obtained within a clutch thus suggesting some level of relatedness. Specifically, the samples of the nests N32 and N33 showed high levels of the relatedness and shared the same maternal haplotype (Fig. 3a, Table 2). Furthermore, the genotype of the mother sampled in Pulpi (nest N33) was compatible with the offspring from the nest in Torrevieja (nest N32) and the two possible fathers inferred for Pulpi nest were also found among the possible father combinations of Torrevieja nest for the genotype of the Pulpi mother. This female, assigned to be of Atlantic origin (Table 2, Fig. 3a) measured 74 cm of curved carapace length (CCL), slightly below of the mean maturation size of Atlantic turtles visiting the Mediterranean of 80 cm CCL54. Considering our results and that the period between the two nests (14 days, Table 1) is within the interesting interval of the species, we hypothesize that these two nests were laid by the same female. The relatedness values among hatchlings of some of the nests of Conigli Beach (Fig. 3b) were generally high (specifically between the nests N8-N9 and N8-N10) suggesting that the same or related females have been laying different clutches in that area.
Figure 3.
Heatmap of the relatedness index among sample pairs of the Mediterranean sporadic nests. Each cell represents the value of Lynch & Ritland relatedness index91 among individual samples as plotted in both the y and x axis. Panel a) includes the values of all the possible sample pairs with the samples of each nest indicated in the diagonal. Panel b) includes the values of all the sample pairs of the nests laid in Conigli Beach (Italy).
Effect of incubation temperature and propagule pressure
A model was performed under different scenarios to test the impact of the incubation temperature and propagule pressure rate (Pr) on colonisation success. Colonisation was possible only at temperatures high enough to produce some females in all scenarios (>28 °C) and was generally faster at warmer temperatures, as the percentage of hatchling females produced increases allowing for descendent females to identify the new area as a suitable nesting site and return as adults to lay their eggs (Fig. 4). However, colonisation decreased again at very high temperatures (33 °C), mainly associated to a decrease of the emergence success due to excess of heating55. As expected, Pr values impacted the colonisation process, by accelerating the time when the first returning females start to contribute to the growing population (Fig. 4). Even at very low Pr values (1 nest per 100 years) colonisation was possible if temperature conditions were appropriate but hundreds of years would be needed to establish a new nesting population (Fig. 4a).
Figure 4.
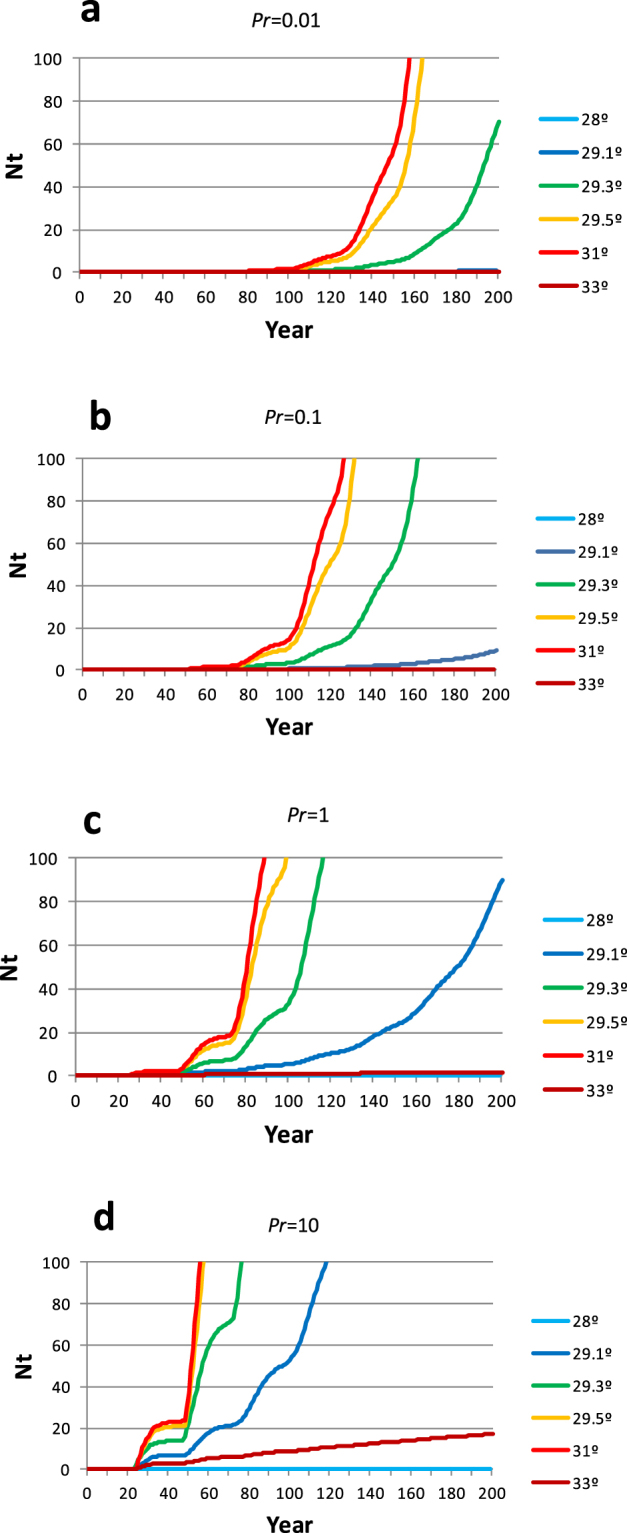
Model of colonisation under different incubation temperatures. The model shows the variation of the annual number of nesting females (Nt) during the colonisation process at different incubation temperatures and propagule pressure rates. Four different propagule pressure rates (Pr) were modelled, (a) 1 sporadic nest per a hundred years (Pr = 0.01), (b) 1 sporadic nest per ten years (Pr = 0.1), (c) 1 sporadic nest per year (Pr = 1) and (d) 10 sporadic nest per year (Pr = 10).
Discussion
Marine turtles have existed for at least 110 million years20,56. Considering that all existing species exhibit at least some degree of philopatry, it is reasonable to conclude that it originated at least several tens of millions years ago, before the current marine turtle families diverged57. The Earth temperature has experienced drastic oscillations during this period58,59, and thus the temporal heterogeneity of the nesting beaches has been necessarily very high. Considering the constraints imposed by TSD and philopatry, it is surprising how this taxon has survived to these oscillations while other lineages went extinct60. Furthermore, in almost all marine turtle species with an oceanic stage, philopatry coexists with a circumtropical distribution61. As a consequence, long dispersal mechanisms must be present to escape from philopatry constraints and allow the colonisation of distant areas. These mechanisms would explain the apparent adaptability of these species to historical temperature changes, their wide distribution and the existence of widespread common haplotypes30. By examining the rare loggerhead sea turtle sporadic nesting events in the western Mediterranean we argue that any delay of the migration from developmental foraging habitats to the adult nesting habitats may result in the establishment of a new population in the vicinity of juvenile foraging grounds, if the conditions of beach temperature are appropriate.
Sporadic nesting events: relicts or colonisers
Our results suggest that the sporadic nests found in the central and western Mediterranean are not remnants of a past population but the result of an ongoing process of colonisation from distant nesting beaches. Many hatchlings were assigned to Mediterranean or Atlantic populations with high probability values while previous studies have shown a deep genetic differentiation between Atlantic and eastern Mediterranean nesting beaches suggesting a profound isolation of these two distant nesting areas46. Furthermore, nests with very different origins were found at close distances, suggesting independent colonisation and not consistent with the hypothesis of an historical population in the western Mediterranean.
The second line of evidence comes from the habitats where these clutches were laid. Beach temperature data derived from temperature recorders deployed at 50 cm below beach surface62, our data collected in some of the reported nesting events (Table 1) and simulations from air temperature data63 indicate that almost all available nesting habitats along the European shore of the western Mediterranean remain below the pivotal temperature and hence are too cold to support a viable population. There are some exceptions, as some of the beaches within the western Mediterranean could potentially host a nesting population (Table 2)63, including southern Spain, southern Italy and the African coast. However, although they may be suitable nesting habitats, they are not of the highest quality63 as egg viability is predicted to be much lower than at regular nesting sites, as our in situ data collection confirmed (Table 1). Thus the expected production of females would be generally low. Furthermore, temperature conditions can be highly variable among years62, and all clutches were, to some extent, manipulated as a consequence of their discovery leading in some cases to artificially increase the proportion of females (Table 1)64. Thus, the expected production of females in some areas could be greater in warmer years as estimated at Conigli Beach in Lampedusa, where nests from two different years were obtained: 2006 was warm enough to produce a significant amount of females, while almost only males were produced in 2002. The disparity of nest temperatures among years was also found in another set of records from the Italian coast65, with an expected higher production of females on warmer years.
As a final line of evidence, beach patrolling was done in all the areas where these sporadic nests were found in the same period of the year and subsequent years, with no female return detected38,62 with the exception of Conigli Beach66,67 and the Thyrrenian64. The conditions in Conigli Beach are more favourable, as indicated by the data collected in situ and by the habitat suitability (Table 1). The repeated nesting found in different years, and the evidence of female returns66,67 suggests that perhaps these locations hosts a very small and new growing population, being in Lampedusa first reported in 197568 and in the Tyrrhenian in 200264. These new populations may be maintained by the females produced in warmer years along with independent recurrent sporadic nesting, probably similar to the nesting events in Calabria69 or in Sicily island70. Our tests of relatedness performed in Conigli nests point in that direction, as some of the nests seem to be related. Finally the clutches from nests N32 and N33 were laid by the same nesting female, within the same nesting season and with a renesting interval normal for the species. Considering all these lines of evidence, we can thus conclude that these sporadic nets are the result of accidental nesting events from individuals originated in distant populations and that mating is possible in foraging areas far away from regular nesting areas.
Long distance colonisation mechanism
Considering our results, we propose a long distance colonisation mechanism for the philopatric loggerhead sea turtle. If the individuals undertaking developmental migrations mature before returning to their nesting areas of origin, they would produce sporadic nesting events near the developmental foraging area, as their nesting beaches of origin would be too far to lay the eggs. Under this hypothesis, it is not surprising that the only female that we could sample and measure as a sporadic nester had an Atlantic origin and a size of 74 cm CCL. This size is slightly below the maturation size of individuals of Atlantic origin developed within the Mediterranean54, despite being large enough to return to her natal area through the Straits of Gibraltar71. Once a sporadic nest is laid, all the females born would be imprinted by the same mechanisms that maintain the philopatry in the species, causing them to return to the new nesting area if they survive to adulthood. This would fix the new nesting location as the place to return in only one generation and without losing philopatry as previously suggested2. This is not unprecedented, as this imprinting mechanism has been the basis of reintroduction projects, like the establishment of a new Kemp’s Ridley nesting area in Texas (USA) by imprinting hatchlings from Rancho Nuevo (Mexico) nesting area72.
Furthermore, the process of colonisation is subject to the production of females and thus it is dependent on temperature. Mean incubation temperatures below 28.3 °C during the middle third of incubation period73, would produce no females to return to the new nesting beaches and the colonisation would fail independently of the number of sporadic nests laid in the new area. However, if the temperature increases, the sporadic nesting events, as those observed in the western Mediterranean would produce more females and potentially allow the establishment of new nesting areas under optimal conditions. Consequently, sporadic nesting nearby developmental feeding areas could act as a dormant mechanism of colonisation that activates when the environmental conditions change. Propagule pressure was long identified as the major single parameter predicting the success of biotic introductions74, and hence isolated nesting events will hardly result into the establishment of new populations of loggerhead turtles. Our results showed that low colonisation rates could potentially establish a new population but hundreds of years would be needed, while faster rates would produce the same effect in only a few decades. Thus, all factors affecting the propagule pressure would have an impact on the probability of colonisation success. For instance, it has been hypothesized that warming of the regular nesting areas would imply a significant population growth through the production of more females, thus increasing the number of females and thus the rate of potential colonisers75. A reduced number of sporadic nests is likely to produce a bottleneck in the potential new population leading to high levels of inbreeding. However, the multiple paternity found in all but two of the nests, in one case even involving parents from very distant nesting areas (nest N13, and among Conigli beach nests), could help to increase the overall genetic variability, reducing the bottleneck effect and thus increasing the potential success of the colonisation process.
Colonisation of the Mediterranean
The Mediterranean loggerhead nesting populations were originated from Atlantic individuals during the Pleistocene before the last glacial maximum (20,000–200,000 years ago) and some of the nesting populations have been suggested to be older than others37,76. In some cases, the genetic structure among populations have been explained by recolonisations within the basin as the species distribution range retracted during the colder phases of the Pleistocene to warmer refugia (such as Libya, Greece and Turkey) and expanded again within the eastern Mediterranean, when the thermal conditions changed. However, this hypothesis hardly explains the genetic structuring of some of the nesting areas in the central Mediterranean69. Previous studies concluded that the high levels of genetic diversity and the presence of one haplotype (CC-A20.1) shared with Atlantic nesting populations in the nesting population of Calabria (Italy) results from an independent colonisation event from Atlantic individuals that migrated into the Mediterranean during the Holocene (<10,000 years)37,69. This has been explained by the fact that central Mediterranean has mean temperatures colder than the eastern basin but warmer than the western basin63. Considering our results, we suggest that the general warming produced during the Holocene could have activated the colonisation process of the central Mediterranean area from sporadic nesting events close to developmental feeding grounds. A similar process could be happening now in Conigli Beach in Lampedusa island, having higher temperatures than all the other reported sporadic nesting sites and probably some of the nests reported already are females born in these areas that are returning after reaching maturity. If Conigli Beach is really an ongoing colonisation, the Atlantic haplotype CC-A9.1 would be the most recent acquired haplotype of the Mediterranean nesting populations.
We thus suggest the hypothesis of a sequential colonisation of the Mediterranean; the eastern Mediterranean was colonized before the last glacial maximum37, the Calabria (Southern Italy) was colonized during the Holocene37 and the central and western Mediterranean may be an ongoing colonisation process (present study), depending on the environmental conditions including global warming28,63,77. Future monitoring of the western Mediterranean potential nesting beaches along the next decades would clarify if we are really facing this ongoing colonisation process and how the origin of the new colonizers impacts on the whole basin.
Currents in the Straits of Gibraltar and the southwestern Mediterranean could play an important role in this potential new colonisation as they trap loggerhead turtles of Atlantic origin within the Mediterranean for long periods, thus probably increasing the chances that they lay a sporadic nest before they return to their beaches of origin71. These currents are reinforced during cold periods78, but if its strength lowers under warmer temperatures, the Atlantic individuals would not be trapped anymore within the western Mediterranan, thus lowering the colonisation pressure in this area. Furthermore, stochastic factors are expected to play an important role in the colonisation process, not in vain we have only detected a minimum of two independent colonisation processes in the Mediterranean in the last 65,000 years37,69. For instance, sea temperature may have an impact in hatchling survival, as it is predicted that the temperature at sea is not going to rise in the same proportion that at the beaches64. Thus, low winter sea surface temperatures may limit the survival and self recruitment of hatchlings.
Conservation implications
The mechanism for long distance expansion of nesting habitat proposed here would be an alternative method for the species to expand its distribution to new suitable distant habitats, as the old nesting habitats become suboptimal. For instance, a warming of Mediterranean sea surface temperatures from east to west has been shown, starting in 1970 and continuing in this century28. Considering our results, a general warming would favour the colonisation of the western Mediterranean during the following decades28, while the eastern Mediterranean nesting areas would become too hot thus decreasing hatchlings survival as predicted in previous studies77. The combination of contiguous range expansion and long distance dispersal mechanisms has proved to be effective in an evolutionary scale, as these animals have survived the drastic climate changes of the last 110 million years. However, whether or not these mechanisms would be fast enough to counterbalance some of the effects of the current climate change is something that remains to be tested. In any case, several potential conservation and research actions may be considered in the light of our results. The first one would be the monitoring of the effect of warming temperatures on sex ratio, inbreeding and nestling survival on current nesting beaches. The detection of sporadic nesting events through extensive monitoring of potential suitable habitats, coupled with protection and conservation of newly colonised sites, might facilitate the possible expansion and long term survival of the species. Furthermore, assisted migration through egg translocation might be an effective action to promote the creation of new populations in more suitable habitats72. The detection of these rare events through extensive monitoring on potential suitable habitats, coupled with its protection and conservation may be crucial to facilitate the possible expansion and long term survival of the species.
Methods
Genetic analysis
Loggerhead turtle sporadic nesting events are very rare and thus their sampling is necessarily opportunistic (Table 1, Fig. 1). When a nesting event occurred, we collected the basic biometric data of the nest and at least one sample per clutch from a dead hatchling and/or embryo found after emergence. When possible, several hatchlings were sampled in order to test for multiple paternities. The collection of samples was conducted in strict accordance with Spanish and European regulations. The Ethics Comitttee of Animal Experimentation of the University of Barcelona stated that the analysed procedures fit the essential ethical rules and the legislation in force according to the article RD2013 of B.O.E from 8th of February 2013. This sort of studies is excluded from the area of application of the legislation, and therefore, the corresponding authorization of this Ethics Comitee is not needed. Furthermore, although the study species is listed in CITES, transportation of samples within the European Union does not require CITES permits. Muscle or skin samples of 120 hatchlings and one female were collected from 18 clutches (Table 2) and stored in 95% ethanol. DNA was extracted using the QIAamp extraction kit (QIAGEN®) following the manufacturer’s instructions.
Individual assignments of all samples to either Atlantic or Mediterranean nesting beaches were done using a combination of a fragment of the mtDNA control region and seven microsatellites46,71. As a first step, we amplified a long (~800 bp) fragment of the mitochondrial DNA (mtDNA) control region of one sample per clutch, using the primers LCM15382 and H95079 as it has been proven to be much more informative49 than the shorter fragment (~380 bp) of the same region used in previous studies44,80. We used the same PCR conditions as in previous studies (e.g.37). Sequences were aligned using BioEdit v7.1.1181 and compared to known loggerhead haplotypes found in the database maintained by the Archie Carr Centre for Sea Turtle Research (http://accstr.ufl.edu/). Published haplotype frequencies on nesting populations37,49,50,82–84 were compared with our samples and the origin of the nesting female was directly determined if it had an exclusive haplotype. Additionally, we genotyped seven microsatellites of all the samples using primers previously used for this species: Cm84, Cc117, Cm72 and Ei885; Cc141 and Cc786; and a modified version80 of Ccar17687 using the same protocols described in the literature80. The combination of the seven microsatellites was checked against the baseline of Atlantic and Mediterranean individuals used in a previous study to perform individual assignments46 using the program STRUCTURE v 2.3.488. This baseline comprised a total of 112 individuals of Mediterranean origin and 56 individuals of Atlantic origin (Supplementary Dataset S1). A previous study46 indicated that the best number of clusters of this baseline was K = 2 and that Atlantic and Mediterranean samples were highly differentiated (FST = 0.029, P < 0.001). Five samples of this baseline were genotyped again for the present study in order to check for changes in allele sizing and thus all microsatellite data was compatible after a correction for allele size changes (data not shown). An input file was prepared including both the baseline and the sporadic nests genotypes. All the samples of the baseline were labelled as belonging either to the Mediterranean (1) or to the Atlantic (2) while all sporadic nests samples were labelled as belonging to an additional group (3). Only samples from the baseline were used as locprior and thus no prior was assumed for the sporadic nests samples. STRUCTURE was run under the assumption of no admixture, considering the differentiation between the Atlantic and Mediterranean samples of the baseline46, performing 1,000,000 repetitions after a 100,000 burn-in period and assuming K = 2. Each assignation was replicated 10 times and the results were averaged using CLUMPP v1.269. Hence, a probability of being either Atlantic or Mediterranean was ascribed to each sample. Samples with assignment values higher than 0.8 to one of the two groups were assumed to be reliable as previous studies recommended46.
Multiple paternity was tested for all clutches sampled for more than two hatchlings, using the microsatellite data and the program GERUD v 2.0. This software allows the reconstruction of parental genotypes from half-sib progeny with unknown parents and infers multiple paternity by identifying more than four different parental alleles from a clucth90. Additionally, we generated the genotypes of all possible mothers and fathers of our offspring and we made individual assignments of up to the 100 most probable parents using the same methodology described above. Furthermore, we calculated the Lynch & Ritland relatedness index91 between all hatchling pairs using GenAlex v 6.592. Values were multiplied by two as indicated in the program to allow the index to vary between −1 and 1. Relatedness values among all sample pairs were used to create a heatmap using ‘ggplots2’93.
Effect of incubation temperature and propagule pressure
The population dynamics in a colonisation process under ideal conditions was modelled using a simplification of the population models developed in the literature94–96 and implemented on an ExcelTM spreadsheet in order to assess the impact of the temperature at incubation and propagule pressure. We defined the propagule pressure rate (Pr) as the number of sporadic clutches laid in a new area per year by individuals from distant populations (colonisers). Considering the number and frequency of the sporadic nests found in the western Mediterranean, we assumed that accidental colonisers lay a single nest before returning to their original nesting area and that the colonisation rate is constant in the whole area. These sporadic colonisers are expected to produce in a year (t) a number of hatchling females (Fc(t)) equivalent to the mean annual number of eggs laid per nest (En), the emergence success (Es) and the expected percentage of females produced per nest (Of) as in equation 1.
| 1 |
The females born in the new nesting area will potentially return in the future as adults to lay their own nests as a result of philopatry through natal imprinting as in equation 2. Thus the annual number of hatchling females produced by the females originally hatched in the new nesting area (Fp(t)) would be:
| 2 |
Which is basically the same as equation 1 but considering N(t) as the number of reproductive females in a given year (t) originated in the new population and Cy the mean number of clutches per female laid each year. As a consequence, the total number of female hatchlings produced at any year (t) in the new nesting area (F(t)) is the sum of the hatchling females produced by the colonisers and those produced by the reproductive females originally hatched in the new population (Equation 3).
| 3 |
At the start of the colonisation process, N(t) value is zero (and so is Fp(t)), as no resident females are found in the new area. Thus, the production of hatchling females in the area would be caused only by the sporadic colonisers until some of the females born in the area settle back there to reproduce. This will happen when the newborn hatchling females reach maturity after Ma years. We thus consider the number of recruited females in any time (R(t)) as the result of the female hatchlings produced Ma years before in the area and that survived to reproduce as adults (Equation 4).
| 4 |
where Sm is the survival rate from hatch to maturity. These recruits will continue to reproduce in the area as a consequence of the philopatry during the following years. Thus, the nesting female population size (N(t)) at any moment, measured as the number of reproductively active females in the population, is the sum of all the females recruited up to Tr years ago that survived to this moment to reproduce (Equation 5).
| 5 |
where Tr is the mean duration of the reproductive period (years) and Sa is the annual survival for adults. As a consequence, the temporal changes in population size from the start of the colonisation can be obtained. In order to test the colonisation process under different scenarios, we used a different combination of parameters that are dependent on the temperature of the middle third of incubation (Table 3). A recent study showed that these reproductive parameters are likely to be independent of population size97 so we assumed that they would be similar in a colonising scenario. Furthermore, four different propagule pressure rates (Pr) were modelled: 1 sporadic nest per a hundred years (Pr = 0.01), 1 sporadic nest per ten years (Pr = 0.1) and 1 sporadic nest per year (Pr = 1) and 10 sporadic nests per year (Pr = 10). Finally, the upper threshold for population size is supposed to be produced by limiting factors related to the nesting area (probably density dependent). As population sizes range from 20 to 687 females in Mediterranean nesting rookeries98, we considered these values as a rough guide for what would be normal population sizes in the Mediterranean after a colonisation.
Table 3.
Summary of the demographic parameters used in the present study.
| Nest temperature | 28 °C | 29.1 °C | 29.3 °C | 29.5 °C | 31.0 °C | 33.0 °C | Reference | |
|---|---|---|---|---|---|---|---|---|
| I d | Incubation duration (days) | 63 | 54 | 53 | 52 | 47 | 43 | 55,73 |
| O f | Percent female in offspring (%) | 0 | 25 | 50 | 75 | 100 | 100 | 73 |
| E s | Emergence success (%) | 90 | 90 | 90 | 90 | 75 | 10 | 55 |
| E n | Mean eggs per nest | 112 | 112 | 112 | 112 | 112 | 112 | 104 |
| C y | Mean number of clutches per year | 3 | 3 | 3 | 3 | 3 | 3 | 104 |
| S m | Survival to maturation (%) | 0.5 | 0.5 | 0.5 | 0.5 | 0.5 | 0.5 | 96 |
| M a | Age of maturation (year)* | 24/29 | 24/29 | 24/29 | 24/29 | 24/29 | 24/29 | 54 |
| S a | Annual survival for adults (%) | 80.9 | 80.9 | 80.9 | 80.9 | 80.9 | 80.9 | 105,106 |
| T r | Duration of reproductive stage (years) | 23–35 | 23–35 | 23–35 | 23–35 | 23–35 | 23–35 | 96,106 |
Six different temperatures during the middle third of the incubation were considered. Some of the parameters were assumed to be constant in the range of temperatures considered. *First value for turtles of Mediterranean origin and second value for turtles of Atlantic origin.
Electronic supplementary material
Acknowledgements
This study was co-funded by project CGL2009-10017 and Zoo de Barcelona. C. C. and M. Pascual are supported by the projects CTM2013-48163 and CTM2017-88080 (AEI/FEDER, UE) of the Spanish Ministry of Economy and Competitivity. C. C. is funded by the Beatriu de Pinós programme of the Generalitat de Catalunya. J. T. is supported by project Prometeo/2011/40 of Conselleria de Educació (Generalitat Valenciana). C. C., M. Pascual and L. C. are part of the research groups 2014SGR-336 and 2014SGR-1364 of the Generalitat de Catalunya. The authors thank staff and volunteers of the Nature Reserve ‘Isola di Lampedusa’ for their help in data recording and samples collection. Beach monitoring in this location was supported by projects LIFE99 NAT/IT/006271 and LIFE03 NAT/IT/000163. We wish also to thank the Conselleria de Infraestructuras, Territorio y Medio Ambiente of the Generalitat Valenciana, and particularly Juan Eymar, for their suport in the study of the two clutches laid in the Valencia Region.
Author Contributions
C.C., L.C., M.Pascual, J.T. and A.M. conceived and designed the study. C.C., J.T., S.H., J.B., P.G., M.Parga and S.P. obtained the samples and in situ data from the sporadic nests. C.C. did the genetic analysis, C.C. and L.C. did the colonisation modelling. L.C., M.Pascual and A.M. contributed with the costs of the genetic analysis and C.C. wrote the manuscript with input and review from all the authors.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
A correction to this article is available online at https://doi.org/10.1038/s41598-018-22718-7.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-19887-w.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Greenwood PJ. Mating systems, philopatry and dispersal in birds and mammals. Animal Behaviour. 1980;28:1140–1162. doi: 10.1016/S0003-3472(80)80103-5. [DOI] [Google Scholar]
- 2.Cury P. Obstinate nature: an ecology of individuals. Thoughts on reproductive behaviour and biodiversity. Can. J. Fish. Aquat. Sci. 1994;51:1664–1673. doi: 10.1139/f94-167. [DOI] [Google Scholar]
- 3.Refsnider, J. M. & Janzen, F. J. Putting eggs in one basket: ecological and evolutionary hypotheses for variation in oviposition-site choice in Annual Review of Ecology, Evolution, and Systematics, Vol. 41 Annual Review of Ecology Evolution and Systematics (eds D. J. Futuyma, H. B. Shafer, & D. Simberloff) 39–57 (Annual Reviews, 2010).
- 4.McNamara JM, Dall SRX. The evolution of unconditional strategies via the ‘multiplier effect’. Ecol. Lett. 2011;14:237–243. doi: 10.1111/j.1461-0248.2010.01576.x. [DOI] [PubMed] [Google Scholar]
- 5.Sheldon BC. Male phenotype, fertility and the pursit of extra-pair copulations by female birds. P Roy Soc B-Biol Sci. 1994;257:25–30. doi: 10.1098/rspb.1994.0089. [DOI] [Google Scholar]
- 6.Kawecki TJ, Ebert D. Conceptual issues in local adaptation. Ecol. Lett. 2004;7:1225–1241. doi: 10.1111/j.1461-0248.2004.00684.x. [DOI] [Google Scholar]
- 7.Chesser RK. Gene diversity and female philopatry. Genetics. 1991;127:437–447. doi: 10.1093/genetics/127.2.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.LePage C, Cury P. How spatial heterogeneity influences population dynamics: Simulations in SeaLab. Adapt. Behav. 1996;4:255–281. doi: 10.1177/105971239600400303. [DOI] [Google Scholar]
- 9.Clutton-Brock TH, Lukas D. The evolution of social philopatry and dispersal in female mammals. Mol. Ecol. 2012;21:472–492. doi: 10.1111/j.1365-294X.2011.05232.x. [DOI] [PubMed] [Google Scholar]
- 10.Ekroos J, Ost M, Karell P, Jaatinen K, Kilpi M. Philopatric predisposition to predation-induced ecological traps: habitat-dependent mortality of breeding eiders. Oecologia. 2012;170:979–986. doi: 10.1007/s00442-012-2378-9. [DOI] [PubMed] [Google Scholar]
- 11.Lewis SE. Roost fidelity of bats - a review. J. Mammal. 1995;76:481–496. doi: 10.2307/1382357. [DOI] [Google Scholar]
- 12.Hoffman JI, Forcada J. Extreme natal philopatry in female Antarctic fur seals (Arctocephalus gazella) Mamm. Biol. 2012;77:71–73. doi: 10.1016/j.mambio.2011.09.002. [DOI] [Google Scholar]
- 13.Foerster RE. The sockeye salmon. Oncorhynchus nerka. Bulletin Fisheries Research Board of Canada No. 1968;162:1–422. [Google Scholar]
- 14.Smith AT, Ivins BL. Colonization in a Pika population -dispersal vs philopatry. Behav. Ecol. Sociobiol. 1983;13:37–47. doi: 10.1007/BF00295074. [DOI] [Google Scholar]
- 15.Lohmann, K. J., Lohmann, C. M. F., Brothers, J. R. & Putman, N. F. Natal homing and imprinting in sea turtles in The biology of sea turtles, volume III (eds J. Wyneken, K. J. Lohmann, & J. A. Musick) 59–78 (CRC Press, 2013).
- 16.Carr A, Carr MH, Meylan AB. The ecology and migrations of sea turtles part 7: the west Caribbean green turtle colony. Bulletin of the American Museum of Natural History. 1978;162:1–46. [Google Scholar]
- 17.Carr A, Ogren L. The ecological migrations of sea turtles, 4. The green turtle in the Caribbean Sea. Bulletin of the American Museum of Natural History. 1960;121:1–48. [Google Scholar]
- 18.Meylan AB, Bowen BW, Avise JC. A genetic test of the natal homing versus social facilitation models for green turtle migration. Science. 1990;248:724–727. doi: 10.1126/science.2333522. [DOI] [PubMed] [Google Scholar]
- 19.Allard MW, Miyamoto MM, Bjorndal KA, Bolten AB, Bowen BW. Support for natal homing in green turtles from mitochondrial DNA sequences. Copeia. 1994;1994:34–41. doi: 10.2307/1446668. [DOI] [Google Scholar]
- 20.Bowen BW, Karl SA. Population genetics and phylogeography of sea turtles. Mol. Ecol. 2007;16:4886–4907. doi: 10.1111/j.1365-294X.2007.03542.x. [DOI] [PubMed] [Google Scholar]
- 21.Dutton PH, et al. Population stock structure of leatherback turtles (Dermochelys coriacea) in the Atlantic revealed using mtDNA and microsatellite markers. Conserv. Genet. 2013;14:625–636. doi: 10.1007/s10592-013-0456-0. [DOI] [Google Scholar]
- 22.Naro-Maciel E, et al. From refugia to rookeries: Phylogeography of Atlantic green turtles. J. Exp. Mar. Biol. Ecol. 2014;461:306–316. doi: 10.1016/j.jembe.2014.08.020. [DOI] [Google Scholar]
- 23.Lohmann, K. J., Witherington, B. E., Lohmann, C. M. F. & Salmon, M. Orientation, navigation and natal beach homing in sea turtles in The biology of sea turtles (eds P.L. Lutz & J. A. Musick) 107–136 (CRC Press, 1997).
- 24.IUCN. Red list of threatened species. Version 2013. 3. http://www.iucnredlist.org/. (2016). [DOI] [PMC free article] [PubMed]
- 25.Fuentes M, Pike DA, Dimatteo A, Wallace BP. Resilience of marine turtle regional management units to climate change. Global Change Biology. 2013;19:1399–1406. doi: 10.1111/gcb.12138. [DOI] [PubMed] [Google Scholar]
- 26.Yntema CL, Mrosovsky N. Sexual differentiation in hatchling loggerheads (Caretta caretta) incubated at different controlled temperatures. Herpetologica. 1980;36:33–36. [Google Scholar]
- 27.Hawkes LA, Broderick AC, Godfrey MH, Godley BJ. Investigating the potential impacts of climate change on a marine turtle population. Global Change Biology. 2007;13:923–932. doi: 10.1111/j.1365-2486.2007.01320.x. [DOI] [Google Scholar]
- 28.Witt MJ, Hawkes LA, Godfrey MH, Godley BJ, Broderick AC. Predicting the impacts of climate change on a globally distributed species: the case of the loggerhead turtle. J. Exp. Biol. 2010;213:901–911. doi: 10.1242/jeb.038133. [DOI] [PubMed] [Google Scholar]
- 29.Hawkes LA, Broderick AC, Godfrey MH, Godley BJ. Climate change and marine turtles. Endang. Species Res. 2009;7:137–154. doi: 10.3354/esr00198. [DOI] [Google Scholar]
- 30.Jensen, M. P., Fitzsimmons, N. N. & Dutton, P. H. Molecular genetics of sea turtles in The biology of sea turtles, volume III (eds J. Wyneken, K. J. Lohmann, & J. A. Musick) 135–162 (CRC Press, 2013).
- 31.Stiebens VA, et al. Living on the edge: how philopatry maintains adaptive potential. P Roy Soc B-Biol Sci. 2013;280:20130305. doi: 10.1098/rspb.2013.0305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schroeder, B., Foley, A. M. & Bagley, D. A. Nesting patterns, reproductive migrations, and adult foraging areas of loggerhead turtles in Loggerhead Sea Turtles (eds A. Bolten & B. Witherington) 114–124 (Smithsonian Books, 2003).
- 33.Margaritoulis, D. Interchange of nesting loggerheads among Greek beaches. in Proceedings of the seventeenth annual sea turtle symposium.NOAA Technical Memorandum Vol. NMFS-SEFSC-415 (eds S. Epperly & J. Braun-McNeill) 225–227 (National Marine Fisheries Service, Southeast Fisheries Science Center, 1998).
- 34.Lebuff CRJ. Unusual nesting relocation in the loggerhead turtle Caretta caretta. Herpetologica. 1974;30:29–31. [Google Scholar]
- 35.Richardson, T. H., Richardson, J. I., Ruckdeschel, C. & Dix, M. W. Remigration patterns of loggerhead sea turtles (Caretta caretta) nesting on Little Cumberland and Cumberland Islands, Georgia. Florida Marine Research Publications, 39–44 (1978).
- 36.Wallace, B. P. et al. Regional Management Units for Marine Turtles: A Novel Framework for Prioritizing Conservation and Research across Multiple Scales. Plos One5 (2010). [DOI] [PMC free article] [PubMed]
- 37.Clusa M, et al. Mitochondrial DNA reveals Pleistocenic colonisation of the Mediterranean by loggerhead turtles (Caretta caretta) J. Exp. Mar. Biol. Ecol. 2013;439:15–24. doi: 10.1016/j.jembe.2012.10.011. [DOI] [Google Scholar]
- 38.Tomás J, et al. Is the Spanish coast within the regular nesting range of the Mediterranean loggerhead sea turtle (Caretta caretta)? Journal of the Marine Biological Association of the United Kingdom. 2008;88:1509–1512. doi: 10.1017/S0025315408001768. [DOI] [Google Scholar]
- 39.Bentivegna F, Treglia G, Hochscheid S. The first report of a loggerhead turtle Caretta caretta nest on the central Tyrrhenian coast (western Mediterranean) Marine Biodiversity Records. 2008;1:1–3. doi: 10.1017/S1755267205001430. [DOI] [Google Scholar]
- 40.Delaugerre M, Cesarini C. Confirmed nesting of the loggerhead turtle in Corsica. Marine Turtle Newsletter. 2004;104:12. [Google Scholar]
- 41.Tomas J, Mons JL, Martin JJ, Bellido JJ, Castillo JJ. Study of the first reported nest of loggerhead sea turtle, Caretta caretta, in the Spanish Mediterranean coast. Journal of the Marine Biological Association of the United Kingdom. 2002;82:1005–1007. doi: 10.1017/S0025315402006537. [DOI] [Google Scholar]
- 42.Sénégas JB, Hochscheid S, Groul JL, Lagarrigue B, Bentivegna F. Discovery of the northernmost loggerhead sea turtle (Caretta caretta) nest. Marine Biodiversity Records. 2008;2:e81. doi: 10.1017/S1755267209000773. [DOI] [Google Scholar]
- 43.Clusa M, et al. Fine-scale distribution of juvenile Atlantic and Mediterranean loggerhead turtles (Caretta caretta) in the Mediterranean Sea. Mar. Biol. 2014;161:509–519. doi: 10.1007/s00227-013-2353-y. [DOI] [Google Scholar]
- 44.Carreras C, et al. Genetic structuring of immature loggerhead sea turtles (Caretta caretta) in the Mediterranean Sea reflects water circulation patterns. Mar. Biol. 2006;149:1269–1279. doi: 10.1007/s00227-006-0282-8. [DOI] [Google Scholar]
- 45.Maffucci F, Kooistra W, Bentiveyna F. Natal origin of loggerhead turtles, Caretta caretta, in the neritic habitat off the Italian coasts, Central Mediterranean. Biol. Conserv. 2006;127:183–189. doi: 10.1016/j.biocon.2005.08.009. [DOI] [Google Scholar]
- 46.Carreras C, et al. Living Together but Remaining Apart: Atlantic and Mediterranean Loggerhead Sea Turtles (Caretta caretta) in Shared Feeding Grounds. J. Hered. 2011;102:666–677. doi: 10.1093/jhered/esr089. [DOI] [PubMed] [Google Scholar]
- 47.Llorente GA, Carretero MA, Pascual X, Perez A. New record of a nesting loggerhead turtle Caretta caretta in western Mediterranean. British Herpetological Society Bulletin. 1992;42:14–17. [Google Scholar]
- 48.Salvador, A. Guia de los anfibios y reptiles españoles. (lnstituto Nacional para la Conservacion e la Naturaleza, Madrid., 1974).
- 49.Shamblin, B. M. et al. Geographic Patterns of Genetic Variation in a Broadly Distributed Marine Vertebrate: New Insights into Loggerhead Turtle Stock Structure from Expanded Mitochondrial DNA Sequences. Plos One9 (2014). [DOI] [PMC free article] [PubMed]
- 50.Shamblin BM, et al. Expanded mitochondrial control region sequences increase resolution of stock structure among North Atlantic loggerhead turtle rookeries. Mar Ecol Prog Ser. 2012;469:145–160. doi: 10.3354/meps09980. [DOI] [Google Scholar]
- 51.Yilmaz C, Turkozan O, Bardakci F, White M, Kararaj E. Loggerhead turtles (Caretta caretta) foraging at Drini Bay in Northern Albania: Genetic characterisation reveals new haplotypes. Acta Herpetol. 2012;7:155–162. [Google Scholar]
- 52.Laurent L, et al. Molecular resolution of marine turtle stock composition in fishery bycatch: a case study in the Mediterranean. Mol. Ecol. 1998;7:1529–1542. doi: 10.1046/j.1365-294x.1998.00471.x. [DOI] [PubMed] [Google Scholar]
- 53.Clusa, M. et al. Potential bycatch impact on distinct sea turtle populations is dependent on fishing ground rather than gear type in the Mediterranean Sea. Mar. Biol. 163 (2016).
- 54.Piovano S, et al. Different growth rates between loggerhead sea turtles (Caretta caretta) of Mediterranean and Atlantic origin in the Mediterranean Sea. Mar. Biol. 2011;158:2577–2587. doi: 10.1007/s00227-011-1759-7. [DOI] [Google Scholar]
- 55.Matsuzawa Y, Sato K, Sakamoto W, Bjorndal KA. Seasonal fluctuations in sand temperature: effects on the incubation period and mortality of loggerhead sea turtle (Caretta caretta) pre-emergent hatchlings in Minabe, Japan. Mar. Biol. 2002;140:639–646. doi: 10.1007/s00227-001-0724-2. [DOI] [Google Scholar]
- 56.Hirayama R. Oldest known sea turtle. Nature. 1998;392:705–708. doi: 10.1038/33669. [DOI] [Google Scholar]
- 57.Naro-Maciel E, Le M, FitzSimmons NN, Amato G. Evolutionary relationships of marine turtles: a molecular phylogeny based on nuclear and mitochondrial genes. Mol. Phylogenet. Evol. 2008;49:659–662. doi: 10.1016/j.ympev.2008.08.004. [DOI] [PubMed] [Google Scholar]
- 58.Emiliani C. Pleistocene temperatures. J. Geol. 1955;63:538–578. doi: 10.1086/626295. [DOI] [Google Scholar]
- 59.Savin SM, Douglas RG, Stehli FG. Tertiary marine paleotemperatures. Geol. Soc. Am. Bull. 1975;86:1499–1510. doi: 10.1130/0016-7606(1975)86<1499:TMP>2.0.CO;2. [DOI] [Google Scholar]
- 60.Raup DM, Sepkoski JJ. Mass extinctions in the marine fossil record. Science. 1982;215:1501–1503. doi: 10.1126/science.215.4539.1501. [DOI] [PubMed] [Google Scholar]
- 61.Bowen BW, et al. Global phylogeography of the ridley sea turtles (Lepidochelys spp.) as inferred from mitochondrial DNA sequences. Genetica (Dordrecht) 1998;101:179–189. doi: 10.1023/a:1018382415005. [DOI] [PubMed] [Google Scholar]
- 62.de Haro A, Capalleras X, Budó J. CARETTA.CAT. Estudi de la viabilitat de la implantació d’una población nidificant de tortuga careta. (Caretta caretta) a Catalunya. Treballs de la Societat Catalana d’Herpetologia. 2012;6:1–44. [Google Scholar]
- 63.Pike DA. Climate influences the global distribution of sea turtle nesting. Glob. Ecol. Biogeogr. 2013;22:555–566. doi: 10.1111/geb.12025. [DOI] [Google Scholar]
- 64.Maffucci, F. et al. Seasonal heterogeneity of ocean warming: a mortality sink for ectotherm colonizers. Scientific Reports6 (2016). [DOI] [PMC free article] [PubMed]
- 65.Bentivegna F, et al. Loggerhead Turtle (Caretta caretta) Nests at High Latitudes in Italy: A Call for Vigilance in the Western Mediterranean. Chelonian Conserv. Bi. 2010;9:283−+. doi: 10.2744/CCB-0862.1. [DOI] [Google Scholar]
- 66.Prazzi, E., Nicolini, G., Piovano, S. & Giacoma, C. La spiaggia del Conigli a Lampedusa: un modello di gestione per la conservazione di Caretta caretta in Atti II Congresso SHI Abruzzo Molise “Testuggini e Tartarughe” (eds L. Di Tizio, L. Brugnola, A. Cameli, & N Di Francesco) 127–133 (Ianeri ed., Pescara, 2013).
- 67.Prazzi E, Nicolini G, Piovano S, Giacoma C. Protezione di Caretta caretta (Reptilia Chelonia) nella riserva naturalle di Lampedusa. Il Naturalista siciliano. 2010;34:265–294. [Google Scholar]
- 68.Giacoma, C. et al. Caretta caretta (Linnaeus, 1758) in Fauna d’Italia Vol. 45 (eds C. Corti et al.) (Ed. Calderini, 2011).
- 69.Garofalo L, Mingozzi T, Mico A, Novelletto A. Loggerhead turtle (Caretta caretta) matrilines in the Mediterranean: further evidence of genetic diversity and connectivity. Mar. Biol. 2009;156:2085–2095. doi: 10.1007/s00227-009-1239-5. [DOI] [Google Scholar]
- 70.Casale P, et al. Exceptional sea turtle nest records in 2011 suggest an underestimated nesting potential in Sicily (Italy) Acta Herpetol. 2012;7:181–188. [Google Scholar]
- 71.Revelles M, et al. Evidence for an asymmetrical size exchange of loggerhead sea turtles between the Mediterranean and the Atlantic through the Straits of Gibraltar. J. Exp. Mar. Biol. Ecol. 2007;349:261–271. doi: 10.1016/j.jembe.2007.05.018. [DOI] [Google Scholar]
- 72.Fontaine C, Shaver D. Head-starting the Kemp’s ridley sea turtle, Lepidochelys kempii, at the NMFS GalvestonLaboratory, 1978–1992: A review. Chelonian Conserv. Bi. 2005;4:838–845. [Google Scholar]
- 73.Mrosovsky N, Kamel S, Rees AF, Margaritoulis D. Pivotal temperature for loggerhead turtles (Caretta caretta) from Kyparissia Bay, Greece. Can. J. Zool.-Rev. Can. Zool. 2002;80:2118–2124. doi: 10.1139/z02-204. [DOI] [Google Scholar]
- 74.Pimm SL. The balance of Nature? Ecological issues in conservation of species and communities. Illinois, USA: University of Chicago Press: Chicago; 1991. [Google Scholar]
- 75.Kallimanis AS. Temperature dependent sex determination andclimate change. Oikos. 2010;119:197–200. doi: 10.1111/j.1600-0706.2009.17674.x. [DOI] [Google Scholar]
- 76.Bowen BW, et al. Population structure of loggerhead turtles (Caretta caretta) in the Northwestern Atlantic Ocean and Mediterranean Sea. Conserv. Biol. 1993;7:834–844. doi: 10.1046/j.1523-1739.1993.740834.x. [DOI] [Google Scholar]
- 77.Pike DA. Forecasting the viability of sea turtle eggs in a warming world. Global Change Biology. 2014;20:7–15. doi: 10.1111/gcb.12397. [DOI] [PubMed] [Google Scholar]
- 78.Cacho I, Grimalt JO, Sierro FJ, Shackleton N, Canals M. Evidence for enhanced Mediterranean thermohaline circulation during rapid climatic coolings. Earth Planet. Sci. Lett. 2000;183:417–429. doi: 10.1016/S0012-821X(00)00296-X. [DOI] [Google Scholar]
- 79.Abreu-Grobois, A. et al. in 26th Annual Symposium onSea Turtle Biology and Conservation. (eds M. G. Frick, A. Panagopoulou, A. Rees, & K. L. Williams)179.
- 80.Carreras C, et al. The genetic structure of the loggerhead sea turtle (Caretta caretta) in the Mediterranean as revealed by nuclear and mitochondrial DNA and its conservation implications. Conserv. Genet. 2007;8:761–775. doi: 10.1007/s10592-006-9224-8. [DOI] [Google Scholar]
- 81.Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series. 1999;41:95–98. [Google Scholar]
- 82.Yilmaz C, Turkozan O, Bardakci F. Genetic structure of loggerhead turtle (Caretta caretta) populations in Turkey. Biochem. Syst. Ecol. 2011;39:266–276. doi: 10.1016/j.bse.2011.08.018. [DOI] [Google Scholar]
- 83.Saied A, et al. Loggerhead turtles nesting in Libya: an important management unit for the Mediterranean stock. Mar Ecol Prog Ser. 2012;450:207–U224. doi: 10.3354/meps09548. [DOI] [Google Scholar]
- 84.Monzon-Arguello C, et al. Population structure and conservation implications for the loggerhead sea turtle of the Cape Verde Islands. Conserv. Genet. 2010;11:1871–1884. doi: 10.1007/s10592-010-0079-7. [DOI] [Google Scholar]
- 85.Fitzsimmons NN, Moritz C, Moore SS. Conservation and dynamics of microsatellite loci over 300 million years of marine turtle evolution. Mol. Biol. Evol. 1995;12:432–440. doi: 10.1093/oxfordjournals.molbev.a040218. [DOI] [PubMed] [Google Scholar]
- 86.Fitzsimmons, N. N. et al. in Proceedings of the International Symposium on Sea turtle Conservation Genetics, NOAA Technical MemorandumNMFS-SEFSC-396. (eds B. W. Bowen & W. N. Witzell)(National Technical Information Service).
- 87.Moore MK, Ball RM. Multiple paternity in loggerhead turtle (Caretta caretta) nests on Melbourne Beach, Florida: a microsatellite analysis. Mol. Ecol. 2002;11:281–288. doi: 10.1046/j.1365-294X.2002.01426.x. [DOI] [PubMed] [Google Scholar]
- 88.Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Jakobsson M, Rosenberg NA. CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinf. 2007;23:1801–1806. doi: 10.1093/bioinformatics/btm233. [DOI] [PubMed] [Google Scholar]
- 90.Jones AG. GERUD 2.0: a computer program for the reconstruction of parental genotypes from half-sib progeny arrays with known or unknown parents. Mol. Ecol. Notes. 2005;5:708–711. doi: 10.1111/j.1471-8286.2005.01029.x. [DOI] [Google Scholar]
- 91.Lynch M, Ritland K. Estimation of pairwise relatedness with molecular markers. Genetics. 1999;152:1753–1766. doi: 10.1093/genetics/152.4.1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Peakall R, Smouse PE. GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research-an update. Bioinf. 2012;28:2537–2539. doi: 10.1093/bioinformatics/bts460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Warnes, G. et al. gplots: various R programming tools for plotting data. http://CRAN.R-project.org/package=gplots. (2016).
- 94.Freedberg S, Wade MJ. Cultural inheritance as a mechanism for population sex-ratio bias in reptiles. Evolution. 2001;55:1049–1055. doi: 10.1554/0014-3820(2001)055[1049:CIAAMF]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 95.Chaloupka, M. Stochastic simulation modeling of loggerhead population dynamics given exposutre to competing mortality risks in the western South Pacific. in Loggerhead Sea Turtles (eds A. Bolten & B. Witherington) 255–273 (Smithsonian Books, 2003).
- 96.Heppell, S., Crowder, L. B., Crouse, D. T., Epperly, S. & Frazer, N. B. Population models for Atlantic loggerheads: past, present and future. In Loggerhead Sea Turtles (eds A. Bolten & B. Witherington) 255–273 (Smithsonian Books, 2003).
- 97.Bell CD, Blumenthal JM, Broderick AC, Godley BJ. Investigating potential for depensation in Marine Turtles: How low can you go? Conserv. Biol. 2010;24:226–235. doi: 10.1111/j.1523-1739.2009.01313.x. [DOI] [PubMed] [Google Scholar]
- 98.Casale, P. & Margaritoulis, D. Sea turtles in the Mediterranean: distribution, threats and conservation priorities. (IUCN/SSC Marine Turtle Specialist Group, 2010).
- 99.Wessel P, Smith W, Scharroo R, Luis J, Wobbe F. EOS, Trans. AGU. 2013. Generic Mapping Tools: Improved Version Released; pp. 409–420. [Google Scholar]
- 100.Mrosovsky N, Baptistotte C, Godfrey MH. Validation of incubation duration as an index of the sex ratio of hatchling sea turtles. Can. J. Zool.-Rev. Can. Zool. 1999;77:831–835. doi: 10.1139/z99-039. [DOI] [Google Scholar]
- 101.Fuller WJ, et al. Importance of spatio-temporal data for predicting the effects of climate change on marine turtle sex ratios. Mar Ecol Prog Ser. 2013;488:267–274. doi: 10.3354/meps10419. [DOI] [Google Scholar]
- 102.Marcovaldi MA, Godfrey MH, Mrosovsky N. Estimating sex ratios of loggerhead turtles in Brazil from pivotal incubation durations. Can. J. Zool.-Rev. Can. Zool. 1997;75:755–770. doi: 10.1139/z97-097. [DOI] [Google Scholar]
- 103.Godfrey MH, Mrosovsky N. Estimating the time between hatching of sea turtles and their emergence from the nest. Chelonian Conserv. Bi. 1997;2:581–585. [Google Scholar]
- 104.Miller, J. D., Limpus, C. J. & Godfrey, M. H. Nest site selection, oviposition, eggs, development, hatching and emergence of loggerhead turtles in Loggerhead Sea Turtles (eds A. Bolten & B. Witherington) 125–143 (Smithsonian Books, 2003).
- 105.Crouse DT, Crowder LB, Caswell H. A Stage-Based Population-Model for Loggerhead Sea-Turtles and Implications for. Conservation. Ecology. 1987;68:1412–1423. [Google Scholar]
- 106.Turtle Expert Working Group, T. E. W. G. T An assessent of the loggerhead turtle population in the western North Atlantic ocean. NOAA Technical Memorandum NMFS SEFSC. 2009;575:131. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.