Abstract
The relationship between O-6-methylguanine-DNA methyltransferase (MGMT) promoter methylation and clinicopathological characteristics of non-small-cell lung carcinoma (NSCLC) has remained controversial and unclear. Therefore, in this study we have undertaken a systematic review and meta-analysis of relevant studies to quantitatively investigate this association. We identified 30 eligible studies investigating 2714 NSCLC patients. The relationship between MGMT hypermethylation and NSCLC was identified based on 20 studies, including 1539 NSCLC patient tissue and 1052 normal and adjacent tissue samples (OR = 4.60, 95% CI = 3.46~6.11, p < 0.00001). MGMT methylation varied with ethnicity (caucasian: OR = 4.56, 95% CI = 2.63~7.92, p < 0.00001; asian: OR = 5.18, 95% CI = 2.03~13.22, p = 0.0006) and control style (autologous: OR = 4.44, 95% CI = 3.32~5.92, p < 0.00001; heterogeneous: OR = 9.05, 95% CI = 1.79~45.71, p = 0.008). In addition, MGMT methylation was observed to be specifically associated with NSCLC clinical stage, and not with age, sex, smoking, pathological types, and differentiation status. Also MGMT methylation did not impact NSCLC patients survival (HR = 1.32, 95% CI = 0.77~2.28, p = 0.31). Our study provided clear evidence about the association of MGMT hypermethylation with increased risk of NSCLC.
Introduction
Lung cancer has been one of the most common causes of cancer-related death in the world1, and its incidence and mortality rates are also much higher than other cancer in China2. It consists of two different histological types, non-small-cell lung carcinoma (NSCLC) and small-cell lung carcinoma (SCLC). Among these two, Chinese patients have usually seen higher incidence and mortality rates of NSCLC in the last two decades3. Despite some advances in the therapeutic options for NSCLC in recent times, its prognosis is still very poor with a 5-year overall survival4. Among the many different reasons for poor outcome, epigenetic modification had played an important role in NSCLC carcinogenesis5,6. For instance, it has been seen that DNA methylation typically is associated with silencing the expression of many tumor suppressor genes in the existing cellular pathways7. Thus, it requires investigation and identification of specific gene methylation patterns that might be helpful for NSCLC diagnosis and can also act as prognostic markers.
The O-6-methylguanine-DNA methyltransferase (MGMT), a specific DNA damage reversal repairs protein, has been demonstrated to protect tissues against the toxic and carcinogenic effects of alkylating agents by removing adducts from O6 position of guanine8,9. Epigenetic silencing of MGMT gene by its promoter methylation at specific CpG islands results in loss of its activity in various cancers, including lung cancer10–12. However, different studies have shown varying level of MGMT promoter methylation frequency in NSCLC3,13,14. But this discrepancy can be attributed to diverse nature of the clinical samples, such as tissues, serum and bronchoalveolar lavage fluid, that were used for this analysis. To be concise, some meta-analysis studies have reported that MGMT methylation is associated with NSCLC incidence15–17, but these meta-analysis were based on few studies that involved small number of diverse samples and thus could lead to an erroneous result. Typically, they indicated quite different rates of MGMT hypermethylation from different samples, and only the samples from tumor tissue and plasma showed higher methylation than control group15,16. Moreover, these studies have also not thoroughly investigated the relationship between MGMT methylation and clinical characteristics of NSCLC. They only reported the risk between MGMT methylation and NSCLC15–17. It is known that different tumor tissue specimens can have some variation, so we in our study have first summarized all published studies which included just the samples from tumor tissues of NSCLC as much as possible, and then performed the systematic review and meta-analysis to quantitatively assess the association of MGMT methylation with incidence and clinical characteristics of NSCLC. With this extensive and careful meta-analysis, we expect to have a better understanding of the role of MGMT methylation in NSCLC.
Results
Eligible studies and their characteristics
Based on the selection criteria, a total of 128 relevant articles were identified. One article was excluded due to duplicative nature, as one another study had similar information. After careful reading of the titles and abstracts, 75 more articles were excluded, as they were either irrelevant or involved just cell or animal studies. The remaining 52 articles were further reviewed in detail, and 22 of them were additionally excluded, as it was observed that tissue specimens were not exclusively were from NSCLC patient’s. In addition, we were unable to extract useful data and some of them were not in English language. Thus, 30 studies3,11,13,15,18–43 qualified the required criteria, as shown in Fig. 1, and were assessed in our final meta-analysis. These studies were published between the year 1999 to 2015, and included a total of 2714 NSCLC patients from different countries including, USA, China, Turkey, Germany, Japan, Serbia, Korea, Hong Kong and Taiwan. All these studies detected MGMT DNA methylation by methylation-specific polymerase chain reaction (MSP), Pyrosequencing or Real-Time MSP (RT-MSP) methods. The NOS scores of all studies varied from 6 to 8 points, thereby indicating high quality. Additional basic characteristics of all the included studies have been shown in Table 1.
Figure 1.
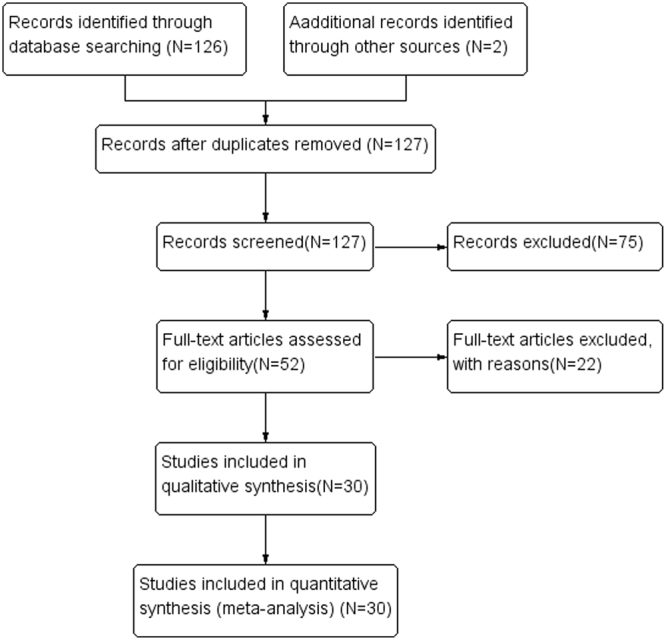
Flow chart depicting the study selection criteria.
Table 1.
Baseline characteristics of eligible studies.
| Study | Year | Country | Ethnicity | Type | Cases Number | Detection Method | Methylation site | Methylation site N(%) | Control style | NOS score |
|---|---|---|---|---|---|---|---|---|---|---|
| Zöchbauer13 | 2001 | USA | Caucasian | NSCLC | 107 | MSP | Promoter, CpG islands | 22(22.56) | A | 8 |
| Zhang3 | 2011 | China | Asian | NSCLC | 78 | MSP | Promoter, CpG islands | 4(5.13) | A | 7 |
| Guo15 | 2015 | China | Asian | NSCLC | 271 | MSP | Promoter, CpG islands | 81(29.89) | A | 7 |
| Feng18 | 2008 | USA | Caucasian | NSCLC | 49 | MSP | Promoter, CpG islands | 6(12.24) | A | 7 |
| Guo11 | 2004 | USA | Caucasian | NSCLC | 20 | MSP | Promoter, CpG islands | 14(70.00) | A | 8 |
| Esteller (1)19 | 1999 | USA | Caucasian | NSCLC | 22 | MSP | Promoter, CpG islands | 6(27.27) | A | 8 |
| Esteller (2)20 | 1999 | USA | Caucasian | NSCLC | 34 | MSP | Promoter, CpG islands | 10(29.41) | A | 8 |
| Ekim21 | 2011 | Turkey | Caucasian | NSCLC | 80 | MSP | Promoter, CpG islands | 51(63.75) | A | 7 |
| Brabender22 | 2003 | Germany | Caucasian | NSCLC | 90 | RT-MSP | Promoter, CpG islands | 34(37.78) | A | 8 |
| Lin23 | 2009 | China | Caucasian | NSCLC | 67 | MSP | Promoter, CpG islands | 1(1.49) | H | 7 |
| Yanagawa24 | 2007 | Japan | Asian | NSCLC | 101 | MSP | Promoter, CpG islands | 14(13.86) | A | 8 |
| Kontic25 | 2012 | Serbia | Caucasian | NSCLC | 65 | MSP | Promoter, CpG islands | 8(12.31) | A | 7 |
| Kim (1)26 | 2005 | Korea | Asian | AC | 72 | MSP | Promoter, CpG islands | 12(16.67) | A | 7 |
| Kim (2)27 | 2005 | Korea | Asian | NSCLC | 61 | MSP | Promoter, CpG islands | 38(62.30) | A | 8 |
| Ishiguro28 | 2013 | USA | Caucasian | NSCLC | 6 | MSP | Promoter, CpG islands | 2(33.33) | A | 8 |
| Vallböhmer29 | 2006 | Germany | Caucasian | NSCLC | 91 | RT-MSP | Promoter, CpG islands | 38(41.76) | A | 8 |
| Safar30 | 2005 | USA | Caucasian | NSCLC | 105 | MSP | Promoter, CpG islands | 11(10.48) | A | 7 |
| Drilon31 | 2014 | USA | Caucasian | NSCLC | 107 | MSP | Promoter, CpG islands | 9(8.41) | A | 7 |
| Pulling32 | 2003 | USA | Caucasian | AC | 237 | MSP | Promoter, CpG islands | 121(51.05) | A | 7 |
| Hayashi33 | 2002 | Japan | Asian | AC | 87 | MSP | Promoter, CpG islands | 31(35.63) | A | 8 |
| Liu34 | 2006 | USA | Caucasian | NSCLC | 121 | MSP | Promoter, CpG islands | 37(30.33) | A | 7 |
| Chan35 | 2002 | Hong Kong | Asian | NSCLC | 75 | MSP | Promoter, CpG islands | 11(14.67) | A | 6 |
| Furonaka36 | 2005 | Japan | Asian | NSCLC | 123 | MSP | Promoter, CpG islands | 47(38.21) | A | 7 |
| Wu37 | 2008 | Taiwan | Asian | NSCLC | 123 | MSP | Promoter, CpG islands | 111(50.45) | A | 7 |
| Harden38 | 2003 | USA | Caucasian | NSCLC | 90 | RT-MSP | Promoter, CpG islands | 14(15.56) | A | 8 |
| Buckingham39 | 2010 | USA | Caucasian | NSCLC | 132 | Pyrosequencing | Promoter, CpG islands | 14(10.61) | A | 7 |
| Topaloglu40 | 2004 | USA | Caucasian | NSCLC | 31 | RT-MSP | Promoter, CpG islands | 12(38.71) | A | 7 |
| Liu41 | 2010 | China | Asian | NSCLC | 98 | MSP | Promoter, CpG islands | 31(31.63) | H | 6 |
| Jin42 | 2010 | China | Asian | NSCLC | 94 | MSP | Promoter, CpG islands | 16(17.02) | A | 6 |
| Kang43 | 2011 | China | Asian | NSCLC | 77 | MSP | Promoter, CpG islands | 26(33.77) | H | 6 |
AC: adenocarcinoma; MSP: methylation-specific polymerase chain reaction; RT-MSP: Real-Time MSP; N: number of total; A: autologous control (the control sample from NSCLC themselves); H: heterogeneous control (the control sample from other individuals).
Correlation analysis between MGMT hypermethylation and NSCLC different clinicopathological features
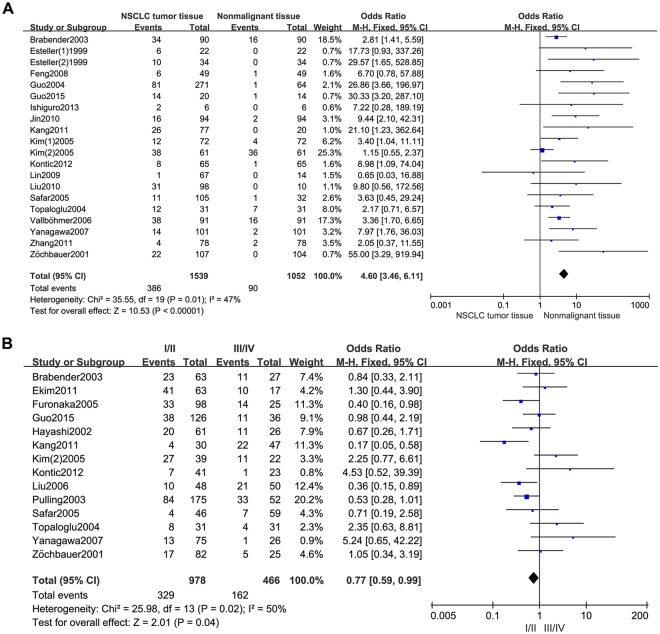
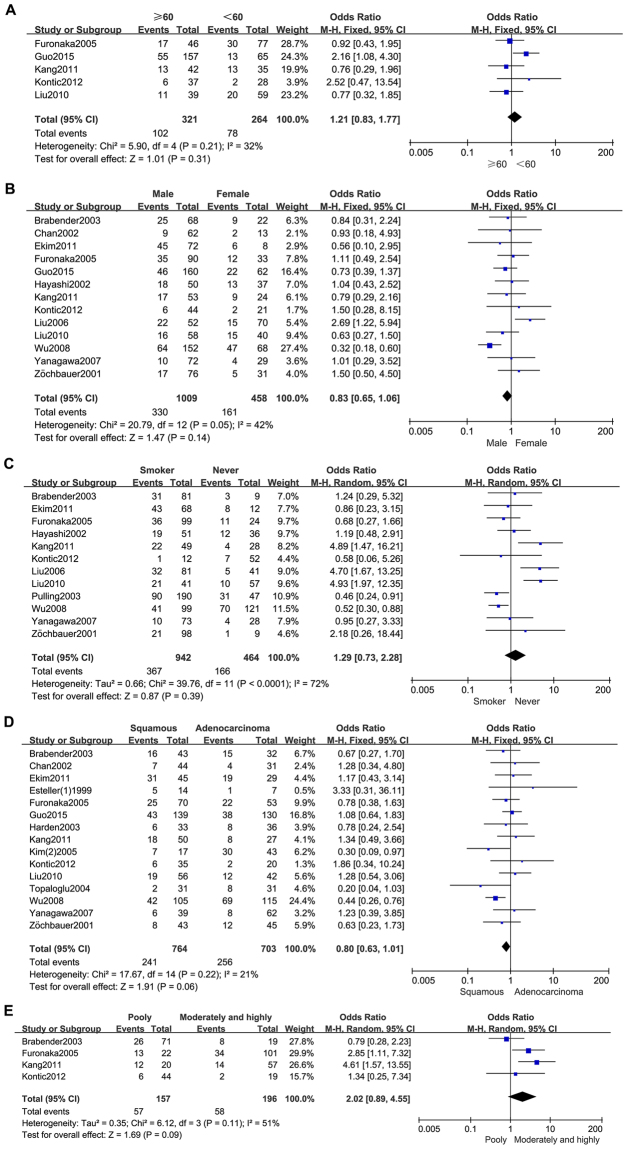
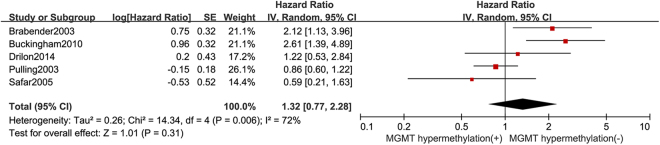
As shown in Table 2 and Fig. 2, Our analysis revealed that NSCLC tissues had significantly higher MGMT promoter hypermethylation than normal and adjacent tissue samples (OR = 4.60, 95%, CI = 3.46~6.11, p < 0.00001, Fig. 2A). The 20 studies including 1539 NSCLC patients’ tissues and 1052 normal and adjacent tissues were involved in the meta-analysis. In an effort to test the correlation between MGMT hypermethylation rate and different staging of NSCLC, we observed that MGMT hypermethylation rate was higher in NSCLC patients with advanced stage than in early stage (OR = 0.77, 95% CI = 0.59~0.99, p = 0.04, Fig. 2B). The meta-analysis was assessed based on 14 studies including 466 NSCLC patients with advanced stage and 978 NSCLC patients in early stage. Our meta-analysis were unable to find the correlation between MGMT hypermethylation rate and rest clinicopathological features, including age (OR = 1.21, 95% CI = 0.83~1.77, p = 0.31, Fig. 3A), sex (OR = 0.83, 95% CI = 0.65~1.06, p = 0.14, Fig. 3B), smoking (OR = 1.29, 95% CI = 0.73~2.28, p = 0.39, Fig. 3C), pathological types (OR = 0.80, 95% CI = 0.63~1.01, p = 0.06, Fig. 3D), differentiation (OR = 2.02, 95% CI = 0.89~4.55, p = 0.09, Fig. 3E). Finally, we also estimated the relationship between overall survival (OS) and the expression of MGMT methylation, by analyzing the data from five observational trails. Four trails were prospective study and one30 was retrospective studies. Since there was high heterogeneity (I2 = 72%, p = 0.006) among these trials, we used random-effects model for statistical adjustment. Our results demonstrated that MGMT hypermethylation in NSCLC did not associate with overall survival (HR = 1.32, 95% CI = 0.77~2.28, p = 0.31, Fig. 4).
Table 2.
Analysis between MGMT hypermethylation and NSCLC different clinicopathological features.
| Analysis | Studies N | Methylation N(+/−) | OR(95% CI) | Method | Heterogeneity | P value | |
|---|---|---|---|---|---|---|---|
| I 2 (%) | P value | ||||||
| NSCLC | 20 | 476/2115 | 4.60 [3.46, 6.11] | Fixed | 47 | 0.01 | <0.00001 |
| age | 5 | 180/405 | 1.21 [0.83, 1.77] | Fixed | 32 | 0.21 | 0.31 |
| sex | 13 | 491/976 | 0.83 [0.65, 1.06] | Fixed | 42 | 0.05 | 0.14 |
| smoking | 12 | 533/873 | 1.29 [0.73, 2.28] | Random | 72 | <0.00001 | 0.39 |
| pathological types | 15 | 497/970 | 0.80 [0.63, 1.01] | Fixed | 21 | 0.22 | 0.06 |
| differentiation | 4 | 115/238 | 2.02 [0.89, 4.55] | Random | 51 | 0.11 | 0.09 |
| clinical stage | 14 | 491/953 | 0.77 [0.59, 0.99] | Random | 50 | 0.02 | 0.04 |
Figure 2.
Forest plot representing the meta-analysis of MGMT methylation in NSCLC and clinical stage. (A) Forest plot of MGMT methylation in NSCLC tissues verse normal tissues. (B) Forest plot showing association of MGMT methylation with clinical stage of NSCLC patients.
Figure 3.
Forest plot representing the meta-analysis of MGMT methylation in clinicopathological features of NSCLC patients. (A) Forest plot showing association of MGMT methylation with age status of NSCLC patients. (B) Forest plot showing association of MGMT methylation with sex status of NSCLC patients. (C) Forest plot showing association of MGMT methylation with smoking status of NSCLC patients. (D) Forest plot showing association of MGMT methylation with different pathological types of NSCLC. (E) Forest plot showing association of MGMT methylation with differentiation status of NSCLC patients.
Figure 4.
Forest plot showing association of MGMT methylation with overall survival of NSCLC patients.
Subgroup and sensitivity analysis
To investigate the influence of other possible factors on the heterogeneity across studies, we conducted subgroup analysis, according to various confounding factors. Specifically, the patients and controls were stratified based on ethnicity, control style and detection method for subgroup analysis, as shown in Table 3. Our results of NSCLC indicated that MGMT hypermethylation varied with ethnicity (Caucasian: OR = 4.56, 95% CI = 2.63~7.92, p < 0.00001; Asian: OR = 5.18, 95% CI = 2.03~13.22, p = 0.0006), control style (Auologous: OR = 4.44, 95% CI = 3.32~5.92, p < 0.00001; Heterogeneous: OR = 9.05, 95% CI = 1.79~45.71, p = 0.008) and detection method (MSP: OR = 6.78, 95% CI = 3.40~13.51, p < 0.00001; RT-MSP: OR = 2.91, 95% CI = 1.87~4.53, p < 0.00001). I2 changed to 27% and 64% in ethnicity subgroup, 50% and 30% in control style subgroup, 53% and 0% in detection method subgroup, compared with 47% of total. The results of subgroup analysis of the association between MGMT hypermethylation and smoking indicated that I2 changed to 66% and 80% in ethnicity subgroup, 52% and 0% in control style subgroup, compared with 72% of total. Therefore, the subgroup analysis implied that the factor of control style influence heterogeneity of the association between MGMT hypermethylation and smoking, ethnicity could not explain the heterogeneity.
Table 3.
Subgroup analysis of the association between MGMT hypermethylation and NSCLC or Smoking.
| Analysis | Studies N | Patients | OR(95% CI) | Method | Heterogeneity | P value | |
|---|---|---|---|---|---|---|---|
| I 2 (%) | P value | ||||||
| NSCLC | |||||||
| Ethnicity | |||||||
| Caucasian | 12 | 1239 | 4.56 [2.63, 7.92] | Fixed | 27 | 0.18 | <0.00001 |
| Asian | 8 | 1352 | 5.18 [2.03, 13.22] | Random | 65 | 0.005 | 0.0006 |
| Control style | |||||||
| Auologous | 17 | 2305 | 4.44 [3.32, 5.92] | Random | 50 | 0.01 | <0.00001 |
| Heterogeneous | 3 | 286 | 9.05 [1.79, 45.71] | Fixed | 30 | 0.24 | 0.008 |
| Detection Method | |||||||
| MSP | 17 | 2167 | 6.78 [3.40, 13.51] | Random | 53 | 0.006 | <0.00001 |
| RT-MSP | 3 | 424 | 2.91 [1.87, 4.53] | Fixed | 0 | 0.80 | <0.00001 |
| Total | 20 | 2591 | 4.60 [3.46, 6.11] | Fixed | 47 | 0.01 | <0.00001 |
| Smoking | |||||||
| Ethnicity | |||||||
| Caucasian | 6 | 700 | 1.18 [0.48, 2.92] | Random | 66 | 0.01 | 0.72 |
| Asian | 6 | 706 | 1.39 [0.61, 3.16] | Random | 80 | 0.0001 | 0.43 |
| Control style | |||||||
| Auologous | 10 | 1231 | 0.92 [0.57, 1.49] | Random | 52 | 0.03 | 0.74 |
| Heterogeneous | 2 | 175 | 4.92 [2.37, 10.19] | Fixed | 0 | 0.99 | <0.0001 |
| Total | 12 | 1406 | 1.29 [0.73, 2.28] | Random | 72 | <0.00001 | 0.39 |
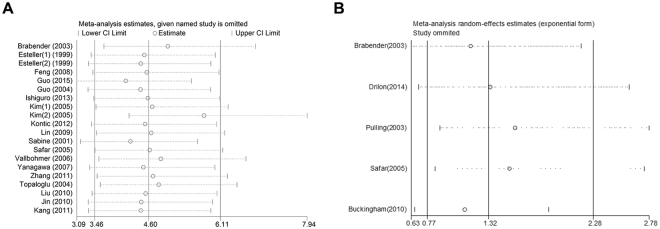
In addition, we also evaluated the sensitivity of our meta-analysis by removing one study at a time and analyzing the remaining studies to assess the stability of the data. The pooled ORs and HR did not significantly alter after the removal of any one study, thereby establishing the stability of our results, as shown in Fig. 5. Interestingly, a moderate heterogeneity was observed (I2 = 47%) in the analysis between MGMT hypermethylation and NSCLC, and deletion of one study by Kim (2) et al.33, significantly reduced the heterogeneity (I2 = 25%).
Figure 5.
Sensitivity analysis by omitting a single study. (A) Sensitivity analysis of the OR coefficients for the association between MGMT methylation and risk of NSCLC. (B) Sensitivity analysis of the HR coefficients for the association between MGMT methylation and overall survival of NSCLC patients.
Publication bias analysis
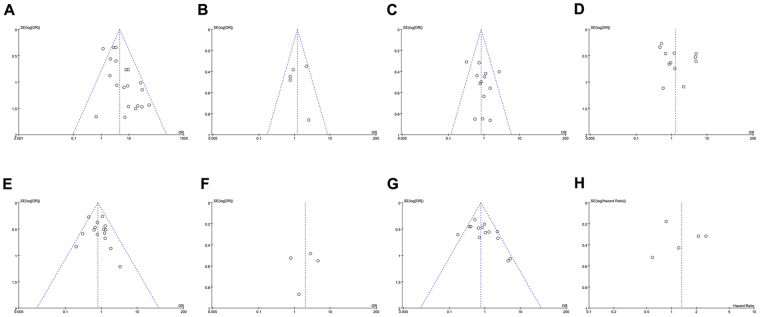
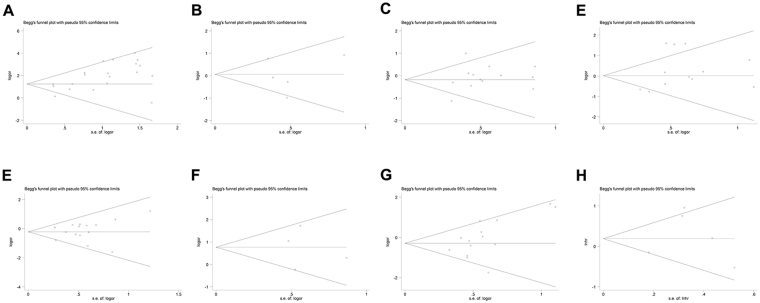
Further, we also evaluated the publication bias of the selected studies by funnel plot and Begg’s test. If the result of funnel plots showed symmetry and p > 0.05 from Begg’s test, indicated that no significant publication bias existed. The funnel plots analysis showed symmetry, as shown in Fig. 6. The Begg’s test also displayed that publication bias was not statistically significant as shown in Fig. 7. These results overall indicated that publication bias had no influence on our meta-analysis.
Figure 6.
Funnel plots analyses to assess the publication bias between MGMT methylation and different NSCLC clinicopathological characteristics; (A) Overall funnel plot from pooled 20 studies, (B) Funnel plot based on age, (C) Funnel plot based on sex status, (D) Funnel plot based on smoking status, (E) Funnel plot based on pathological types, (F) Funnel plot based on differentiation status, (G) Funnel plot based on clinical stage status, and (H) Funnel plot based on NSCLC overall survival.
Figure 7.
Begg’s funnel plot analyses to assess the publication bias between MGMT methylation and different NSCLC clinicopathological characteristics; (A) Overall funnel plot from pooled 20 studies, (B) Funnel plot based on age, (C) Funnel plot based on sex status, (D) Funnel plot based on smoking status, (E) Funnel plot based on pathological types, (F) Funnel plot based on differentiation status, (G) Funnel plot based on clinical stage status, and (H) Funnel plot based on NSCLC overall survival.
Discussion
MGMT is a DNA repair gene located on a human chromosome band 10q26, and encodes a high-efficiency DNA repair protein to protect cells and tissues from disintegration by ubiquitination-dependent proteolysis, by removing alkyl groups from the O6 position of guanine nucleotide9,44,45. During the early stage of carcinogenesis, epigenetic and genetic alterations are common events, and silencing of this MGMT gene by its promoter methylation is one of the major mechanism for carcinogenesis in tumor tissues of various cancers, including NSCLC10,12,46. In recent years, some studies have reported about MGMT methylation in NSCLC, however the persuasive evidence about its role in NSCLC and clinical significance is not very convincing. Thus, we undertook this meta-analysis to identify the association between MGMT promoter methylation and different clinicopathological characteristics of NSCLC.
Our overall pooled data demonstrated that (1) the frequency of MGMT methylation in NSCLC tissue was much higher than normal tissue samples; (2) MGMT methylation was not correlated with clinicopathological characteristics like age, sex, smoking, pathological types, and differentiated status; (3) MGMT methylation played an important role in the staging and was higher in advanced staged (III and IV) NSCLC tissue than in early staged (I and II) tissue samples; and (4) MGMT methylation could not be a prognostic factor for NSCLC prognosis. MGMT gene promoter methylation is a frequent event in NSCLC tissues showed that the MGMT gene promoter hypermethylation is associated with formation and development of NSCLC. Inactivation of the gene of MGMT play an important role in tumor aberrant progression. Our these observation were consistent with the previously published meta-analyses reports17,47, which had also reported similar observations. These findings helped us to speculate that it could be a factor for the NSCLC incidence or its risk. Based on the current technology, DNA methylation patterns of any gene can be detected from all kinds of body fluids. Since DNA methylation is an early event in the tumor initiation and is best-characterized as epigenetic alteration, and has been shown to contribute towards carcinogenesis, it could act as an alternative biomarker48,49. In this reference, one can say based on our findings that MGMT methylation can have a potential role for the NSCLC diagnosis. In addition, many other studies have reported that number of methylated genes were associated with the clinical characteristics3,13,24,34, however our meta-analyses indicated that MGMT promoter methylation was not associated with any clinical characteristics in NSCLC, except staging. It suggested that the increased ability of proliferation and invasion of NSCLC cells may be associated with MGMT hypermethylation. Inactivation of gene of MGMT could contribute tumor progression. In contrast, the study by Huang et al. has reported that MGMT gene methylation was associated with smoking behavior in NSCLC50. This different observation can be attributed to less number of studies that were analyzed in that study, as only 8 studies including 817 patients were selected. However, our data is based on the analysis of 12 studies, which included 1304 patients. In addition, similar to our observation of MGMT gene methylation not associated with worse NSCLC survival, the study by Chen et al. also confirmed similar results51.
Importantly, we observed a moderate heterogeneity (I2 = 47%) in our met-analysis. But after deletion of one study conducted by Kim (2) et al.27, the heterogeneity significantly declined (I2 = 25%). Now, it was not very clear about the possible reasons of why the results in their study were so different, but we could speculate that may be the method of MGMT methylation detection was a bit different. As we all know that heterogeneity can be due to different characteristics of the patients, including the ethnicity and control style. To minimize the influence of these confounding factors, we performed subgroup analysis and observed that heterogeneity significantly reduced in several subgroups. High heterogeneity was also observed in smoking (I2 = 72%, p < 0.0001) and NSCLC overall survival (I2 = 72%, p = 0.006) meta-analysis. We conducted subgroup analysis and sensitivity analysis in the same way, then we observed that heterogeneity significantly reduced in several subgroups. When using the samples from heterogeneous control, the pooled OR of MGMT methylation in smoking patients was much higher than that in no smoking patients. When using the samples from autologous control, the result was opposite. The reason might be high concentration of MGMT methylation in samples of autologous control. This maybe partly explain the heterogeneity of our study. In sensitivity analysis, the pooled ORs and HR did not significantly alter after the removal of any one study. It suggested the robustness of our results.
However, we have to note that there were some limitations of our meta-analysis. First, our study selection criteria was restrictive to all English language studies only, and it is highly possible that studies published in other language or unpublished data and studies could tilt the overall conclusion. Second, the result could also be influenced by selection and information biases. Negative results were not as conclusive as the positive results. Bias caused by unpublished articles is the same reason for all the meta-analysis. We could collect data as comprehensive as possible. Write letter to the author for their manuscript. The best way to control the publication bias is to register all the clinical studies, and build database. Third, different studies using varying methods to detect the level of MGMT gene methylation can also effect the overall assessment and thus a universal method of methylation detection should be standardized.
In summary, our meta-analysis revealed that MGMT gene methylation was higher in NSCLC tissue samples than normal. Also advanced stage NSCLC patients showed higher methylation than early stage patients. Finally, it would be suffice to say that MGMT methylation is indeed associated with an increased NSCLC risk, and thus has the potential to be a good “biomarker” for NSCLC diagnosis in the future. More importantly, additional large-scale studies would be required to further clarify the value of MGMT methylation in clinical use for NSCLC diagnosis/risk assessment.
Methods
Search strategy
We systematically searched the Pubmed, Cochrane library, Embase and China National Knowledge Infrastructure (CNKI) databases to identify the relevant studies between, January 1, 1997 to August 10, 2016. The following search terms were used: (“lung”) and (“cancer” or “tumor” or “neoplasm” or “carcinoma”) and (“methylation”) and (“MGMT” or “O6-methylguanine-DNA methyltransferase gene”). In addition, we also manually searched the reference lists from the relevant retrieved articles and reviews.
Selection criteria
The studies were selected for the meta-analysis based on the following specific selection criteria. The eligible studies included; (1) NSCLC patients specimens evaluated for MGMT methylation, (2) Studies reporting the relationship between MGMT methylation level and clinicopathological parameters or prognosis in NSCLC patients, (3) studies either having the direct information about the hazard ratio (HR) and 95% confidence interval (CI) for survival, or with a sufficient data where these can be calculated, and (4) studies with definitive detection method of MGMT methylation. However, the studies were excluded if they; (1) were only letters, reviews, editorials, expert opinions, case reports, meeting records or conference abstracts, (2) were not written in English, (3) lacked the information about clinicopathological parameters or sufficient data for the estimation of HR with 95% CI, (4) had NSCLC tissue specimens other than the serum, plasma, pleural effusion, sputum and Bronchoalveolar lavage, (5) were studies conducted on cells or animal only, and (6) were duplicate publications.
Data extraction
The data from these selected studies was independently extracted and reviewed by two authors, Wang and Chen, according to the predefined criteria from eligible studies. The key characteristics of each study recorded were as follows; first author name, year of publication, country, ethnicity, number of cases, source of sample, MGMT methylation detection method and methylation site and frequency. In addition, the extracted information also included clinicopathological parameters of patients, like age, gender, smoking, histological type of cancer, differentiated status, cancer stage (tumor node metastasis, TNM) and prognosis. All these data for study characteristics and clinical responses have been summarized in a table format.
Quality assessment
To assure the high quality of our research, all included studies were systematically and independently assessed according to the Newcastle-Ottawa scale (NOS) criteria. The studies were scored by two authors as follows: (1) subject selection, 0~4 points, (2) comparability of subject, 0~2 points, (3) clinical outcome, 0~3 points. The NOS scores ranged from 0 to 9 with a score of ≥7 indicating good quality. All the disagreements were resolved by discussion and consensus with a third author, Yong Li.
Statistical analysis
Meta-analyses were performed using Review Manager 5.3 (Cochrane Collaboration, Oxford, UK) and STATA 12.0 (Stata Corporation, TX, USA) statistical software. The frequency of MGMT methylation was compared in different clinicopathological parameters and odds ratios (OR), HR and 95% CI were calculated. The pooled OR represented the actual association between MGMT methylation and clinicopathological features. HR implied the hazard of mortality for prognosis (p < 0.05 were considered statistically significant). If the value of HR and 95% CI was not directly provided in any study, then we analyzed the K-M curves using Engauge Digitizer version 4.1 software to calculate HR with 95% CI52. The heterogeneity among different studies was estimated by the Cochran’s Q test (p < 0.05 indicated significant heterogeneity) and I2 statistic (0~25%, low heterogeneity; 25~50%, moderate heterogeneity; 50~75%, high heterogeneity; 75~100%, extreme high heterogeneity. According According to the Cochrane handbook, heterogeneity could be accepted if the I2 ≤ 50%.)53,54. If the I2 value was ≥50%, then random effects model was used for meta-analysis, otherwise the fixed effects model was used. Subgroup and sensitivity analysis were performed if a statistically significant heterogeneity was observed in the meta-analysis. The publication bias was assessed by funnel plot55,56 and Begg’s test57, (p < 0.05 indicated significant publication bias).
Acknowledgements
This study was financially supported in part by grants from the National Natural Science Foundation of China (81560379).
Author Contributions
L.C., Y.W., and Y.L. contributed to design, collect data and literature evaluation. F.L., L.X., F.P., N.Z., B.F., Z.Z., Y.S., J.L., R.W., C.W. wrote the main manuscript text. S.Y. prepared Figs 1–7. All authors reviewed the manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Lin Chen and Yong Wang contributed equally to this work.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Jemal A, et al. Cancer statistics, 2009. CA Cancer J Clin. 2009;59:225–249. doi: 10.3322/caac.20006. [DOI] [PubMed] [Google Scholar]
- 2.Chen W, et al. Report of cancer incidence and mortality in China, 2010. Ann Transl Med. 2014;2:61. doi: 10.3978/j.issn.2305-5839.2014.04.05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Siegel R, et al. Cancer treatment and survivorship statistics. CA Cancer J Clin. 2012;62:220–241. doi: 10.3322/caac.21149. [DOI] [PubMed] [Google Scholar]
- 4.Si LB, et al. Potential use of microRNA-200c as a prognostic marker in non-small cell lung cancer. Oncol Lett. 2017;14:4325–4330. doi: 10.3892/ol.2017.6667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Risch A, Plass C. Lung cancer epigenetics and genetics. Int J Cancer. 2008;123:1–7. doi: 10.1002/ijc.23605. [DOI] [PubMed] [Google Scholar]
- 6.Ghavifekr FM, Farshdousti HM, Shanehbandi D, Baradaran B. DNA methylation pattern as important epigenetic criterion in cancer. Genet Res Int. 2013;31:317569. doi: 10.1155/2013/317569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Esteller M. Epigenetics in cancer. N Engl J Med. 2008;358:1148–1159. doi: 10.1056/NEJMra072067. [DOI] [PubMed] [Google Scholar]
- 8.Srivenugopal KS, Ali-Osman F. The DNA repair protein, O(6)-methylguanine-DNA methyltransferase is a proteolytic target for the E6 human papillomavirus oncoprotein. Oncogene. 2002;21:5940–5945. doi: 10.1038/sj.onc.1205762. [DOI] [PubMed] [Google Scholar]
- 9.Belinsky SA, et al. Plutonium targets the p16 gene for inactivation by promoter hypermethylation in human lung adenocarcinoma. Carcinogenesis. 2004;25:1063–1067. doi: 10.1093/carcin/bgh096. [DOI] [PubMed] [Google Scholar]
- 10.Fujii M, et al. Aberrant DNA methylation profile in pleural fluid for differential diagnosis of malignant pleural mesothelioma. Cancer Sci. 2012;103:510–514. doi: 10.1111/j.1349-7006.2011.02180.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guo M, et al. Promoter hypermethylation of resected bronchial margins: a field defect of changes. Clin Cancer Res. 2004;10:5131–6. doi: 10.1158/1078-0432.CCR-03-0763. [DOI] [PubMed] [Google Scholar]
- 12.Zou XP, et al. Promoter hypermethylation of multiple genes in early gastric adenocarcinoma and precancerous lesions. Hum Pathol. 2009;40:1534–1542. doi: 10.1016/j.humpath.2009.01.029. [DOI] [PubMed] [Google Scholar]
- 13.Zöchbauer-Müller S, et al. Aberrant promoter methylation of multiple genes in non-small cell lung cancers. Cancer Res. 2001;61:249–255. [PubMed] [Google Scholar]
- 14.Guo M, et al. CHFR methylation strongly correlates with methylation of DNA damage repair and apoptotic pathway genes in non-small cell lung cancer. Discov Med. 2015;19:151–158. [PubMed] [Google Scholar]
- 15.Yang Z, Li F. O-6-methylguanine-DNA methyltransferase gene promoter methylation and lung cancer risk: A meta-analysis. J Cancer Res Ther. 2016;12:C233–C236. doi: 10.4103/0973-1482.200745. [DOI] [PubMed] [Google Scholar]
- 16.Fang N, Gu J, Wei H, You J, Zhou QA. meta-analysis of Association between MGMT gene promoter methylation and non-small cell lung cancer. Zhongguo Fei Ai Za Zhi. 2014;17:601–605. doi: 10.3779/j.issn.1009-3419.2014.08.04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gu C, et al. Association between MGMT promoter methylation and non-small cell lung cancer: a meta-analysis. PLoS One. 2013;8:e72633. doi: 10.1371/journal.pone.0072633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Feng Q, et al. DNA methylation in tumor and matched normal tissues from non-small cell lung cancer patients. Cancer Epidemiol Biomarkers Prev. 2008;17:645–654. doi: 10.1158/1055-9965.EPI-07-2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Esteller M, et al. Detection of aberrant promoter hypermethylation of tumor suppressor genes in serum DNA from non-small cell lung cancer patients. Cancer Res. 1999;59:67–70. [PubMed] [Google Scholar]
- 20.Esteller M, Hamilton SR, Burger PC, Baylin SB, Herman JG. Inactivation of the DNA repair gene O6-methylguanine-DNA methyltransferase by promoter hypermethylation is a common event in primary human neoplasia. Cancer Res. 1999;59:793–797. [PubMed] [Google Scholar]
- 21.Ekim M, et al. Determination of O6-methylguanine DNA methyltransferase promoter methylation in non-small cell lung cancer. Genet Test Mol. Biomarkers. 2011;15:357–360. doi: 10.1089/gtmb.2010.0211. [DOI] [PubMed] [Google Scholar]
- 22.Brabender J, et al. Quantitative O(6)-methylguanine DNA methyltransferase methylation analysis in curatively resected non-small cell lung cancer: associations with clinical outcome. Clin Cancer Res. 2003;9:223–227. [PubMed] [Google Scholar]
- 23.Lin Q, et al. RASSF1A, APC, ESR1, ABCB1 and HOXC9, but not p16INK4A, DAPK1, PTEN and MT1G genes were frequently methylated in the stage I non-small cell lung cancer in China. J Cancer Res Clin Oncol. 2009;135:1675–1684. doi: 10.1007/s00432-009-0614-4. [DOI] [PubMed] [Google Scholar]
- 24.Yanagawa N, et al. Promoter hypermethylation of RASSF1A and RUNX3 genes as an independent prognostic prediction marker in surgically resected non-small cell lung cancers. Lung Cancer. 2007;58:131–138. doi: 10.1016/j.lungcan.2007.05.011. [DOI] [PubMed] [Google Scholar]
- 25.Kontic M, et al. Aberrant promoter methylation of CDH13 and MGMT genes is associated with clinicopathologic characteristics of primary non-small-cell lung carcinoma. Clin Lung Cancer. 2012;13:297–303. doi: 10.1016/j.cllc.2011.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim YT, et al. Prognostic implication of aberrant promoter hypermethylation of CpG islands in adenocarcinoma of the lung. J Thorac Cardiovasc Surg. 2005;130:1378. doi: 10.1016/j.jtcvs.2005.06.015. [DOI] [PubMed] [Google Scholar]
- 27.Kim YT, Lee SH, Sung SW, Kim JH. Can aberrant promoter hypermethylation of CpG islands predict the clinical outcome of non-small cell lung cancer after curative resection. Ann Thorac Surg. 2005;79:1180–1188. doi: 10.1016/j.athoracsur.2004.09.060. [DOI] [PubMed] [Google Scholar]
- 28.Ishiguro K, et al. Expression of O(6)-Methylguanine-DNA Methyltransferase Examined by Alkyl-Transfer Assays, Methylation-Specific PCR and Western Blots in Tumors and Matched Normal Tissue. J Cancer Ther. 2013;4:919–931. doi: 10.4236/jct.2013.44103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vallböhmer D, et al. DNA methyltransferases messenger RNA expression and aberrant methylation of CpG islands in non-small-cell lung cancer: association and prognostic value. Clin Lung Cancer. 2006;8:39–44. doi: 10.3816/CLC.2006.n.031. [DOI] [PubMed] [Google Scholar]
- 30.Safar AM, et al. Methylation profiling of archived non-small cell lung cancer: a promising prognostic system. Clin Cancer Res. 2005;11:4400–4405. doi: 10.1158/1078-0432.CCR-04-2378. [DOI] [PubMed] [Google Scholar]
- 31.Drilon A, et al. A prospective study of tumor suppressor gene methylation as a prognostic biomarker in surgically resected stage I to IIIA non-small-cell lung cancers. J Thorac Oncol. 2014;9:1272–1277. doi: 10.1097/JTO.0000000000000256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pulling LC, et al. Promoter hypermethylation of the O6-methylguanine-DNA methyltransferase gene: more common in lung adenocarcinomas from never-smokers than smokers and associated with tumor progression. Cancer Res. 2003;63:4842–4848. [PubMed] [Google Scholar]
- 33.Hayashi H, et al. Inactivation of O6-methylguanine-DNA methyltransferase in human lung adenocarcinoma relates to high-grade histology and worse prognosis among smokers. Jpn J Cancer Res. 2002;93:184–189. doi: 10.1111/j.1349-7006.2002.tb01257.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu Y, Lan Q, Siegfried JM, Luketich JD, Keohavong P. Aberrant promoter methylation of p16 and MGMT genes in lung tumors from smoking and never-smoking lung cancer patients. Neoplasia. 2006;8:46–51. doi: 10.1593/neo.05586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chan EC, et al. Aberrant promoter methylation in Chinese patients with non-small cell lung cancer: patterns in primary tumors and potential diagnostic application in bronchoalevolar lavage. Clin Cancer Res. 2002;8:3741–3746. [PubMed] [Google Scholar]
- 36.Furonaka O, et al. Aberrant methylation and loss of expression of O-methylguanine-DNA methyltransferase in pulmonary squamous cell carcinoma and adenocarcinoma. Pathol Int. 2005;55:303–309. doi: 10.1111/j.1440-1827.2005.01830.x. [DOI] [PubMed] [Google Scholar]
- 37.Wu JY, et al. Association of O6-methylguanine-DNA methyltransferase (MGMT) promoter methylation with p53 mutation occurrence in non-small cell lung cancer with different histology, gender, and smoking status. Ann Surg Oncol. 2008;15:3272–3277. doi: 10.1245/s10434-008-0078-9. [DOI] [PubMed] [Google Scholar]
- 38.Harden SV, et al. Gene promoter hypermethylation in tumors and lymph nodes of stage I lung cancer patients. Clin Cancer Res. 2003;9:1370–1375. [PubMed] [Google Scholar]
- 39.Buckingham L, et al. PTEN, RASSF1 and DAPK site-specific hypermethylation and outcome in surgically treated stage I and II nonsmall cell lung cancer patients. Int J Cancer. 2010;126:1630–1639. doi: 10.1002/ijc.24896. [DOI] [PubMed] [Google Scholar]
- 40.Topaloglu O, et al. Detection of promoter hypermethylation of multiple genes in the tumor and bronchoalveolar lavage of patients with lung cancer. Clin Cancer Res. 2004;10:2284–2288. doi: 10.1158/1078-0432.CCR-1111-3. [DOI] [PubMed] [Google Scholar]
- 41.Liu Y, Wang J, Miao LJ, Wu YM, Wu YJ. Promoter hypermethylation of MGMT gene and expression of K-ras in human non-small cell lung cancer. J Medical. FORUM. 2010;31:1–4. [Google Scholar]
- 42.Jin YT, et al. Association of Abnormal Methylation of CpG Islands in Promoter Domains of Multiple Tumor Suppressor Genes with Non-Small Cell Lung Cancer. Chin. J Clin Oncol. 2010;37:1109–1114. [Google Scholar]
- 43.Kang CY, Tao ZM, Xiao H, Zhang XZ, Xue YX. Investigation on the relationship between the promoter hypermethylation of MGMT and clinicopathologic feature in NSCLC tissues. Chin J Laboratory Diagn. 2011;15:81–82. [Google Scholar]
- 44.Silber JR, Bobola MS, Blank A, Chamberlain MC. O(6)-methylguanine-DNA methyltransferase in glioma therapy: promise and problems. Biochim Biophys Acta. 2012;1826:71–82. doi: 10.1016/j.bbcan.2011.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Christmann M, Verbeek B, Roos WP, Kaina B. O(6)-Methylguanine-DNA methyltransferase (MGMT) in normal tissues and tumors: enzyme activity, promoter methylation and immunohistochemistry. Biochim Biophys Acta. 2011;1816:179–190. doi: 10.1016/j.bbcan.2011.06.002. [DOI] [PubMed] [Google Scholar]
- 46.Candiloro IL, Dobrovic A. Detection of MGMT promoter methylation in normal individuals is strongly associated with the T allele of the rs16906252 MGMT promoter single nucleotide polymorphism. Cancer Prev Res (Phila). 2009;2:862–867. doi: 10.1158/1940-6207.CAPR-09-0056. [DOI] [PubMed] [Google Scholar]
- 47.Citron M, et al. O6-methylguanine-DNA methyltransferase in human normal and malignant lung tissues. Cancer Invest. 1993;11:258–263. doi: 10.3109/07357909309024850. [DOI] [PubMed] [Google Scholar]
- 48.Das PM, Singal R. DNA methylation and cancer. J Clin Oncol. 2004;22:4632–4642. doi: 10.1200/JCO.2004.07.151. [DOI] [PubMed] [Google Scholar]
- 49.Ma X, Wang YW, Zhang MQ, Gazdar AF. DNA methylation data analysis and its application to cancer research. Epigenomics. 2013;5:301–316. doi: 10.2217/epi.13.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Huang T, et al. Meta-analyses of gene methylation and smoking behavior in non-small cell lung cancer patients. Sci Rep. 2015;5:8897. doi: 10.1038/srep08897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen C, et al. Prognosis value of MGMT promoter methylation for patients with lung cancer: a meta-analysis. Int J Clin Exp Pathol. 2015;8:11560–11564. [PMC free article] [PubMed] [Google Scholar]
- 52.Tierney JF, Stewart LA, Ghersi D, Burdett S, Sydes MR. Practical methods for incorporating summary time-to-event data into meta-analysis. Trials. 2007;8:16. doi: 10.1186/1745-6215-8-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zintzaras E, Ioannidis JP. HEGESMA: genome search meta-analysis and heterogeneity testing. Bioinformatics. 2005;21:3672–3673. doi: 10.1093/bioinformatics/bti536. [DOI] [PubMed] [Google Scholar]
- 54.Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ. 2003;327:557–560. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Peters JL, Sutton AJ, Jones DR, Abrams KR, Rushton L. Comparison of two methods to detect publication bias in meta-analysis. JAMA. 2006;295:676–680. doi: 10.1001/jama.295.6.676. [DOI] [PubMed] [Google Scholar]
- 56.Bax L, et al. More than numbers: the power of graphs in meta-analysis. Am J Epidemiol. 2009;169:249–255. doi: 10.1093/aje/kwn340. [DOI] [PubMed] [Google Scholar]
- 57.Begg CB, Berlin JA. Publication bias and dissemination of clinical research. J Natl Cancer Inst. 1989;87:107–115. doi: 10.1093/jnci/81.2.107. [DOI] [PubMed] [Google Scholar]