Abstract
Please cite this paper as: Koul PA., et al. (2011) Pandemic and seasonal influenza viruses among patients with acute respiratory illness in Kashmir (India). Influenza and Other Respiratory Viruses 5(6), e521–e527.
Background With the emergence of pandemic influenza A (2009A/H1N1) virus in India, we sought to determine the prevalence and clinical presentations of seasonal and pandemic influenza viruses among acute respiratory illness (ARI) patients from Srinagar, a temperate climate area in northern India, during the peak winter season.
Methods Combined throat and nasal swabs, obtained from 194 (108 male) presenting with ARI from January to March 2010 (Week 53‐week 10), were tested by RT‐PCR for influenza A and B, including 2009A/H1N1 viruses. HA1 gene of selected 2009A/H1N1‐positive samples was sequenced, and phylogenetic analysis was carried out.
Results Twenty‐one (10·8%, age 15–80 years, median age 40 years) patients tested positive for influenza viruses: 13 (62%) for 2009A/H1N1 virus, 6 (28·5%) for seasonal influenza A (H3N2), and 2 (9·5%) for influenza B. Twelve of the 13 patients with 2009A/H1N1 presented with febrile ARI, and eight had associated comorbidities. All of the patients recovered. Phylogenetic analysis of HA gene (n = 8) revealed that all strains from Srinagar clustered in 2009A/H1N1 clade seven along with the other 2009A/H1N1 strains from India. Amino acid substitutions in the HA protein defining clade seven (P83S, S203T, and I321V) were found in almost all isolates from Srinagar.
Conclusions Both seasonal and 2009A/H1N1 viruses appear to be associated with ARI in Srinagar. The 2009A/H1N1 in Srinagar is genetically similar to globally circulating clade 7 strains, with unique signature sequences in the HA gene. Further investigations into ascertain the role of these mutations in possible alteration of the virulence and transmissibility of the virus are needed.
Keywords: HA gene, influenza‐like illness, seasonal flu, Swine flu
Introduction
Influenza is a vaccine‐preventable disease and a major cause of morbidity and mortality in developed and developing countries. 1 , 2 In temperate regions of the Northern and Southern Hemispheres, influenza activity has been well described with its typical seasonality 3 and annual epidemics associated with excess deaths from influenza. Few data are available to assess the epidemiology and burden of influenza in tropical and developing countries. 4 Improved understanding of the temporal and geographic circulation of influenza viruses and the impact of influenza among populations living in tropical and subtropical regions is essential for the development of influenza prevention and control strategies.
A novel influenza A (H1N1) virus emerged in mid‐April 2009 and spread rapidly among humans worldwide. 5 , 6 As of August 2010, worldwide, more than 214 countries and overseas territories or communities had reported laboratory‐confirmed cases of 2009A/H1N1, including over 18 449 deaths; 7 2009A/H1N1 has been reported from various parts of India, 8 , 9 and as of October 10, 2010, 1 93 404 samples have been tested at various government and few private laboratories, and 44 889 (23·2%)among them tested positive for 2009A/H1N1, with cumulative 2606 deaths related to the disease. 10 The Kashmir province of the northern Indian state of Jammu and Kashmir has a temperate climate, and respiratory illnesses constitute the bulk of hospital visits and admissions during the winter and spring months, either in the form of acute respiratory illnesses or as exacerbations of chronic lung diseases. Even as many of these illnesses are generally believed to be of a viral etiology, no data exist about the contribution of influenza viral infection in respiratory illness as no formal surveillance data from this part of the country are available.
Genetic analysis of the 2009A/H1N1 virus revealed a unique reassortment of gene segments from North American and Eurasian swine lineages and ancestral genes derived from avian species and humans. 11 , 12 For example, HA of 2009A/H1N1 virus shows a close relationship to that of the North American influenza A virus, and NA shows a close relationship to that of Eurasian influenza A virus of swine origin. 11 , 12 The recent spread of HA of 2009A/H1N1 virus implies multiple events of reassortment, creating a population of viruses with an ancient and diverse NA gene and a much less diverse HA gene. Understanding the evolution of genetic diversity of 2009A/H1N1 is critical for studying the molecular mechanisms involved in the emergence, global spread and resistance mutations in pandemic H1N1 strains circulating worldwide. This study was designed to assess the contribution of influenza viruses, specifically of the 2009A/H1N1, on clinical presentations in patients with acute respiratory illness attending the inpatient and outpatient department of a major referral hospital in Srinagar, Kashmir (India). In addition, we provide data on genetic diversity of the pandemic virus in the northern region of India.
Material and methods
Study patients
Sheri‐Kashmir Institute of Medical Sciences is a 650‐bedded facility and constitutes the main tertiary care cum referral center for the respiratory cases for the valley of Kashmir. One hundred and ninety‐four patients with acute respiratory illness (ARI) that presented to the Internal & Pulmonary Medicine Department of Sheri‐Kashmir Institute of Medical Sciences, Srinagar, over a period of 10 weeks from January 01 to March 14, 2010 (week53‐week10) were enrolled for the study. The various age groups included 0–15 years (n = 5, I male), 16–30 years (n = 48, 18 male), 31–45 years (n = 39, 22 male), 45–60 years (n = 33, 17 male), and >60 years (n = 69, 38 male with seven patients being >75 years). The patients were interviewed by a team of 2–3 medical residents, and the answers were recorded in a predefined questionnaire. Informed consent was obtained from all the participants for inclusion in the survey, and the survey study was approved by the hospital ethics committee.
Influenza virus testing
Combined throat and nasal swabs were collected in viral transport medium and stored at 4°C for a maximum of 2–3 days before being transported to the Microbiology Laboratory at All India Institute for Medical Sciences, New Delhi, for analysis. All the samples were tested by real‐time RT‐PCR for the detection of influenza viruses including 2009A/H1N1 virus using the Centers for Disease Control and Prevention protocol. 13 All seasonal influenza A‐positive samples were further subtyped using primers and probes for A/H1and A/H3. 13
Sequencing and phylogenetic analysis
Haemagglutinin 1 (HA‐1) gene was sequenced following dideoxynucleotides chain termination method as described previously, 14 in automated 3100 DNA sequencer (Applied Biosystem, Foster City, USA) using Big‐Dye Terminator Chemistry. Neighbor‐joining (N‐J) tree was generated using pairwise gap deletion and maximum composite likelihood using Tamura‐Nei nucleotide model in MEGA version 4. 15 Sequences of strains from Srinagar were compared with published cognate sequences of corresponding genes, including those from India. 16 All the eight strains from Srinagar are submitted to GISAID Influenza Database, with accession numbers from EPI286637, EPI286638, and EPI286646‐51.
Statistical analysis
Statistical analysis of the data was performed using SPSS version 11.5 statistical software. Data have been expressed as Mean ± SD. Categorical variables were compared using Fisher’s exact/chi‐square test and continuous variables by employing Student’s t‐test. A P‐value of <0·05 was considered significant.
Results
Of the 194 patients (age 9–85 years; median 50; 108 male) recruited for the study, 86 (44%) required hospitalization, whereas 108 were from outpatient clinics. Various clinical diagnoses included acute respiratory tract infection (n = 156) and acute exacerbation of underlying lung disease (COPD; n = 38) with evidence of respiratory failure in 13 and sepsis syndrome in 2. Fifteen patients had community acquired pneumonia, and one had a bronchopneumonia. Associated medical illnesses included chronic lung disease (n = 60), hypertension (n = 58), heart failure (n = 22), diabetes (n = 18), hypothyroidism (n = 8), past CVA (n = 3), GI malignancy (n = 2), multiple myeloma (n = 1), and non‐Hodgkin’s lymphoma (n = 1). The comorbidities were found mostly in the age group of >60 years.
Twenty‐one (10·8%; median age 40; 13 male) of the 194 patients tested positive for influenza viruses by real‐time PCR. Further subtyping revealed that 13 (62%; age range 15–75; three were >65 years) were positive for 2009A/H1N1, 6 (28·5%) for seasonal influenza (A/H3), and 2 (9·5%) for influenza B. While we observed highest rate of positivity for 2009A/H1N1 in the first week of Jan (week 53), there was a steady decline in positivity thereafter. Interestingly, there was cocirculation of primarily A/H3N2 throughout the study period, with sporadic cases of influenza B (Figure 1).
Figure 1.
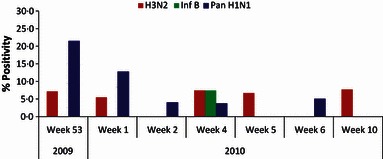
Week‐wise prevalence of seasonal, 2009A/H1N1, and influenza B at a tertiary care hospital, Sheri‐Kashmir Institute of Medical Sciences, Srinagar, India from January 1 to March 14, 2010 (week 53 of 2009 to week 10 of 2010).
Seven of the eight patients with seasonal influenza presented with febrile acute respiratory illnesses, and four were aged >62 (median 65 years). Of the eight patients with seasonal influenza, two had radiologically documented infiltrates and one developed a necrotizing pneumonia. Two of these patients were hospitalized, whereas 6 were from the outpatient clinics. Comorbidities in these patients included hypertension (n = 2), chronic lung disease (n = 2), and diabetes and a previous cerebrovascular accident in one case each.
Twelve of the 13 patients with 2009A/H1N1 presented with febrile ARI, whereas one was afebrile (Table 1). The patients presented with symptoms of fever (n = 12), chills (n = 11), rigors (n = 8), cough (n = 12), shortness of breath (n = 11), sore throat (n = 7), nasal discharge (n = 11), headache (n = 10), body aches (n = 7), malaise (n = 6), and vomiting and diarrhea in two cases each. Three 2009/A/H1N1‐positive patients developed radiographically confirmed lobar pneumonia, and three had heterogeneous pulmonary infiltrates (bilateral in one). Of the 13 patients with 2009A/H1N1, five patients were hospitalized, three with evidence of clinical and radiological evidence of lung consolidation, and one with renal failure; the remaining eight patients had a milder disease and were managed on outpatient basis only. One of the patients with 2009A/H1N1 infection required ventilatory assistance for respiratory failure (later recovered). Underlying comorbidities in patients with 2009/A/H1N1 included COPD and asthma (two cases each) and GI malignancy, heart failure, multiple myeloma, and post‐tubercular fibrosis (one case each). Comparative analysis of clinical presentations among influenza positive and influenza negative revealed a significantly higher frequency of clinical nasal discharge and a history of contact with a person with respiratory illness in past 1 week, in the influenza‐positive patients (Table 2). Overall, there was minimal difference in clinical presentations among those having 2009A/H1N1 or seasonal influenza positives (data not shown). Patients with febrile ARI suspected to have 2009A/H1N1 infection with severe disease with/without comorbidities (n = 6) and the sick hospitalized patients (n = 2) with seasonal influenza received oseltamivir treatment.
Table 1.
Demographic and clinical presentations of patients with 2009A/H1N1 infection in Kashmir, India
| Pt ID | Age/Sex | In/Outpatient | Symptoms | Comorbidity | Isolate Name | Segment Id |
|---|---|---|---|---|---|---|
| 42 | 75/M | IP | F,C,E,SOB,ST | COPD,CHF | A/India‐Srinagar/3/2010 H1N1 | EPI286646 |
| 3 | 65/F | IP | F,R,CH,ND,C,E,SOB, D,H | COPD | A/India‐Srinagar/2/2010 H1N1 | EPI286638 |
| 55 | 65/M | IP | F,ND,C,SOBSOB,H, V | GI Malignancy | A/India‐Srinagar/4/2010 H1N1 | EPI286647 |
| 101 | 55/M | OP | F,R,CH,ND,CSOB,H | _ | – | – |
| 10 | 44/M | OP | F,R, CH,ND,SOB | – | – | – |
| 59* | 40/M | OP | ND, C,E,SOB,SOB,H | _ | A/India‐Srinagar/8/2010 H1N1 | EPI286651 |
| 36* | 39/M | OP | F,R,CH,ND, C,E,,SOB, ST,V | Asthma, Post TB fibrosis | A/India‐Srinagar/1/2010 H1N1 | EPI286637 |
| 13* | 28/M | OP | F,R,CH,ND, C,SOB, ST, H | – | – | – |
| 33* | 28/M | OP | F,R,CH,ND, C,ESOB, ST | – | A/India‐Srinagar/6/2010 H1N1 | EPI286649 |
| 73* | 25/M | OP | F,R,CH,ND,C,H | – | A/India‐Srinagar/7/2010 H1N1 | EPI286650 |
| 53* | 23/F | IP | F,C,E, SOB,ST | – | – | – |
| 20 | 20/M | OP | F,R,ND,C, E, ST, C,E,H | ABPA/Asthma | A/India‐Srinagar/5/2010 H1N1 | EPI286648 |
| 155* | 15/F | IP | F,R,CH,ND, C,SOB,ST,H | Multiple Myeloma | – | – |
F, Fever; R, Rigors; CH, Chills; ND, Nasal discharge; C, Cough; E, Expectoration; SOB, Shortness of breath; ST, Sore Throat; D, Diarrhea; V, Vomiting; H, Headache; COPD, Chronic obstructive pulmonary disease; ABPA, Allergic bronchopulmonary aspergillosis; CHF, Congestive heart failure, TB, Tuberculosis. *denotes patients having a history of contact with another case of acute respiratory illness in past 7 days at home/workplace.
Table 2.
Comparison of clinical symptoms among influenza‐positive and influenza‐negative cases in Kashmir, India
| Clinical feature | Influenza negative N = 173 n (%) | Influenza positive N = 21 n (%) | P value |
|---|---|---|---|
| Contact in last 7 days | 64 (37) | 14 (67) | 0·009 |
| Nasal discharge on examination | 44 (25) | 13 (62) | 0·0005 |
| Fever | 136 (79) | 17 (81) | 0·80 |
| Chills | 113 (65) | 17(81) | 0·15 |
| Rigors | 36 (21) | 7 (33) | 0·19 |
| Cough | 141 (82) | 18 (86) | 0·63 |
| Sputum production | 101 (58) | 13 (62) | 0·76 |
| Sore throat | 57 (33) | 8 (38) | 0·64 |
| Nasal discharge | 103 (60) | 16 (76) | 0·14 |
| Nasal stuffiness | 81 (47) | 14 (67) | 0·08 |
| Body ache | 115 (67) | 12 (57) | 0·39 |
| Malaise | 40 (23) | 5 (24) | 1·00 |
| Vomiting | 25 (14) | 3 (14) | 1·00 |
| Breathlessness | 96 (56) | 15 (71) | 0·16 |
| Current smoker | 59 (34) | 5 (24) | 0·34 |
| Respiratory cases at home | 37 (21) | 7 (33) | 0·21 |
| Underlying OAD | 45 (26) | 6 (29) | 0·80 |
| Cyanosis | 41 (24) | 2 (10) | 0·17 |
| Accessory muscle usage | 52 (30) | 11(52) | 0·04 |
| Crepitations | 68 (39) | 8 (38) | 0·91 |
| Rhonchi/Wheeze | 48 (28) | 6 (29) | 0·94 |
| Hepatomegaly | 16 (9) | 2(10) | 1·00 |
| Pneumonia/Infiltrates on radiograph | 53 (31) | 9 (43) | 0·25 |
| Duration of symptoms (mean days, range) | 5·31 + 2·91 (1–15) | 4·81 ± 2·97 (1–15) | 0·67 |
Sequence and phylogenetic analysis
Recent phylogenetic studies, based on concatenated whole genomes, have shown that the early diversification of the 2009A/H1N1 viruses resulted in seven lineages, named clades 1–7 with defined spatial patterns. 17 Phylogenetic and evolutionary analysis of 2009/AH1N1 strains from Srinagar (n = 8) was carried out on the basis of HA1 nucleotide sequences (Figure 2). Viral sequences from Srinagar clustered with 2009A/H1N1 strains collected worldwide as well with other strains from India belonging to clade 7. 16 , 17 Few of the strains from Srinagar clustered with the Asian strains, with A/India‐Srinagar/3/2010 showing similarities to (A/Quebec/144147/2009); A/India‐Srinagar/4/2010 and A/India‐Srinagar/7/2010 showing similarities to (A/EHIME/33/2010), and A/India‐Srinagar/1/2010, A/India‐Srinagar/2/2010, and A/India‐Srinagar/6/2010 showing similarities to (A/Daegu/1873/2009) (Figure 2). Unlike cocirculation of clades 5, 6, and 7, in some parts of India and South America, 16 , 18 all isolates from Srinagar clustered in clade 7 (Figure 2).
Figure 2.
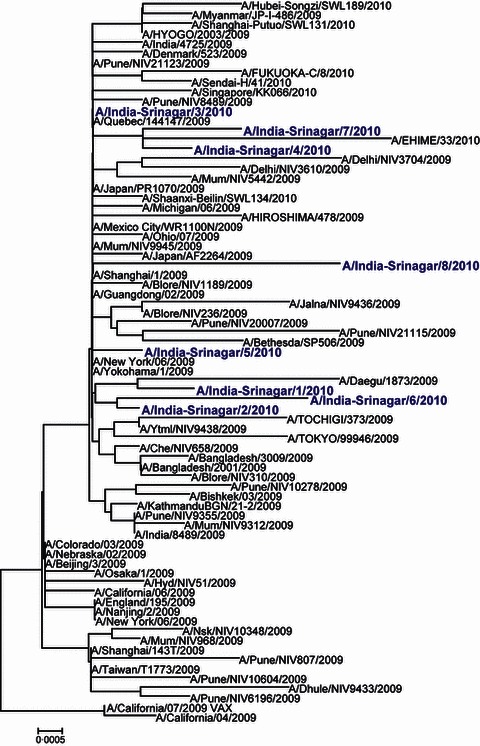
Phylogenetic analysis of HA1 region of the HA gene of pandemic 2009A/H1N1 strains isolated from Srinagar, India. The representative strains from 2009/A/California/H1N1 strain and the 2010 vaccine strain A/California/7/2009, India, and from other parts of the world were used to generate the phylogenetic tree using neighbor‐joining method.
To identify characteristic amino acid substitutions in 2009A/H1N1 strains, the HA protein from Srinagar was compared with that of the prototype isolates (A/California/04/2009) and vaccine isolate (A/California/07/2009). Amino acid substitutions defining clade 7 were found in all Srinagar isolates (P83S, S203T, and I321V), as well as other isolates from India. 16 The later substitution (I321V) was not present in two of the Srinagar isolates (A/India‐Srinagar/1/2010; A/India‐Srinagar/6/2010); however, both of the strains still clustered in clade 7. Analysis of HA structure from Srinagar revealed a highly conserved 190‐helix (residues 184–191), a 220‐loop (residues 218–225), and a 130‐loop (residues 131–135), as well other highly conserved residues: Tyr91, Trp150, His180, and Tyr192. However, minor point mutations were observed in few isolates, including A/India‐Srinagar/8/2010 with three point mutations (S183C, A256G, and M257K) and A/India‐Srinagar/7/2010 with two mutations (V19I and L44I) when compared to A/California/04/2009. Few additional point mutations were observed in strains from Srinagar, including V19I (A/India‐Srinagar/4/2010), D97N (A/India‐Srinagar/5/2010), and T197S (A/India‐Srinagar/1/2010); however, their significance remains to be determined.
Discussion
Since the emergence of 2009A/H1N1 in April 2009, the virus has spread worldwide. Globally, 2009A/H1N1 transmission remains most active in parts of South Asia and in limited areas of tropical South and Central America. In Asia, the most active areas of 2009A/H1N1 influenza virus transmission currently are in parts of India and, to a much lesser extent, in parts of Nepal and Bhutan. 9 In India, the initial cases were seen in travelers from North America; however, the virus soon became entrenched in the cities and communities, and indigenous transmission was observed. 10 The Kashmir province has temperate climate, and influenza‐like illnesses here are so ubiquitous that nearly everyone has influenza‐like illness at one time or the other during the months of fall and winter. Our data demonstrate that both 2009A/H1N1 and seasonal influenza viral infections are associated with subset of patients with acute respiratory infections in Kashmir.
In addition to confirming the presence of influenza in the valley of Kashmir, several important observations emerge from the study. The overall prevalence of influenza in Srinagar was lower than that observed in other cities in north or west of India. 9 , 10 , 19 This could be because of sample bias as the sample collection started late in the course of pandemic in Srinagar, as well samples were collected in a tertiary care hospital where patients present late during symptomatic phase. Another limitation of our study is that the current survey was primarily conducted in an adult clinic with limited subjects from younger age group. However, a limited surveillance conducted by the Ministry of Health & Family Welfare for 2009A/H1N1 infection in the same period did not find any pH1N1 positives in the age groups of <15 years (personal communication).
The median time of presentation of the patients from the onset of the symptoms was 4 days; however, 35 patients had symptoms for >8 days (range 8–15 days; median 10 days). Most patients with influenza infections are known to shed the virus from 1 day before the onset of symptoms until 5–7 days after the onset of symptoms. 1 Thus, a relatively late collection of samples (>8 days after initial onset of symptoms) for testing in up to 18% of patients could have potentially reduced the rate of positivity among these subjects with ARI. Median time of collection of samples in 2009A/H1N1‐positive patients was 3 days but two cases (aged 65 and 75, both with underlying COPD) had their samples collected beyond 8 days of the onset of illness, suggesting a relatively prolonged shedding of the virus. Further, although viral etiology was previously only suspected in exacerbations of COPD in Kashmir, we were able to confirm the presence of influenza (seasonal and pandemic) in subset (10·8%) of these patients.
We observed cocirculation of both seasonal and 2009A/H1N1 influenza viruses during the peak of winter in Kashmir. These data are in accordance with worldwide circulation of 2009A/H1N1, which cocirculated with seasonal influenza strains. 9 , 10 , 19 In addition, both seasonal and 2009A/H1N1 viruses were present among patients >62 years of age in Srinagar. Influenza is a prevalent viral infection that can cause severe or fatal disease in the elderly and those with underlying illness. 2 , 5 In temperate climates of Europe and North America, winter time seasonal influenza epidemics often result in dramatic increases in hospitalizations and deaths especially in elderly populations. 4 , 20 , 21 The detection of seasonal as well as 2009A/H1N1 among elderly patients in the current study suggests that the elderly population in Kashmir too remains at high risk for influenza‐related infection and complications. Indeed, a recent study conducted among COPD patients at SKIMS has shown 10 patients >60 years of age to be positive for A/2009/H1N1, further supporting influenza virus infections among elderly patients (Koul et al., unpublished).
Despite 12 of the 21 influenza‐positive cases in current study having evidence of respiratory failure, all experienced a full recovery. The severity of disease with seasonal and 2009A/H1N1 infection varies from study to study. 6 , 9 , 22 Recent data from India suggest that 2009A/H1N1 virus was associated with more severe disease outcomes in terms of both hospitalization and mortality, 19 the severity being lower than that reported for ‘Spanish flu, 1918’ but much higher than reported for other pandemics of 20th century. 5 , 8 , 22 While high case fatality rates have been reported from many countries, 19 , 22 , 23 , 24 the 2009A/H1N1 pandemic has generally been characterized by mild and self‐limiting disease in the majority of cases. 4 , 10 Limited data suggest that the overall intensity and severity of the current regional epidemics in India do not yet appear to exceed what was observed during the first wave in 2009; however, it is too early to make a complete assessment of the situation as the regional epidemics are still evolving. 9
Phylogenetic analysis of the HA 1 gene from 2009A/H1N1 isolates from Kashmir revealed similarities to 2009A/H1N1 clade 7. This is in agreement with recent results obtained in other parts of the world, where clade 7 was the most commonly detected clade, although cocirculation of clades 5, 6, and 7 has been observed in some parts of the world. 16 , 17 , 18 Amino acid substitutions in the HA protein characteristic for clade 7 (P83S, S203T and I321V) were identified in all Srinagar isolates, with two exception where I321I was present in two of the isolates. It is important to note that substitutions P83S and S203T are located in the antigenic sites E and D, respectively, whereas substitution I321V is located outside the major defined HA antigenic epitope. 18 None of these substitutions are found in the vaccine strain A/California/7/2009. 25 Specific mutations in the HA (D222G and Q293H) have been found to be associated with fatal outcome in certain cases; 26 , 27 however, none of these mutations were observed in isolates from the current study patients who experienced full recovery. Likewise, although a number of mutations have been reported in 2009A/H1N1 viruses, they have not affected virus antigenicity and pathogenicity. 5 , 26 , 28 Functional significance of these mutations, if any, remains to be determined.
The highly conserved nature of 2009A/H1N1 virus seen during the first wave of pandemic is in agreement with circulating 2009A/H1N1 viruses across the world. Sixteen months after the first pandemic viruses were isolated from Mexico and United States in April 2009, the virus is still antigenically homogeneous. However, as the HA continues to circulate in the human population, its HA antigenic sites are likely be targeted by antibody‐mediated selection pressure, which may affect antigenicity or virulence as this virus evolves. HA variation may also be driven by neuraminidase inhibitor use as well as antibody‐mediated selection. HA mutations affecting enzymatic activity may appear in viruses that generate neuraminidase inhibitor resistance mutations. 29 However, no patients in our study who received oseltamivir were assessed for neuraminidase inhibitor resistance. We conclude that 2009/A/H1N1 infection (with phylogenetic similarity to California 2009/A/H1N1) was seen in a subset of patients with ARI as well as exacerbations of COPD in Srinagar, the summer capital of the northern Indian state of Jammu & Kashmir.
Disclaimer
The findings and conclusions of this report are those of the authors and do not necessarily represent the views of the Center for Disease Control and Prevention.
References
- 1. Thompson WW, Shay DK, Weintraub E et al. Mortality associated with influenza and respiratory syncytial virus in the United States. JAMA 2003; 289:179–186. [DOI] [PubMed] [Google Scholar]
- 2. Nicholson KG, McNally T, Silverman M, Simons P, Stockton JD, Zambon MC. Rates of hospitalisation for influenza, respiratory syncytial virus and human metapneumovirus among infants and young children. Vaccine 2006; 24:102–108. [DOI] [PubMed] [Google Scholar]
- 3. Finkelman BS, Viboud C, Koelle K, Ferrari MJ, Bharti N, Grenfell BT. Global patterns in seasonal activity of influenza A/H3N2, A/H1N1, and B from 1997 to 2005: viral coexistence and latitudinal gradients. PLoS ONE 2007; 2: e1296. doi:10.1371/journal.pone.0001296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Leo YS, Lye DC, Chow A. Influenza in the tropics. Lancet Infect Dis 2009; 9:457–458. [DOI] [PubMed] [Google Scholar]
- 5. Fraser C, Donelly CA, Cauchemez S et al. WHO Rapid Assessment Collaboration. Pandemic potential of a strain of influenza A (H1N1): early findings. Science 2009; 324:1557–1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dawood FS, Jain S, Finelli L et al Novel Swine‐Origin Influenza A (H1N1) Virus Investigation Team. Emergence of a novel swine origin influenza A (H1N1) virus in humans. N Engl J Med 2009; 360:2605–2615. [DOI] [PubMed] [Google Scholar]
- 7. World Health Organization . Pandemic (H1N1) 2009 –Weekly update 112. [cited 2010 Sept 28]. Available from http://www.who.int/csr/don/2010_08_06/en/index.html.
- 8. Mukherjee A, Roy T, Agrawal AS et al. Prevalence and epidemiology of pandemic H1N1 strains in hospitals of eastern India. J Public Health Epidemiol 2010; 2:171–174. [Google Scholar]
- 9. Gurav YK, Pawar SD, Chaddha MS et al. Pandemic Influenza A (H1N1) 2009 outbreak in a residential school at Panchgani, Maharashtra, India. Indian J Med Res 2010; 132:67–71. [PubMed] [Google Scholar]
- 10. Pandemic Influenza A (H1N1) . Ministry of Health and Family Welfare, Government of India. Update on pandemic Influenza A, 2010 [updated 2010 Aug 15; cited 2010 Sep 28]. Available from http://mohfw‐h1n1.nic.in/documents/PDF/SituationalUpdatesArchives/August2010/Situational20Updates20on2015.08.2010.pdf.
- 11. Garten RJ, Davis CT, Russel CA et al. Antigenic and genetic characteristics of swine‐origin 2009 A (H1N1) influenza viruses circulating in humans. Science 2009; 325:197–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zimmer SM, Burke DS. Historical perspective—emergence of influenza A (H1N1) viruses. N Engl J Med 2009; 361:279–285. [DOI] [PubMed] [Google Scholar]
- 13. World Health Organization. CDC protocol of real‐time RTPCR for influenza A (H1N1). Geneva. 2009.[cited 2010 Sep 28]. Available from http://www.who.int/csr/resources/publications/swineflu/CDCrealtimeRTPCRprotocol_20090428.pdf.
- 14. Agarwal A, Sarkar M, Ghosh M et al. Genetic characterization of circulating seasonal influenza A viruses (2005–2009) revealed introduction of oseltamivir resistant H1N1 strain during 2009 in eastern India. Infect Genet Evol 2010; 10:1188–1198. [DOI] [PubMed] [Google Scholar]
- 15. Tamura K, Dudley J, Nei M, Kumar S. MEGA4: molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol 2007; 24:1596–1599. [DOI] [PubMed] [Google Scholar]
- 16. Potdar VA, Chadha MS, Jadav SM, Mullick J, Cherian SS, Mishra AC. Genetic characterization of the influenza a pandemic (H1N1) 2009 virus isolates from India. PLoS ONE 2009; 5: e9693. doi:10.1371/journal.pone.0009693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sarkar M, Agarwal AS, Sharma Dey R et al. Molecular characterization and comparative analysis of pandemic H1N1/2009 strains with co‐circulating seasonal H1N1/2209 strains from Eastern India. Arch Virol 2010; 156:207–217. [DOI] [PubMed] [Google Scholar]
- 18. Goni N, Moratorio G, Ramas V, Coppola L, Chiparelli H, Cristina J. Phylogenetic analysis of pandemic 2009 influenza A virus circulating in the South American region: genetic relationships and vaccine strain match. Arch Virol. 2010. doi: 10.1007/s00705‐010‐0825‐7. [DOI] [PubMed] [Google Scholar]
- 19. Mishra AC, Chadha MS, Choudhary ML, Potdar VA. Pandemic Influenza (H1N1) 2009 is associated with severe disease in India. PLoS ONE 2010; 5: e10540. doi:10.1371/journal.pone.0010540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Dushoff J, Plotkin JB, Viboud C, Earn DJ, Simonsen L. Mortality due to Influenza in the United States.: an annualized regression approach using multiple‐cause mortality dat a. Am J Epidemiol 2006; 163:181–187. [DOI] [PubMed] [Google Scholar]
- 21. Simmerman J, Uyeki T. The burden of influenza in East and South‐East Asia: a review of the English language literature. Influenza Other Respi Viruses 2008; 2:81–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Belshe RB. The origins of pandemic influenza—lessons from the 1918 virus. N Engl J Med 2009; 353:2209–2211. [DOI] [PubMed] [Google Scholar]
- 23. Miller E, Hoschler K, Hardelid P, Stanford E, Andrews N, Zambon M. Incidence of 2009 pandemic influenza A H1N1infection in England: a cross‐sectional serologicalstudy. Lancet 2010; 375:1100. [DOI] [PubMed] [Google Scholar]
- 24. Wilson N, Baker MG. The emerging influenza pandemic: estimating the case fatality ratio. Euro Surveill 2009; 14:pii=19255. [PubMed] [Google Scholar]
- 25. World Health Organization. Candidate H1N1 pandemic vaccine viruses 2010. [updated 2010 May 24; cited 2010 Sep 28] Available from http://www.who.int/csr/disease/swineflu/guidance/vaccines/candidates/en/.
- 26. Glinsky GV. Genomic analysis of pandemic (H1N1) 2009 reveals association of increasing disease severity with emergence of novel hemagglutinin mutations. Cell Cycle 2010; 9:958–970. [DOI] [PubMed] [Google Scholar]
- 27. Kilander A, Rykkvin R, Dudman SG, Hungnes O. Observed association between the HA1 mutation D222G in the 2009 pandemic influenza A(H1N1) virus and severe clinical outcome, Norway 2009–2010. Euro Surveill bullet 2010;15 PMID: 20214869 [DOI] [PubMed] [Google Scholar]
- 28. Padlan EA. The pandemic 2009 (H1N1) swine influenza virus is mild compared to the pandemic 1918 (H1N1) virus because of a proline‐to‐serine substitution in the receptor‐binding site of its hemagglutinin – a hypothesis. Med Hypotheses 2009; doi: 10.1016/j.mehy.2009.09.034 [DOI] [PubMed] [Google Scholar]
- 29. Hensley SE, Das SR, Gibbs JS, Bailey AL, Schmidt LM et al. Influenza A Virus Hemagglutinin antibody escape promotes neuraminidase antigenic variation and drug resistance. PLoS ONE 2011; 6:e15190. [DOI] [PMC free article] [PubMed] [Google Scholar]


